Genetic Variations in Prostaglandin E2 Pathway Identified as Susceptibility Biomarkers for Gastric Cancer in an Intermediate Risk European Country
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. Genotype Frequencies and Risk Estimates
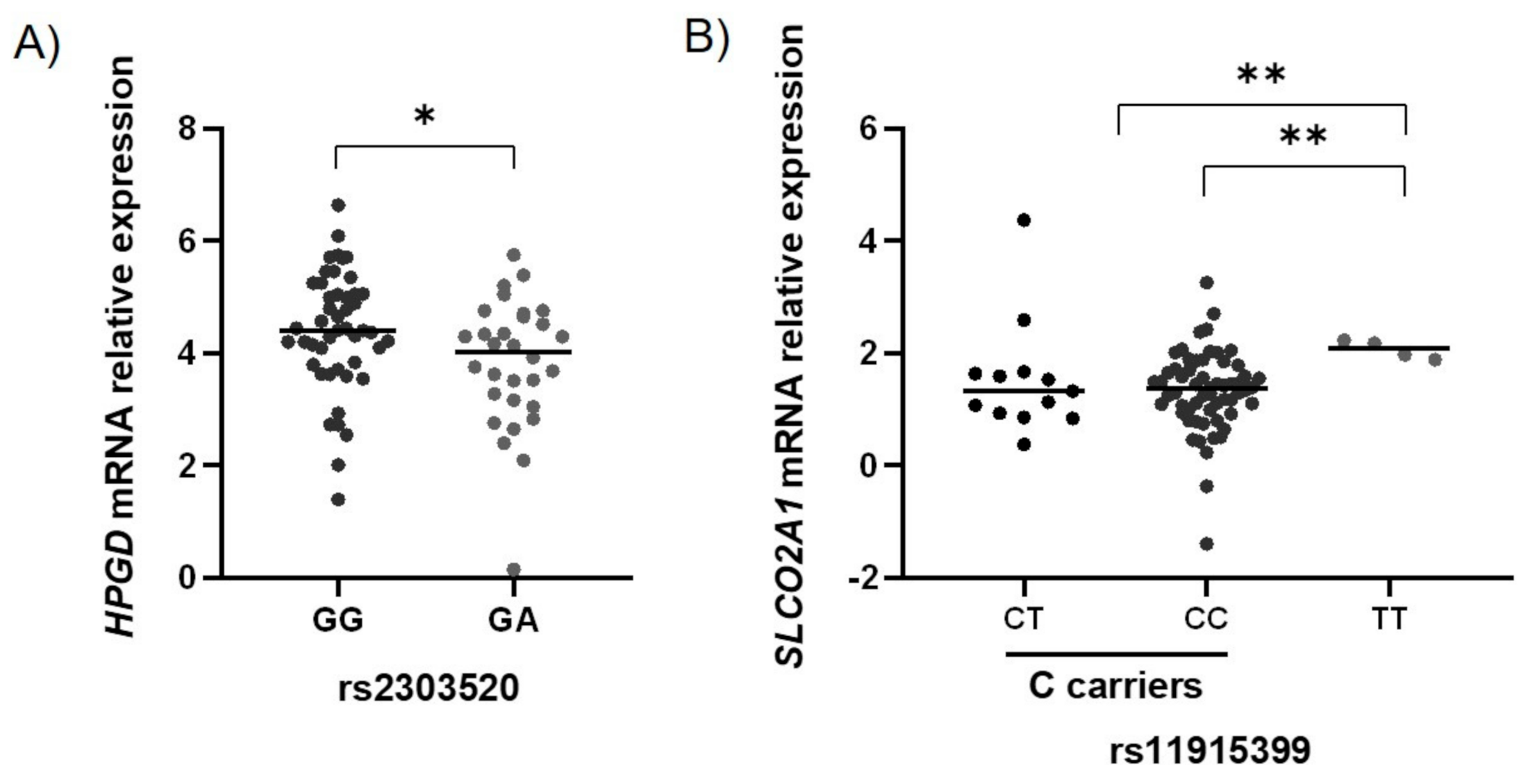
2.3. Functional Characterization of the GC-Associated Biomarkers
2.4. Haplotype Analysis
2.5. Gene-“Environment” Interaction Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection and Processing
4.3. Genetic Polymorphisms Selection and Characterization
4.4. Reverse Transcription Reaction
4.5. Real-Time PCR
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 15-PGDH | 15-hydroxyprostaglandin dehydrogenase |
| ABCC4 | ATP binding cassette subfamily c member 4 |
| B2M | beta-2-microglobulin |
| CASP8 | caspase 8 |
| cDNA | complementary deoxyribonucleic acid |
| CEU | Utah residents with Northern and Western European ancestry |
| CI | confidence interval |
| CT | cycle threshold |
| CVC | cross-variation consistency |
| EM | expectation-maximization algorithm |
| FDR | false discovery rate |
| FFPE | formalin-fixed paraffin-embedded |
| GC | gastric cancer |
| GSTP1 | glutathione s-transferase pi 1 |
| GUSB | glucuronidase beta |
| HPGD | 15-hydroxyprostaglandin dehydrogenase |
| HPRT1 | hypoxanthine phosphoribosyltransferase 1 |
| HWE | Hardy-Weinberg equilibrium |
| IPO8 | importin 8 |
| MDR | multifactor dimensionality reduction |
| mRNA | messenger ribonucleic acid |
| MRP4 | multidrug resistance protein 4 |
| MTX1 | metaxin 1 |
| MUC1 | mucin 1, cell surface-associated |
| OD | optical density |
| PCR | polymerase chain reaction |
| PGE2 | prostaglandin E2 |
| PGT | prostaglandin transporter |
| PKLR | pyruvate kinase L/R |
| PLCE1 | phospholipase C epsilon 1 |
| PPIA | peptidylprolyl isomerase A |
| PRKAA1 | protein kinase AMP-activated catalytic subunit alpha |
| PSCA | prostate stem cell antigen |
| PTGS2 | prostaglandin-endoperoxide synthase 2 |
| RPL29 | ribosomal protein L29 |
| RT | reverse transcription |
| SLCO2A1 | solute carrier organic anion transporter family member 2A1 |
| SPSS | statistical package for the social sciences |
| TGFBR2 | transforming growth factor-beta receptor 2 |
| TNF | tumor necrosis factor-alpha |
References
- Gao, Y.; Zhang, K.; Xi, H.; Cai, A.; Wu, X.; Cui, J.; Li, J.; Qiao, Z.; Wei, B.; Chen, L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: A meta-analysis. Oncotarget 2016, 8, 6330–6340. [Google Scholar] [CrossRef] [PubMed]
- Maconi, G.; Manes, G.; Porro, G.-B. Role of symptoms in diagnosis and outcome of gastric cancer. World J. Gastroenterol. 2008, 14, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Areia, M.; Spaander, M.C.; Kuipers, E.J.; Dinis-Ribeiro, M. Endoscopic screening for gastric cancer: A cost-utility analysis for countries with an intermediate gastric cancer risk. United Eur. Gastroenterol. J. 2018, 6, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; DuBois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Greenhough, A.; Smartt, H.J.M.; Moore, A.E.; Roberts, H.R.; Williams, A.C.; Paraskeva, C.; Kaidi, A. The COX-2/PGE 2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30, 377–386. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Chen, E.P.; Smyth, E.M. COX-2 and PGE2-dependent immunomodulation in breast cancer. Prostaglandins Other Lipid Mediat. 2011, 96, 14–20. [Google Scholar] [CrossRef]
