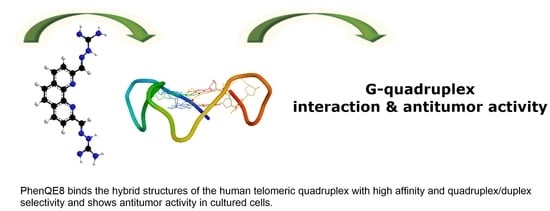
PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex †
Abstract
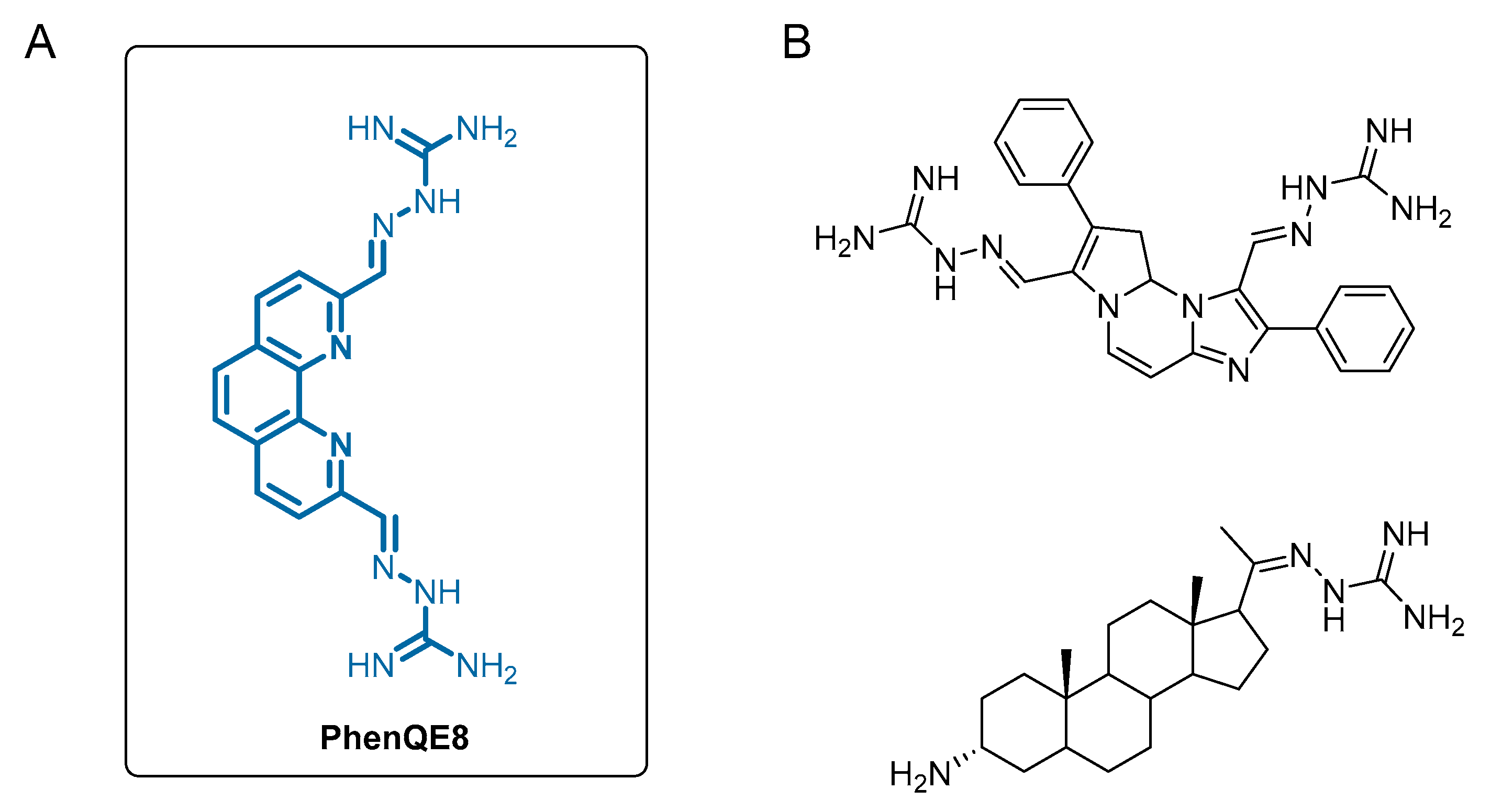
- A novel G-quadruplex DNA ligand, PhenQE8, based on 1,10-phenanthroline and containing two guanyl hydrazone groups was synthesized with good yields.
