Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis
Abstract
1. Introduction
2. Results
2.1. Univariate Analysis of Normal and MCF-7 Cells Gives Little Information for Objective Discrimination
2.2. Application of Multivariate Statistical Methods to Discriminate Cancer Cells
2.2.1. Principal Components Analysis
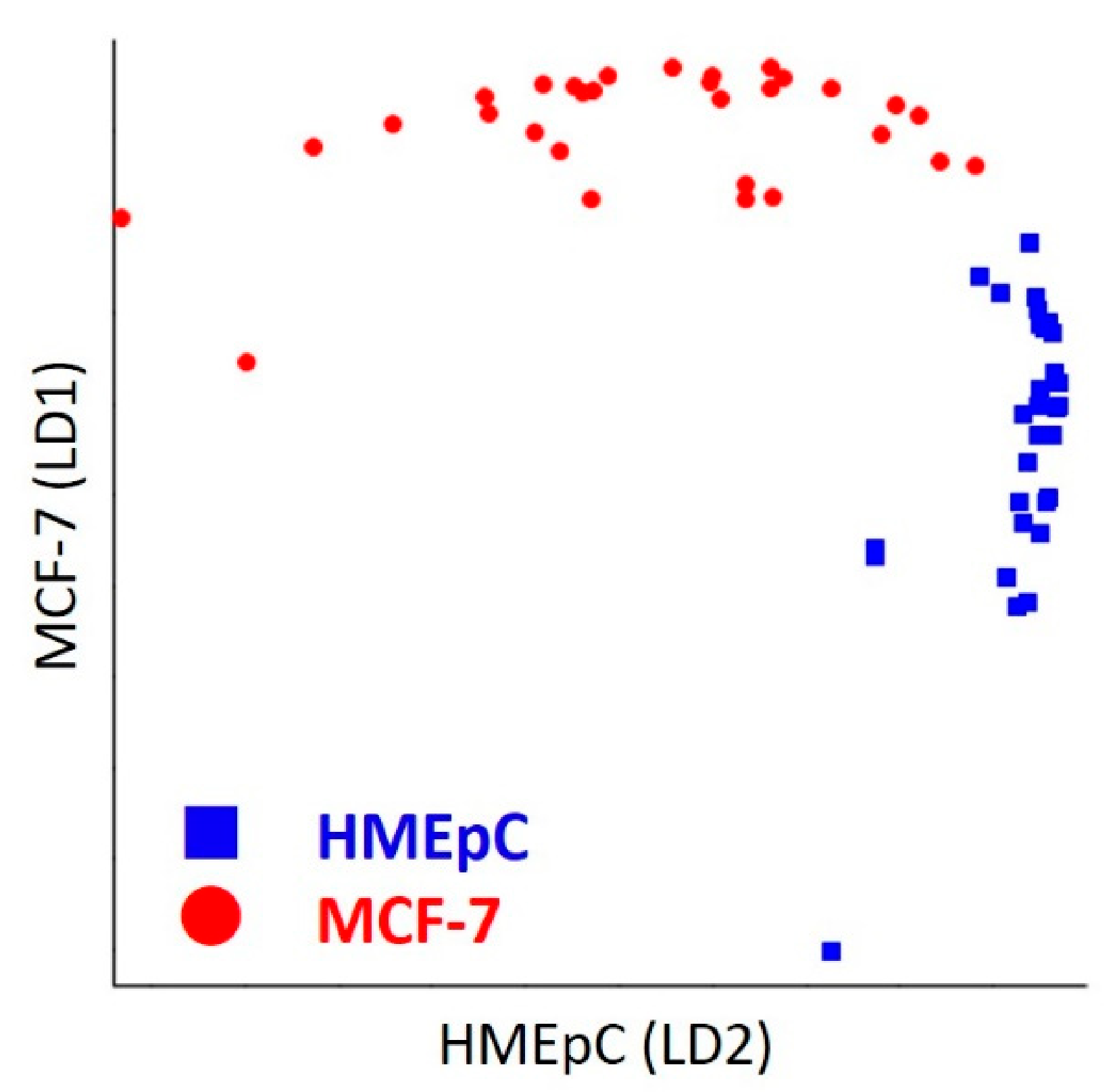
2.2.2. Linear Discriminant Analysis
2.2.3. Support Vector Machine Analysis
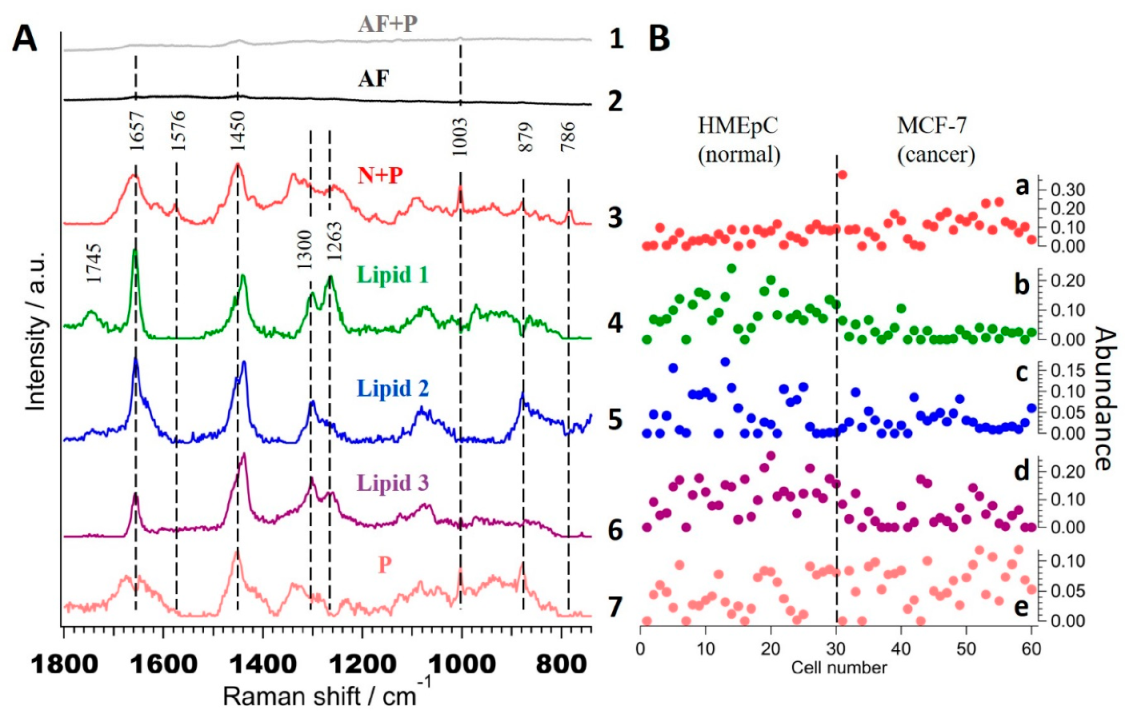
2.2.4. Multivariate Curve Resolution Analysis
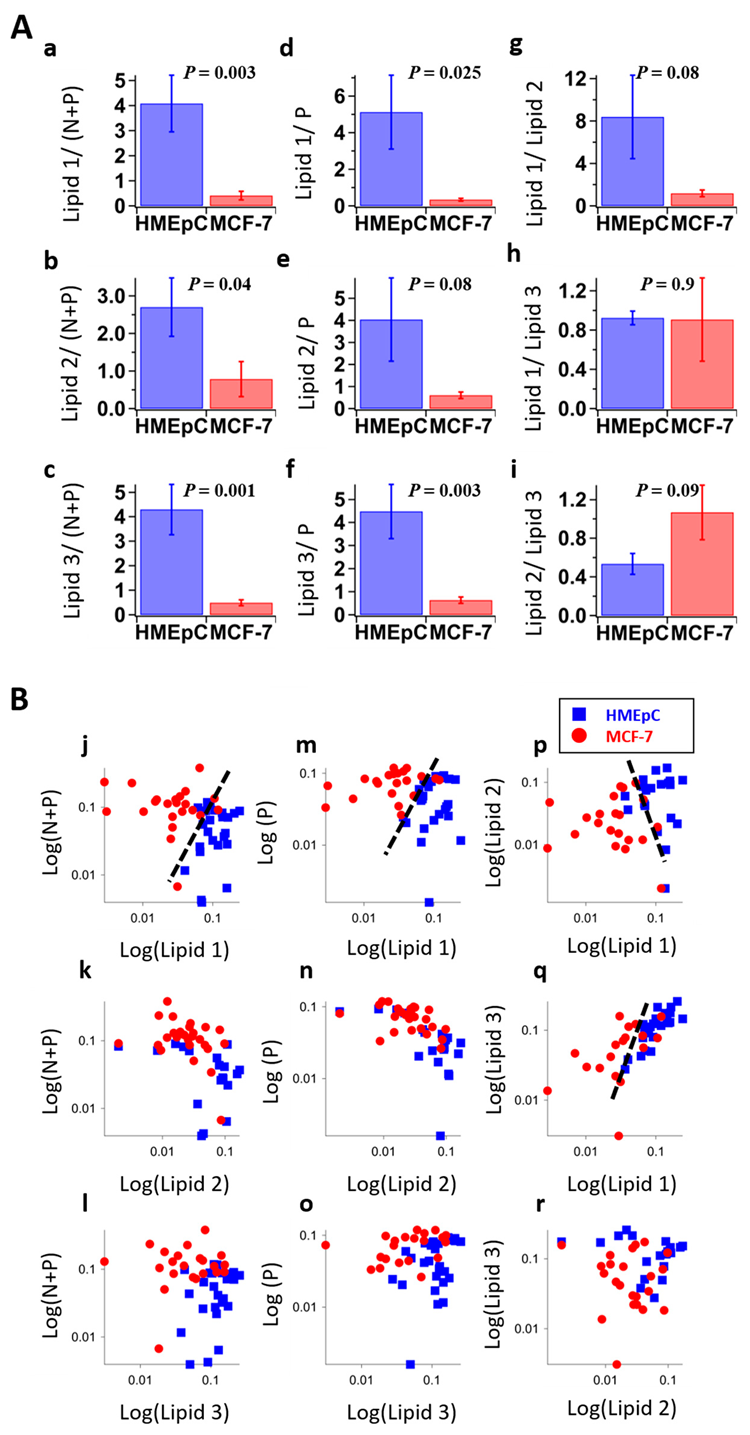
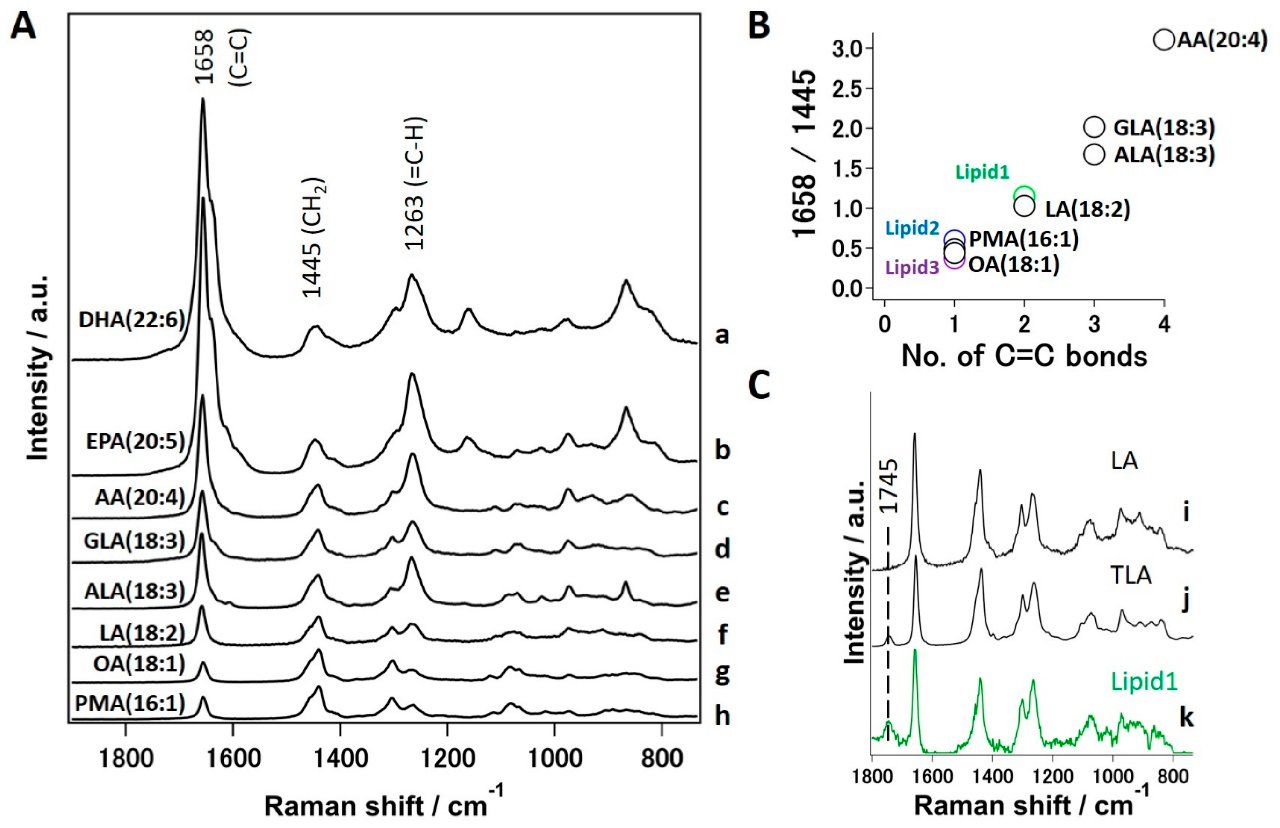
2.3. Molecular Assignment of MCR-ALS Extracted Lipid Components
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Raman Microspectroscopy
4.3. Data Analysis
4.3.1. Discriminant Analysis
4.3.2. MCR-ALS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | Arachidonic acid |
| ALA | α-linolenic acid |
| DHA | Docosahexaenoic acid |
| EPA | Eicosapentaenoic acid |
| GLA | γ-linolenic acid |
| HMEpC | Human mammary epithelial cells |
| LA | Linoleic acid |
| LDA | Linear discriminant analysis |
| MA | Multivariate analysis |
| MCF-7 | Michigan cancer foundation-7 |
| MCR-ALS | Multivariate curve resolution-alternating least squares |
| OA | Oleic acid |
| PC | Principal components |
| PCA | Principal components analysis |
| PMA | Palmitoleic acid |
| PUFA | Polyunsaturated fatty acids |
| RS | Raman Spectroscopy |
| SVD | Singular value decomposition |
| SVM | Support vector machine |
| TLA | Trilinoleic acid |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.; Gera, P.; Malik, A.; Nair, S.; Chaturvedi, P.; Murali Krishna, C. Raman exfoliative cytology for prognosis prediction in oral cancers: A proof of concept study. J. Biophotonics 2019, 12, e201800334. [Google Scholar] [CrossRef]