- Kasper, H.U.; Konze, E.; Dienes, H.P.; Stippel, D.L.; Schirmacher, P.; Kern, M. COX-2 expression and effects of COX-2 inhibition in colorectal carcinomas and their liver metastases. Anticancer Res. 2010, 30, 2017–2023. [Google Scholar]
- Mrena, J.; Wiksten, J.-P.; Kokkola, A.; Nordling, S.; Ristimäki, A.; Haglund, C. COX-2 is associated with proliferation and apoptosis markers and serves as an independent prognostic factor in gastric cancer. Tumor Biol. 2010, 31, 1–7. [Google Scholar] [CrossRef]
- Gomes, R.N.; da Costa, S.F.; Colquhoun, A. Eicosanoids and cancer. Clinics 2018, 73. [Google Scholar] [CrossRef]
- Nakanishi, T.; Tamai, I. Roles of Organic Anion Transporting Polypeptide 2A1 (OATP2A1/SLCO2A1) in Regulating the Pathophysiological Actions of Prostaglandins. AAPS J. 2017, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Verdi, D.; Pooley, K.A.; Nitti, D. Genetic variation and gastric cancer risk: A field synopsis and meta-analysis. Gut 2015, 64, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Pereira, C.; Farinha, M.; Medeiros, R.; Dinis-Ribeiro, M. Prostaglandin E2 pathway is dysregulated in gastric adenocarcinoma in a Caucasian population. Int. J. Mol. Sci. 2020, 21, 7680. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Queiros, S.; Galaghar, A.; Sousa, H.; Pimentel-Nunes, P.; Brandao, C.; Moreira-Dias, L.; Medeiros, R.; Dinis-Ribeiro, M. Genetic variability in key genes in prostaglandin E2 pathway (COX-2, HPGD, ABCC4 and SLCO2A1) and their involvement in colorectal cancer development. PLoS ONE 2014, 9, e92000. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.D.; Silva, C.; Rocha, A.; Nogueira, C.; Castro-Poças, F.; Araujo, A.; Matos, E.; Pereira, C.; Medeiros, R.; Lopes, C. Predictive clinical model of tumor response after chemoradiation in rectal cancer. Oncotarget 2017, 8, 58133–58151. [Google Scholar] [CrossRef]
- Pereira, C.; Queiros, S.; Galaghar, A.; Sousa, H.; Marcos-Pinto, R.; Pimentel-Nunes, P.; Brandao, C.; Medeiros, R.; Dinis-Ribeiro, M. Influence of genetic polymorphisms in prostaglandin E2 pathway (COX-2/HPGD/SLCO2A1/ABCC4) on the risk for colorectal adenoma development and recurrence after polypectomy. Clin. Transl. Gastroenterol. 2016, 7, e191. [Google Scholar] [CrossRef]
- Luo, M.-X.; Long, B.-B.; Li, F.; Zhang, C.; Pan, M.-T.; Huang, Y.-Q.; Chen, B. Roles of Cyclooxygenase-2 gene −765G>C (rs20417) and −1195G>A (rs689466) polymorphisms in gastric cancer: A systematic review and meta-analysis. Gene 2019, 685, 125–135. [Google Scholar] [CrossRef]
- Huang, T.; Shu, Y.; Cai, Y.-D. Genetic differences among ethnic groups. BMC Genom. 2015, 16, 1093. [Google Scholar] [CrossRef]
- Spielman, R.S.; Bastone, L.A.; Burdick, J.T.; Morley, M.; Ewens, W.J.; Cheung, V.G. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007, 39, 226–231. [Google Scholar] [CrossRef]
- Zmigrodzka, M.; Rzepecka, A.; Krzyzowska, M.; Witkowska-Pilaszewicz, O.; Cywinska, A.; Winnicka, A. The cyclooxygenase-2/prostaglandin E2 pathway and its role in the pathogenesis of human and dog hematological malignancies. J. Physiol. Pharmacol. 2018, 69. [Google Scholar] [CrossRef]