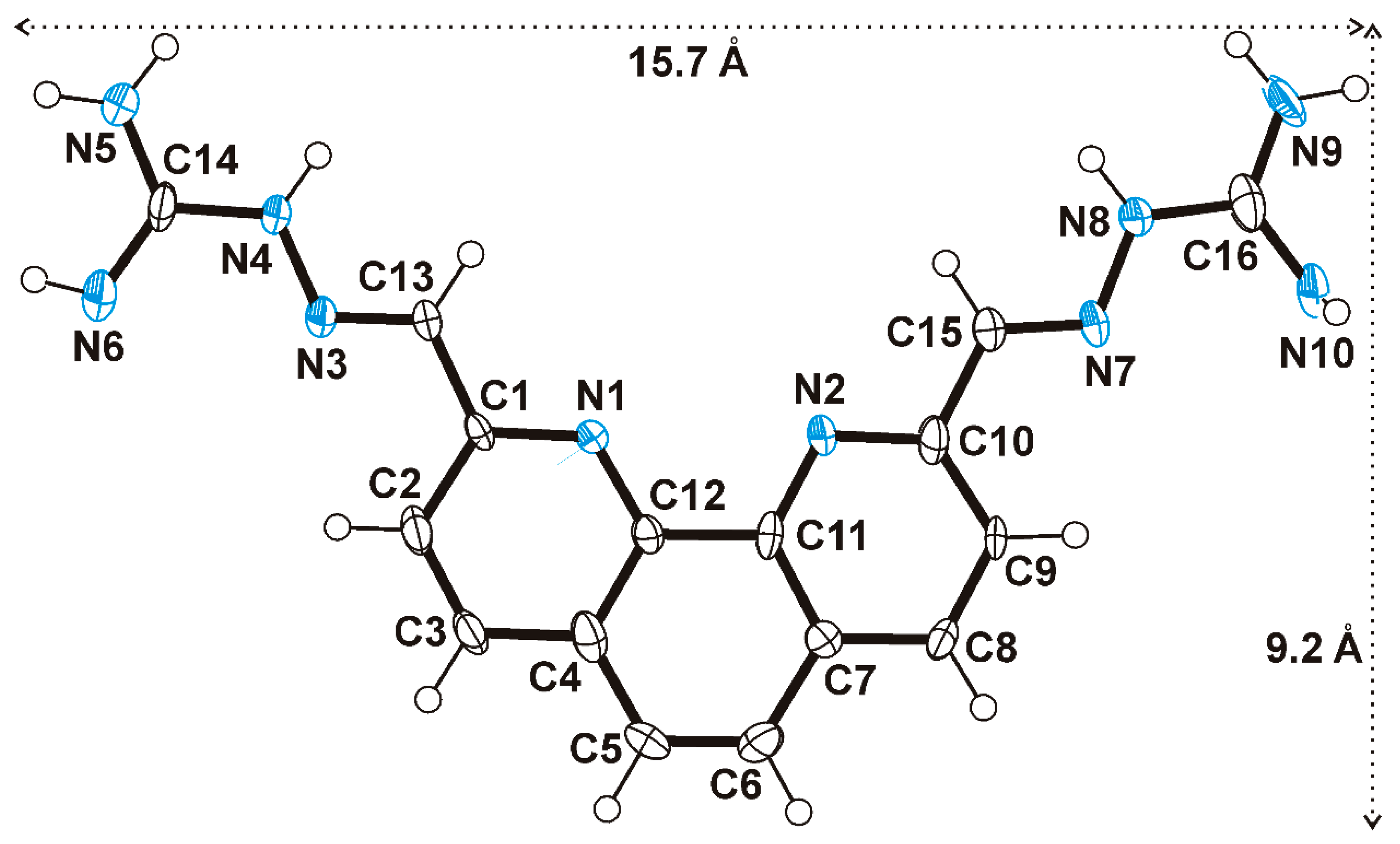
- PhenQE8 can adopt an extended conformation with one molecular dimension expanding ~16 Å.
- PhenQE8 can recognize the hybrid (mixed) structures of the telomeric quadruplex with good binding affinity.
- PhenQE8 shows high quadruplex/duplex DNA binding selectivity.
- PhenQE8 exhibits antitumor activity in cell culture and presents lower cytotoxicity in healthy human fibroblasts and normal prostate cells.
1. Introduction
2. Results and Discussion
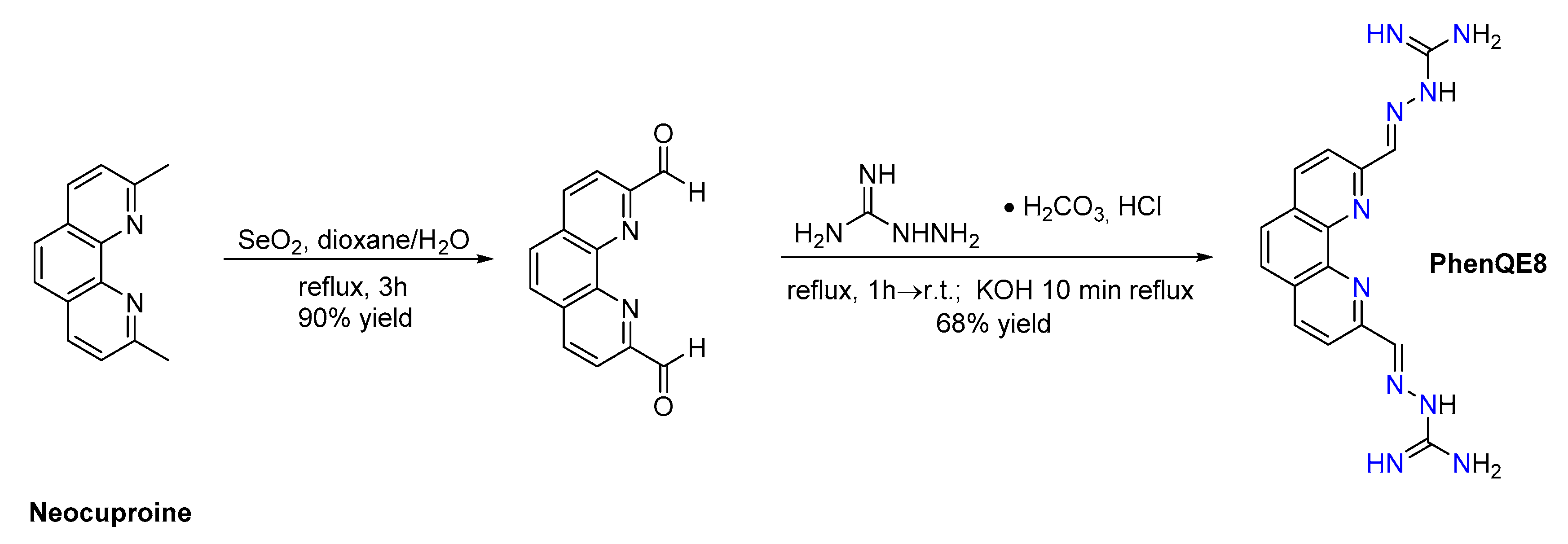
2.1. Synthesis and Structural Study of PhenQE8
2.2. Quadruplex and Duplex DNA Interactions
2.2.1. FRET DNA Melting Assays
2.2.2. Circular Dichroism
2.2.3. Noncompetition and Competition Equilibrium Dialysis
2.2.4. CT DNA Viscometric Titration
2.2.5. Antitumor Activity in Cultured Cells
3. Conclusions
4. Materials and Methods
4.1. General Methods
4.2. Synthesis and Structural Characterization
4.3. DNA Interactions and Antitumor Activity
4.3.1. Fret DNA Melting Assays
4.3.2. Circular Dichroism Spectra
4.3.3. Equilibrium Dialysis
4.3.4. Viscometric Titration
4.3.5. Cell Culture and MTT Colorimetric Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 41. [Google Scholar] [CrossRef]
- Tian, T.; Chen, Y.Q.; Wang, S.R.; Zhou, X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Chem 2018, 4, 1314–1344. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Lan, L. The DNA secondary structures at telomeres and genome instability. Cell Biosci. 2020, 10, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Valton, A.-L.; Prioleau, M.-N. G-Quadruplexes in DNA Replication: A Problem or a Necessity? Trends Genet. 2016, 32, 697–706. [Google Scholar] [CrossRef]
- Bugaut, A.; Balasubramanian, S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Sengupta, A.; Ganguly, A.; Chowdhury, S. Promise of G-quadruplex structure binding ligands as epigenetic modifiers with anti-cancer effects. Molecules 2019, 24. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Sharma, S.; Chowdhury, S. Non-duplex G-Quadruplex Structures Emerge as Mediators of Epigenetic Modifications. Trends Genet. 2019, 35, 129–135. [Google Scholar] [CrossRef]
- Neidle, S. Therapeutic Applications of Quadruplex Nucleic Acids, 1st ed.; Academic Press: London, UK, 2012. [Google Scholar]