- Hemanth, N.; Suguru, U.; Naoki, O.; Yoshikazu, K.; Masahiro, A.; Hiro-o, H.; Tatsuyuki, Y. Towards the development of a non-bioptic diagnostic technique for eosinophilic esophagitis using raman spectroscopy. Vib. Spectrosc. 2016, 85, 7–10. [Google Scholar]
- Iwasaki, K.; Noothalapati, H.; Yamamoto, T. Chapter 15—Recent advances in raman spectroscopy of proteins for disease diagnosis. In Vibrational Spectroscopy in Protein Research; Ozaki, Y., Baranska, M., Lednev, I.K., Wood, B.R., Eds.; Academic Press: London, UK, 2020; pp. 435–459. [Google Scholar]
- Tolstik, T.; Marquardt, C.; Matthaus, C.; Bergner, N.; Bielecki, C.; Krafft, C.; Stallmach, A.; Popp, J. Discrimination and classification of liver cancer cells and proliferation states by raman spectroscopic imaging. Analyst 2014, 139, 6036–6043. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, G.R.; Orr, L.E.; Christie-Brown, J.; McCarthy, K.; Rose, S.; Thomas, M.; Stone, N. Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising raman spectroscopy and multivariate analysis. Analyst 2013, 138, 3900–3908. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Kaneko, A.; Tanaka, Y.; Ishikawa, T.; Noothalapati, H.; Yamamoto, T. Visualizing wax ester fermentation in single euglena gracilis cells by raman microspectroscopy and multivariate curve resolution analysis. Biotechnol. Biofuels 2019, 12, 128. [Google Scholar] [CrossRef]
- Noothalapati, H.; Sasaki, T.; Kaino, T.; Kawamukai, M.; Ando, M.; Hamaguchi, H.O.; Yamamoto, T. Label-free chemical imaging of fungal spore walls by raman microscopy and multivariate curve resolution analysis. Sci. Rep. 2016, 6, 27789. [Google Scholar] [CrossRef]
- Laor, D.; Sade, D.; Shaham-Niv, S.; Zaguri, D.; Gartner, M.; Basavalingappa, V.; Raveh, A.; Pichinuk, E.; Engel, H.; Iwasaki, K.; et al. Fibril formation and therapeutic targeting of amyloid-like structures in a yeast model of adenine accumulation. Nat. Commun. 2019, 10, 62. [Google Scholar] [CrossRef]