- Hashemi Goradel, N.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in cancer: A review. J. Cell. Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Omori, K.; Murata, T. Role of prostaglandins in tumor microenvironment. Cancer Metastasis Rev. 2018, 37, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Rosenberg, D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 2013, 35, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Gagnon-Bartsch, J.A.; Tan, I.B.; Earle, S.; Ruff, L.; Pettinger, K.; Ylstra, B.; van Grieken, N.; Rha, S.Y.; Chung, H.C.; et al. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut 2015, 64, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, T.; Tohnai, N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002, 68–69, 95–114. [Google Scholar] [CrossRef]
- Pereira, C.; Sousa, H.; Silva, J.; Brandao, C.; Elgueta-Karstegl, C.; Farrell, P.J.; Medeiros, R.; Dinis-Ribeiro, M. The -1195G allele increases the transcriptional activity of cyclooxygenase-2 gene (COX-2) in colon cancer cell lines. Mol. Carcinog. 2014, 53 (Suppl. 1), E92–E95. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Zhao, H.; Chen, C.; Ali, M.A. COX-2 gene rs689466 polymorphism is associated with increased risk of colorectal cancer among Caucasians: A meta-analysis. World J. Surg. Oncol. 2020, 18, 192. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Liu, T.; Tang, W.; Ma, Z. Cyclooxygenase-2 -1195G>A (rs689466) polymorphism and cancer susceptibility: An updated meta-analysis involving 50,672 subjects. Int. J. Clin. Exp. Med. 2015, 8, 12448–12462. [Google Scholar]
- Zamudio, R.; Pereira, L.; Rocha, C.D.; Berg, D.E.; Muniz-Queiroz, T.; Sant Anna, H.P.; Cabrera, L.; Combe, J.M.; Herrera, P.; Jahuira, M.H.; et al. Population, epidemiological, and functional genetics of gastric cancer candidate genes in Peruvians with predominant Amerindian ancestry. Dig. Dis. Sci. 2016, 61, 107–116. [Google Scholar] [CrossRef]
- Rigau, M.; Juan, D.; Valencia, A.; Rico, D. Intronic CNVs and gene expression variation in human populations. PLoS Genet. 2019, 15, e1007902. [Google Scholar] [CrossRef]
- Lomelin, D.; Jorgenson, E.; Risch, N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010, 20, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Van Steen, K. The search for gene-gene interactions in genome-wide association studies: Challenges in abundance of methods, practical considerations, and biological interpretation. Ann. Transl. Med. 2018, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.-Y.; Braunschweig, T.; Hewitt, S.M. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn. Mol. Pathol. 2006, 15, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.-Y.; Braunschweig, T.; Williams, R.; Guerrero, N.; Hoffmann, K.M.; Kwon, M.; Song, Y.K.; Libutti, S.K.; Hewitt, S.M. Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue. J. Histochem. Cytochem. 2008, 56, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Antonov, J.; Goldstein, D.R.; Oberli, A.; Baltzer, A.; Pirotta, M.; Fleischmann, A.; Altermatt, H.J.; Jaggi, R. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab. Investig. 2005, 85, 1040–1050. [Google Scholar] [CrossRef]
- Walter, R.F.H.; Mairinger, F.D.; Wohlschlaeger, J.; Worm, K.; Ting, S.; Vollbrecht, C.; Schmid, K.W.; Hager, T. FFPE tissue as a feasible source for gene expression analysis—A comparison of three reference genes and one tumor marker. Pathol. Res. Pract. 2013, 209, 784–789. [Google Scholar] [CrossRef]