- Monchaud, D. Foreword: Biological relevance and therapeutic applications of DNA-and RNA-quadruplexes: Double helix versus quadruple helix. Biol. Relev. Ther. Appl. DNA RNA Quadruplexes 2015. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.-L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In The Alkali Metal Ions: Their Role for Life, Metal Ions in Life Sciences; Springer: Cham, Switzerland, 2016; pp. 203–258. [Google Scholar]
- Harkness, R.W.; Mittermaier, A.K. G-quadruplex dynamics. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Bončina, M.; Hamon, F.; Islam, B.; Teulade-Fichou, M.P.; Vesnaver, G.; Haider, S.; Lah, J. Dominant Driving Forces in Human Telomere Quadruplex Binding-Induced Structural Alterations. Biophys. J. 2015, 108, 2903–2911. [Google Scholar] [CrossRef] [PubMed]
- Comez, L.; Bianchi, F.; Libera, V.; Longo, M.; Petrillo, C.; Sacchetti, F.; Sebastiani, F.; D’Amico, F.; Rossi, B.; Gessini, A.; et al. Polymorphism of human telomeric quadruplexes with drugs: A multi-technique biophysical study. Phys. Chem. Chem. Phys. 2020, 22, 11583–11592. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef]
- Gaynutdinov, T.I.; Neumann, R.D.; Panyutin, I.G. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: Effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008, 36, 4079–4087. [Google Scholar] [CrossRef]
- Lin, C.; Wu, G.; Wang, K.; Onel, B.; Sakai, S.; Shao, Y.; Yang, D. Molecular Recognition of the Hybrid-2 Human Telomeric G-Quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures. Angew. Chem. Int. Ed. 2018, 57, 10888–10893. [Google Scholar] [CrossRef]
- Lin, C.; Yang, D. Human telomeric G-quadruplex structures and G-quadruplex-interactive compounds. In Methods in Molecular Biology; Springer Nature: Cham, Switzerland, 2017; Volume 1587. [Google Scholar]
- Bao, H.L.; Liu, H.S.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef]
- D’Atri, V.; Gabelica, V. DNA and RNA telomeric G-quadruplexes: What topology features can be inferred from ion mobility mass spectrometry? Analyst 2019, 144, 6074–6088. [Google Scholar] [CrossRef]
- Manna, S.; Panse, C.H.; Sontakke, V.A.; Sangamesh, S.; Srivatsan, S.G. Probing Human Telomeric DNA and RNA Topology and Ligand Binding in a Cellular Model by Using Responsive Fluorescent Nucleoside Probes. ChemBioChem 2017, 18, 1604–1615. [Google Scholar] [CrossRef]
- Poudel, L.; Steinmetz, N.F.; French, R.H.; Parsegian, V.A.; Podgornik, R.; Ching, W.Y. Implication of the solvent effect, metal ions and topology in the electronic structure and hydrogen bonding of human telomeric G-quadruplex DNA. Phys. Chem. Chem. Phys. 2016, 18, 21573–21585. [Google Scholar] [CrossRef]