- Hemanth, N.; Shinsuke, S. Exploring metabolic pathways in vivo by a combined approach of mixed stable isotope-labeled raman microspectroscopy and multivariate curve resolution analysis. Anal. Chem. 2014, 86, 7828–7834. [Google Scholar]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer by using raman spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 12371–12376. [Google Scholar] [CrossRef]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar] [CrossRef] [PubMed]
- Manfait, M.; Jeannesson, P.; Jardillier, J.C.; Ginot, L.; Alix, A.J.P. Raman-spectroscopy of cancer-cells—A new approach to the study of the drug-cell interactions. Ann. Biol. Clin. 1982, 40, 394. [Google Scholar]
- Nabiev, I.R.; Morjani, H.; Manfait, M. Selective analysis of antitumor drug-interaction with living cancer-cells as probed by surface-enhanced raman-spectroscopy. Eur. Biophys. J. 1991, 19, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.J.; Redd, D.C.; Gansler, T.S.; McCreery, R.L. Characterization of human breast biopsy specimens with near-ir raman spectroscopy. Anal. Chem. 1994, 66, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.J.; McCreery, R.L.; Redd, D.C. Raman spectroscopy of normal and diseased human breast tissues. Anal. Chem. 1995, 67, 777–783. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Notingher, I.; Verrier, S.; Romanska, H.; Bishop, A.E.; Polak, J.M.; Hench, L.L. In Situ characterisation of living cells by raman spectroscopy. J. Spectrosc. 2002, 16, 43–51. [Google Scholar] [CrossRef]
- Vici, P.; Pizzuti, L.; Natoli, C.; Gamucci, T.; Di Lauro, L.; Barba, M.; Sergi, D.; Botti, C.; Michelotti, A.; Moscetti, L.; et al. Triple positive breast cancer: A distinct subtype? Cancer Treat. Rev. 2015, 41, 69–76. [Google Scholar] [CrossRef]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using raman spectroscopy. Cancer Res. 2002, 62, 5375–5380. [Google Scholar]