- Trybek, T.; Kowalik, A.; Góźdź, S.; Kowalska, A. Telomeres and telomerase in oncogenesis (review). Oncol. Lett. 2020, 20, 1015–1027. [Google Scholar] [CrossRef]
- Xu, Y.; Goldkorn, A. Telomere and telomerase therapeutics in cancer. Genes 2016, 7. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing novel G-quadruplex ligands: From interaction with nucleic acids to interfering with nucleic acid–protein interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. A Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.-P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Neidle, S. Design Principles for Quadruplex-binding Small Molecules. In Therapeutic Applications of Quadruplex Nucleic Acids; Academic Press: London, UK, 2012. [Google Scholar]
- Hemalatha, C.N.; Vijey Aanandhi, M. G-Quadruplex ligands as stabilizer targeting telomerase enzyme as anti cancer agents. Asian J. Pharm. Clin. Res. 2017, 10, 50–53. [Google Scholar] [CrossRef][Green Version]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 5351–5355. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, S.; Dotsenko, I.A.; Samoshin, V.V.; Xue, L. Arylsulfanyl groups—Suitable side chains for 5-substituted 1,10-phenanthroline and nickel complexes as G4 ligands and telomerase inhibitors. J. Inorg. Biochem. 2017, 173, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.C.; Borch, J.; Ulven, T. Design, synthesis and evaluation of 4,7-diamino-1,10-phenanthroline G-quadruplex ligands. Bioorg. Med. Chem. 2009, 17, 8241–8246. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, Y.; Liu, J.; Zhou, J.; Li, C.; Wei, C. Promoting the formation and stabilization of human telomeric G-quadruplex DNA, inhibition of telomerase and cytotoxicity by phenanthroline derivatives. Org. Biomol. Chem. 2011, 9, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, L.; Zhang, N.; Liu, Y.; Zheng, W.; Chang, A.; Wang, F.; Li, S.; Shangguan, D. A Bis(methylpiperazinylstyryl)phenanthroline as a Fluorescent Ligand for G-Quadruplexes. Chem. A Eur. J. 2016, 22, 6037–6047. [Google Scholar] [CrossRef]
- Ang, D.L.; Kelso, C.; Beck, J.L.; Ralph, S.F.; Harman, D.G.; Aldrich-Wright, J.R. A study of Pt(II)–phenanthroline complex interactions with double-stranded and G-quadruplex DNA by ESI–MS, circular dichroism, and computational docking. J. Biol. Inorg. Chem. 2020, 25. [Google Scholar] [CrossRef]
- Luo, X.J.; Qin, Q.P.; Li, Y.L.; Liu, Y.C.; Chen, Z.F.; Liang, H. Three platinum(II) complexes of 2-(methoxy-phenyl)-imidazo-[4,5-f]-[1,10] phenanthroline: Cell apoptosis induction by sub-G1 phase cell cycle arrest and G-quadruplex binding properties. Inorg. Chem. Commun. 2014, 46, 176–179. [Google Scholar] [CrossRef]
- Rigo, R.; Bianco, S.; Musetti, C.; Palumbo, M.; Sissi, C. Molecular Basis for Differential Recognition of G-Quadruplex versus Double-Helix DNA by Bis-Phenanthroline Metal Complexes. ChemMedChem 2016, 1–9. [Google Scholar] [CrossRef]
- Duskova, K.; Sierra, S.; Arias-Pérez, M.-S.; Gude, L. Human telomeric G-quadruplex DNA interactions of N-phenanthroline glycosylamine copper(II) complexes. Bioorg. Med. Chem. 2016, 33–41. [Google Scholar] [CrossRef]
- Saczewski, F.; Balewski, Ł. Biological activities of guanidine compounds. Expert Opin. Ther. Pat. 2009, 19, 1417–1448. [Google Scholar] [CrossRef]
- Luedtke, N.W.; Baker, T.J.; Goodman, M.; Tor, Y. Guanidinoglycosides: A Novel Family of RNA Ligands. J. Am. Chem. Soc. 2000, 122, 12035–12036. [Google Scholar] [CrossRef]
- Alzeer, J.; Vummidi, B.R.; Roth, P.J.C.; Luedtke, N.W. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew. Chem. Int. Ed. 2009, 48, 9362–9365. [Google Scholar] [CrossRef] [PubMed]
- Ilyinsky, N.S.; Shchyolkina, A.K.; Borisova, O.F.; Mamaeva, O.K.; Zvereva, M.I.; Azhibek, D.M.; Livshits, M.A.; Mitkevich, V.A.; Balzarini, J.; Sinkevich, Y.B.; et al. Novel multi-targeting anthra [2,3-b]thiophene-5,10-diones with guanidine-containing side chains: Interaction with telomeric G-quadruplex, inhibition of telomerase and topoisomerase i and cytotoxic properties. Eur. J. Med. Chem. 2014, 85, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Morigi, R.; Pagano, B.; Pagano, A.; Ohnmacht, S.; De Magis, A.; Tiang, Y.-P.; Capranico, G.; Locatelli, A.; Graziadio, A.; et al. Toward the Development of Specific G-Quadruplex Binders: Synthesis, Biophysical, and Biological Studies of New Hydrazone Derivatives. J. Med. Chem. 2016, 59, 5706–5720. [Google Scholar] [CrossRef] [PubMed]
- Brassart, B.; Gomez, D.; De Cian, A.; Paterski, R.; Montagnac, A.; Qui, K.H.; Temime-Smaali, N.; Trentesaux, C.; Mergny, J.L.; Gueritte, F.; et al. A new steroid derivative stabilizes G-quadruplexes and induces telomere uncapping in human tumor cells. Mol. Pharmacol. 2007, 72, 631–640. [Google Scholar] [CrossRef]