- Chowdary, M.V.; Kumar, K.K.; Kurien, J.; Mathew, S.; Krishna, C.M. Discrimination of normal, benign, and malignant breast tissues by raman spectroscopy. Biopolymers 2006, 83, 556–569. [Google Scholar] [CrossRef]
- Pichardo-Molina, J.L.; Frausto-Reyes, C.; Barbosa-Garcia, O.; Huerta-Franco, R.; Gonzalez-Trujillo, J.L.; Ramirez-Alvarado, C.A.; Gutierrez-Juarez, G.; Medina-Gutierrez, C. Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med. Sci. 2007, 22, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Lyng, F.M.; Traynor, D.; Nguyen, T.N.Q.; Meade, A.D.; Rakib, F.; Al-Saady, R.; Goormaghtigh, E.; Al-Saad, K.; Ali, M.H. Discrimination of breast cancer from benign tumours using raman spectroscopy. PLoS ONE 2019, 14, e0212376. [Google Scholar]
- Hedegaard, M.; Krafft, C.; Ditzel, H.J.; Johansen, L.E.; Hassing, S.; Popp, J. Discriminating isogenic cancer cells and identifying altered unsaturated fatty acid content as associated with metastasis status, using k-means clustering and partial least squares-discriminant analysis of raman maps. Anal. Chem. 2010, 82, 2797–2802. [Google Scholar] [CrossRef] [PubMed]
- Haka, A.S.; Volynskaya, Z.; Gardecki, J.A.; Nazemi, J.; Lyons, J.; Hicks, D.; Fitzmaurice, M.; Dasari, R.R.; Crowe, J.P.; Feld, M.S. In vivo margin assessment during partial mastectomy breast surgery using raman spectroscopy. Cancer Res. 2006, 66, 3317–3322. [Google Scholar] [CrossRef] [PubMed]
- Bitar, R.A.; Martinho, H.a.S.; Tierra-Criollo, C.J.; Ramalho, Z.L.N.; Netto, M.M.; Martin, A.A. Biochemical analysis of human breast tissues using fourier-transform raman spectroscopy. J. Biomed. Opt. 2006, 11, 054001. [Google Scholar] [CrossRef] [PubMed]
- Brozek-Pluska, B.; Musial, J.; Kordek, R.; Bailo, E.; Dieing, T.; Abramczyk, H. Raman spectroscopy and imaging: Applications in human breast cancer diagnosis. Analyst 2012, 137, 3773–3780. [Google Scholar] [CrossRef] [PubMed]