- Battogtokh, G.; Choi, Y.S.; Kang, D.S.; Park, S.J.; Shim, M.S.; Huh, K.M.; Cho, Y.Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Mitochondria-targeting drug conjugates for cytotoxic, anti-oxidizing and sensing purposes: Current strategies and future perspectives. Acta Pharm. Sin. B 2018, 8, 862–880. [Google Scholar] [CrossRef]
- Sibrian-Vazquez, M.; Nesterova, I.V.; Jensen, T.J.; Vicente, M.G.H. Mitochondria targeting by guanidine- and biguanidine-porphyrin photosensitizers. Bioconjug. Chem. 2008, 19, 705–713. [Google Scholar] [CrossRef]
- Chandler, C.J.; Deady, L.W.; Reiss, J.A. Synthesis of some 2,9-Disubsituted-1,10-phenanthrolines. J. Heterocycl. Chem. 1981, 18, 599–601. [Google Scholar] [CrossRef]
- Sierra, S. Síntesis de Ligandos de ADN Cuádruple-G Derivados de 1,10-Fenantrolina. Estudios de Interacción y Actividad Biológica. Ph.D. Thesis, Universidad de Alcalá, Madrid, Spain, 2017. [Google Scholar]
- Amidi, S.; Esfahanizadeh, M.; Tabib, K.; Soleimani, Z.; Kobarfard, F. Rational Design and Synthesis of 1-(Arylideneamino)-4-aryl-1H-imidazole-2-amine Derivatives as Antiplatelet Agents. ChemMedChem 2017, 12, 962–971. [Google Scholar] [CrossRef]
- De Cian, A.; Guittat, L.; Kaiser, M.; Saccà, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.P.; Lacroix, L.; Mergny, J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef]
- Kieltyka, R.; Englebienne, P.; Fakhoury, J.; Autexier, C.; Moitessier, N.; Sleiman, H.F. A Platinum Supramolecular Square as an Effective G-Quadruplex Binder and Telomerase Inhibitor. J. Am. Chem. Soc. 2008, 130, 10040–10041. [Google Scholar] [CrossRef]
- Randazzo, A.; Spada, G.P.; Da Silva, M.W. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013, 330, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef] [PubMed]
- del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Karsisiotis, A.I.; Hessari, N.M.A.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba Da Silva, M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Marchand, A.; Granzhan, A.; Iida, K.; Tsushima, Y.; Ma, Y.; Nagasawa, K.; Teulade-Fichou, M.P.; Gabelica, V. Ligand-induced conformational changes with cation ejection upon binding to human telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2015, 137, 750–756. [Google Scholar] [CrossRef]
- White, E.W.; Tanious, F.; Ismail, M.A.; Reszka, A.P.; Neidle, S.; Boykin, D.W.; Wilson, W.D. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007, 126, 140–153. [Google Scholar] [CrossRef]