- Marro, M.; Nieva, C.; de Juan, A.; Sierra, A. Unravelling the metabolic progression of breast cancer cells to bone metastasis by coupling raman spectroscopy and a novel use of mcr-als algorithm. Anal. Chem. 2018, 90, 5594–5602. [Google Scholar] [CrossRef]
- Sixian, Y.; Haohua, T.; Youbo, Z.; Yuan, L.; Eric, J.C.; Marina, M.; Stephen, A.B. Raman spectroscopic analysis reveals abnormal fatty acid composition in tumor micro- and macroenvironments in human breast and rat mammary cancer. Sci. Rep. 2016, 6, 32922. [Google Scholar]
- Kuo, C.Y.; Ann, D.K. When fats commit crimes: Fatty acid metabolism, cancer stemness and therapeutic resistance. Cancer Commun. 2018, 38, 47. [Google Scholar] [CrossRef]
- Yi, M.; Li, J.; Chen, S.; Cai, J.; Ban, Y.; Peng, Q.; Zhou, Y.; Zeng, Z.; Peng, S.; Li, X.; et al. Emerging role of lipid metabolism alterations in cancer stem cells. J. Exp. Clin. Cancer Res. 2018, 37, 118. [Google Scholar] [CrossRef]
- Peck, B.; Schulze, A. Lipid desaturation—The next step in targeting lipogenesis in cancer? FEBS J. 2016, 283, 2767–2778. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dubois, R.N. Prostaglandins and cancer. Gut 2006, 55, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Menter, D.G.; Dubois, R.N. Prostaglandins in cancer cell adhesion, migration, and invasion. Int. J. Cell Biol. 2012, 2012, 723419. [Google Scholar] [CrossRef]
- Ma, X.; Yang, Q.; Wilson, K.T.; Kundu, N.; Meltzer, S.J.; Fulton, A.M. Promoter methylation regulates cyclooxygenase expression in breast cancer. Breast Cancer Res. 2004, 6, R316–R321. [Google Scholar] [CrossRef]
- Noothalapati, H.; Iwasaki, K.; Yoshimoto, C.; Yoshikiyo, K.; Nishikawa, T.; Ando, M.; Hamaguchi, H.O.; Yamamoto, T. Imaging phospholipid conformational disorder and packing in giant multilamellar liposome by confocal raman microspectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 187, 186–190. [Google Scholar] [CrossRef]