- Garbett, N.C.; Ragazzon, P.A.; Chaires, J.O.B. Circular dichroism to determine binding mode and affinity of ligand-dna interactions. Nat. Protoc. 2007, 2. [Google Scholar] [CrossRef]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of human telomere G-quadruplex in the presence of a model drug along the thermal unfolding pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. G4-FID: A fluorescent DNA probe displacement assay for rapid evaluation of quadruplex ligands. Methods Mol. Biol. 2010, 608, 257–271. [Google Scholar] [CrossRef]
- Chaires, J.B. Structural selectivity of drug-nucleic acid interactions probed by competition dialysis. Top. Curr. Chem. 2005, 253, 33–53. [Google Scholar] [CrossRef]
- Suh, D.; Chaires, J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995, 3, 723–728. [Google Scholar] [CrossRef]
- Cohen, G.; Eisenberg, H. Viscosity and sedimentation study of sonicated DNA–proflavine complexes. Biopolymers 1969, 8, 45–55. [Google Scholar] [CrossRef]
- Fairley, T.A.; Tidwell, R.R.; Donkor, I.; Naiman, N.A.; Ohemeng, K.A.; Lombardy, R.J.; Bentley, J.A.; Cory, M. Structure, DNA Minor Groove Binding, and Base Pair Specificity of Alkyl- and Aryl-Linked Bis(amidinobenzimidazoles) and Bis(amidinoindoles). J. Med. Chem. 1993, 36, 1746–1753. [Google Scholar] [CrossRef]
- Bailly, C.; Arafa, R.K.; Tanious, F.A.; Laine, W.; Tardy, C.; Lansiaux, A.; Colson, P.; Boykin, D.W.; Wilson, W.D. Molecular determinants for DNA minor groove recognition: Design of a bis-guanidinium derivative of ethidium that is highly selective for AT-Rich DNA sequences. Biochemistry 2005, 44, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- Nagle, P.S.; McKeever, C.; Rodriguez, F.; Nguyen, B.; Wilson, W.D.; Rozas, I. Unexpected DNA affinity and sequence selectivity through core rigidity in guanidinium-based minor groove binders. J. Med. Chem. 2014, 57, 7663–7672. [Google Scholar] [CrossRef] [PubMed]
- Nagle, P.S.; Quinn, S.J.; Kelly, J.M.; O’Donovan, D.H.; Khan, A.R.; Rodriguez, F.; Nguyen, B.; Wilson, W.D.; Rozas, I. Understanding the DNA binding of novel non-symmetrical guanidinium/2-aminoimidazolinium derivatives. Org. Biomol. Chem. 2010, 8, 5558–5567. [Google Scholar] [CrossRef] [PubMed]
- McKeever, C.; Kaiser, M.; Rozas, I. Aminoalkyl derivatives of guanidine diaromatic minor groove binders with antiprotozoal activity. J. Med. Chem. 2013, 56, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Arafa, K.; Sedky, N.K.; Alakhdar, A.A.; Arafa, R.K. Triaryl dicationic DNA minor-groove binders with antioxidant activity display cytotoxicity and induce apoptosis in breast cancer: Dicationic Compounds as Anticancer and Apoptotic Inducing Agents. Chem. Biol. Interact. 2020, 324, 5558–5567. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, H.E.; Kotlyar, V.; Nudelman, A. NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997, 62, 7512–7515. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 9–18. [Google Scholar] [CrossRef]
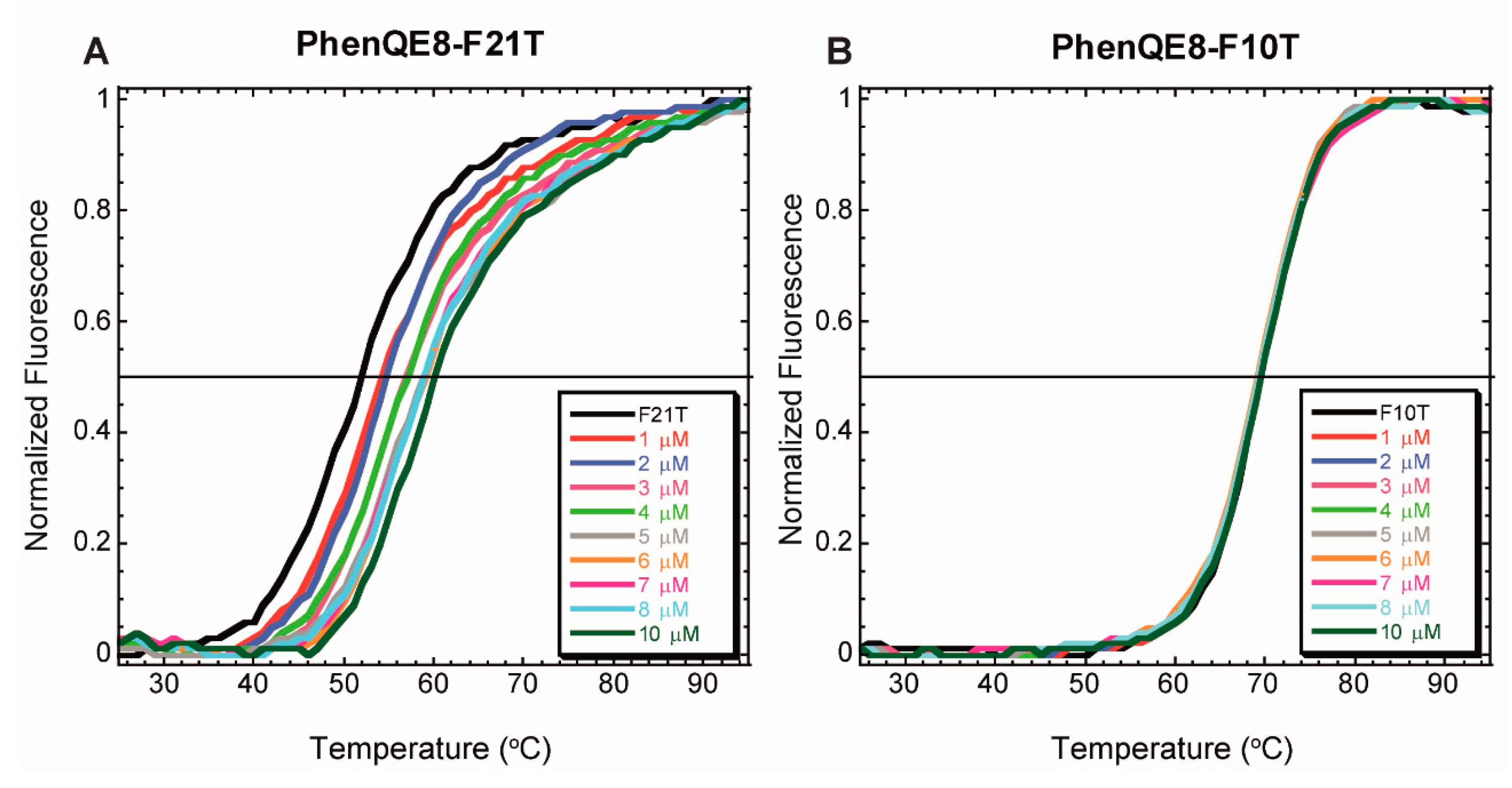
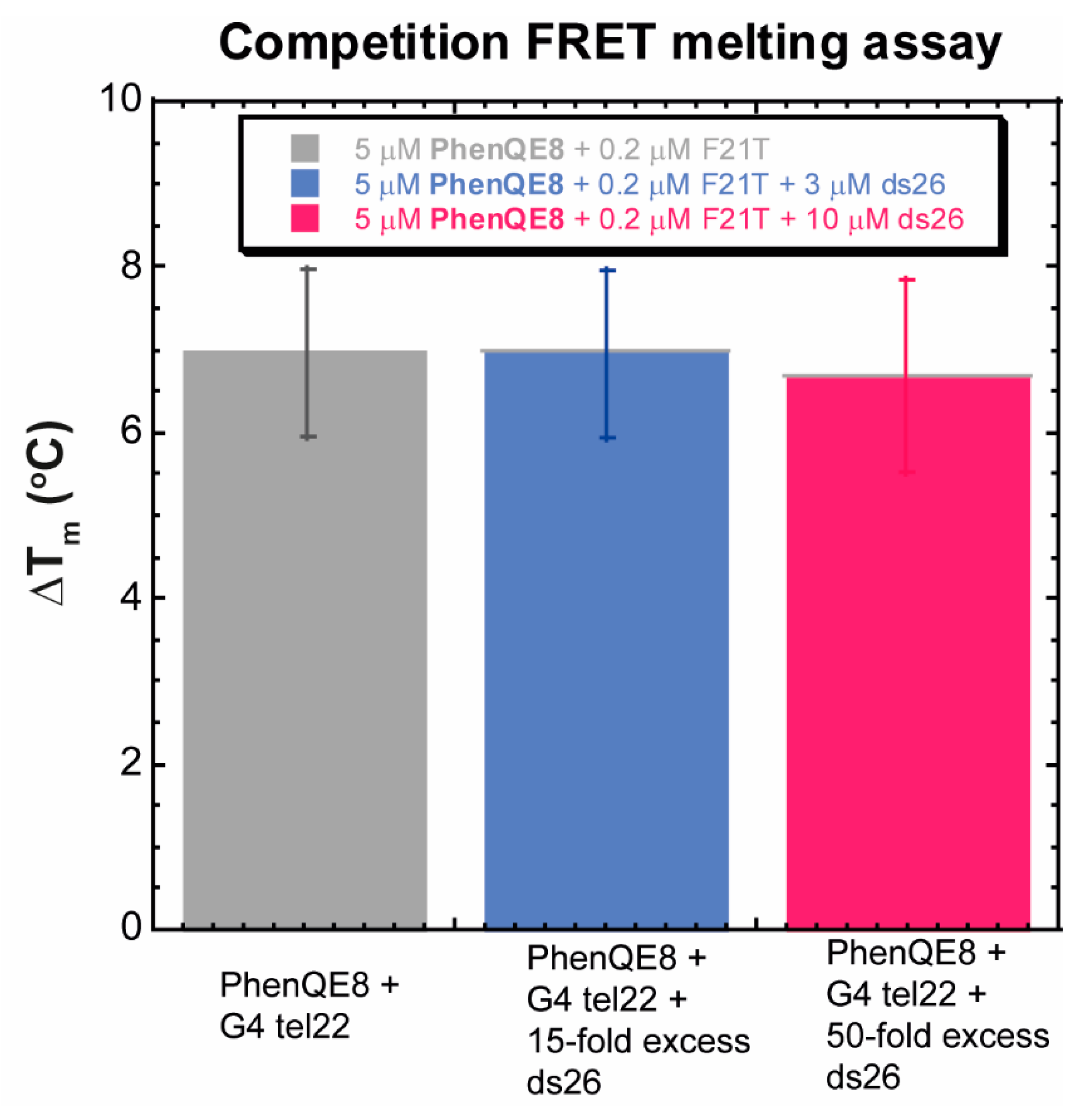

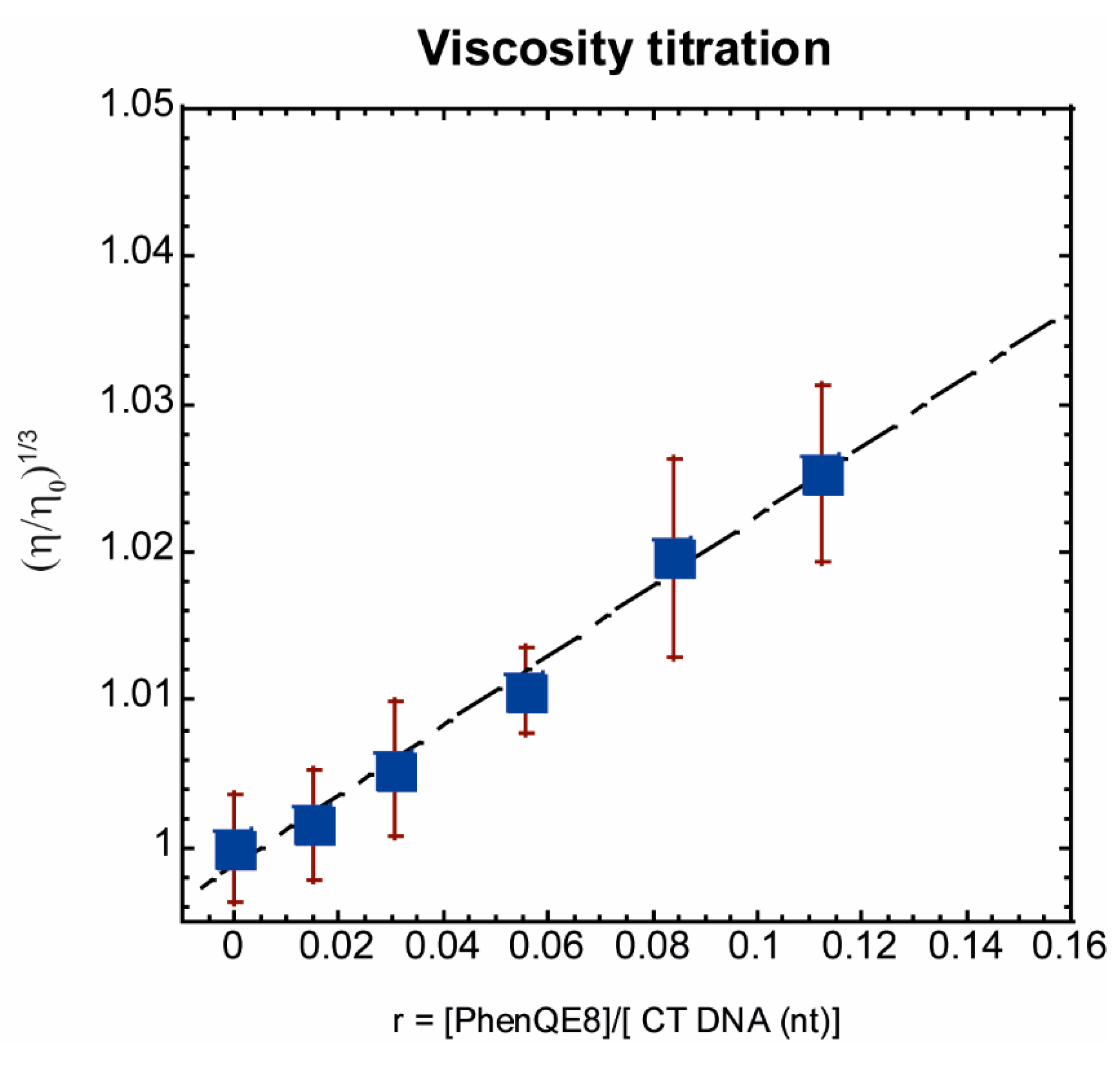
| PhenQE8 + Tel22 | PhenQE8 + ds17 | PhenQE8 + Tel22 + ds17 | PhenQE8 + CT | |
|---|---|---|---|---|
| DNA | Kapp(M−1) × 10−6 | Kapp(M−1) × 10−6 | Kapp(M−1) × 10−6 | Kapp(M−1) × 10−6 |
| Tel22 (Q) | 2.3 ± 0.5 | -- | 1.35 ± 0.26 | -- |
| ds17 (D) | -- | 0.029 ± 0.004 | 0.015 ± 0.005 | -- |
| CT dsDNA | -- | -- | -- | 0.058 ± 0.013 |
| PC-3 | HeLa | MCF-7 | HFF-1 | RWPE-1 | |
|---|---|---|---|---|---|
| Compound | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) | IC50 (μM) |
| PhenQE8 | 5.6 ± 0.3 | 16.2 ± 1.6 | 52.7 ± 1.0 | 170 ± 23 | 38.8 ± 0.5 |
| 360A | 2.5 ± 0.8 | 8.5 ± 1.4 | 44.9 ± 9.2 | 30.6 ± 2.3 | 9.9 ± 1.0 |
| cisplatin | 12.8 ± 0.9 | 18.0 ± 1.9 | 96.5 ± 6.7 | 36.6 ± 4.4 | 16.4 ± 1.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gratal, P.B.; Quero, J.G.; Pérez-Redondo, A.; Gándara, Z.; Gude, L. PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex. Int. J. Mol. Sci. 2021, 22, 749. https://doi.org/10.3390/ijms22020749
Gratal PB, Quero JG, Pérez-Redondo A, Gándara Z, Gude L. PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex. International Journal of Molecular Sciences. 2021; 22(2):749. https://doi.org/10.3390/ijms22020749
Chicago/Turabian StyleGratal, Patricia B., Julia G. Quero, Adrián Pérez-Redondo, Zoila Gándara, and Lourdes Gude. 2021. "PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex" International Journal of Molecular Sciences 22, no. 2: 749. https://doi.org/10.3390/ijms22020749
APA StyleGratal, P. B., Quero, J. G., Pérez-Redondo, A., Gándara, Z., & Gude, L. (2021). PhenQE8, a Novel Ligand of the Human Telomeric Quadruplex. International Journal of Molecular Sciences, 22(2), 749. https://doi.org/10.3390/ijms22020749