- Noothalapati, H.; Ikarashi, R.; Iwasaki, K.; Nishida, T.; Kaino, T.; Yoshikiyo, K.; Terao, K.; Nakata, D.; Ikuta, N.; Ando, M.; et al. Studying anti-oxidative properties of inclusion complexes of alpha-lipoic acid with gamma-cyclodextrin in single living fission yeast by confocal raman microspectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 197, 237–243. [Google Scholar] [CrossRef]
- Noothalapati, H.; Iwasaki, K.; Yamamoto, T. Biological and medical applications of multivariate curve resolution assisted raman spectroscopy. Anal. Sci. 2017, 33, 15–22. [Google Scholar] [CrossRef]


| HMEpC | MCF-7 | ||
|---|---|---|---|
| Predicted | HMEpC | 30 | 1 |
| MCF-7 | 0 | 29 |
| Actual | |||
|---|---|---|---|
| HMEpC | MCF-7 | ||
| Predicted | HMEpC | 30 | 0 |
| MCF-7 | 0 | 30 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwasaki, K.; Araki, A.; Krishna, C.M.; Maruyama, R.; Yamamoto, T.; Noothalapati, H. Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis. Int. J. Mol. Sci. 2021, 22, 800. https://doi.org/10.3390/ijms22020800
Iwasaki K, Araki A, Krishna CM, Maruyama R, Yamamoto T, Noothalapati H. Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis. International Journal of Molecular Sciences. 2021; 22(2):800. https://doi.org/10.3390/ijms22020800
Chicago/Turabian StyleIwasaki, Keita, Asuka Araki, C Murali Krishna, Riruke Maruyama, Tatsuyuki Yamamoto, and Hemanth Noothalapati. 2021. "Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis" International Journal of Molecular Sciences 22, no. 2: 800. https://doi.org/10.3390/ijms22020800
APA StyleIwasaki, K., Araki, A., Krishna, C. M., Maruyama, R., Yamamoto, T., & Noothalapati, H. (2021). Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis. International Journal of Molecular Sciences, 22(2), 800. https://doi.org/10.3390/ijms22020800