- Mueller, J.C.; Lõhmussaar, E.; Mägi, R.; Remm, M.; Bettecken, T.; Lichtner, P.; Biskup, S.; Illig, T.; Pfeufer, A.; Luedemann, J.; et al. Linkage disequilibrium patterns and tagSNP transferability among European populations. Am. J. Hum. Genet. 2005, 76, 387–398. [Google Scholar] [CrossRef][Green Version]
- Service, S.; Sabatti, C.; Freimer, N. Tag SNPs chosen from HapMap perform well in several population isolates. Genet. Epidemiol. 2007, 31, 189–194. [Google Scholar] [CrossRef]
- Ribas, G.; González-Neira, A.; Salas, A.; Milne, R.L.; Vega, A.; Carracedo, B.; González, E.; Barroso, E.; Fernández, L.P.; Yankilevich, P.; et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum. Genet. 2006, 118, 669–679. [Google Scholar] [CrossRef]
- Gu, S.; Pakstis, A.J.; Li, H.; Speed, W.C.; Kidd, J.R.; Kidd, K.K. Significant variation in haplotype block structure but conservation in tagSNP patterns among global populations. Eur. J. Hum. Genet. 2007, 15, 302–312. [Google Scholar] [CrossRef]
- de Bakker, P.I.W.; Burtt, N.P.; Graham, R.R.; Guiducci, C.; Yelensky, R.; Drake, J.A.; Bersaglieri, T.; Penney, K.L.; Butler, J.; Young, S.; et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat. Genet. 2006, 38, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: Berlin/Heidelberg, Germany, 2018; p. 1032. [Google Scholar] [CrossRef]
- Pereira, C.; Sousa, H.; Ferreira, P.; Fragoso, M.; Moreira-Dias, L.; Lopes, C.; Medeiros, R.; Dinis-Ribeiro, M. -765G > C COX-2 polymorphism may be a susceptibility marker for gastric adenocarcinoma in patients with atrophy or intestinal metaplasia. World J. Gastroenterol. 2006, 12, 5473–5478. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.L.; Fink, S.P.; Lutterbaugh, J.D.; Elston, R.C.; Veigl, M.L.; Markowitz, S.D.; Li, L. Genetic variation in 15-hydroxyprostaglandin dehydrogenase and colon cancer susceptibility. PLoS ONE 2013, 8, e64122. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, F.; Liu, X.; Yuan, D.; Zi, Y.; He, X.; He, R. Detection and clinical significance of COX-2 gene SNPs in gastric cancer. Cell Biochem. Biophys. 2015, 72, 657–660. [Google Scholar] [CrossRef] [PubMed]
- Hoeft, B.; Linseisen, J.; Beckmann, L.; Müller-Decker, K.; Canzian, F.; Hüsing, A.; Kaaks, R.; Vogel, U.; Jakobsen, M.U.; Overvad, K.; et al. Polymorphisms in fatty acid metabolism-related genes are associated with colorectal cancer risk. Carcinogenesis 2009, 31, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Sorby, L.A.A.; Andersen, S.N.; Bukholm, I.R.K.; Jacobsen, M.B. Evaluation of suitable reference genes for normalization of real-time reverse transcription PCR analysis in colon cancer. J. Exp. Clin. Cancer Res. 2010, 29. [Google Scholar] [CrossRef]
- Rho, H.W.; Lee, B.C.; Choi, E.S.; Choi, I.J.; Lee, Y.S.; Goh, S.H. Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qPCR. BMC Cancer 2010, 10, 240. [Google Scholar] [CrossRef]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Cases (n = 260) | Controls (n = 476) | p Value | ||
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | ||||
| Mean ± SD | 69.87 ± 0.60 | 57.98 ± 0.23 | < 0.001 | |
| Median (min-max) | 70 (50–92) | 58 (50–69) | ||
| Sex, n (%) | ||||
| Male | 152 (58.5) | 312 (65.5) | 0.057 | |
| Female | 108 (41.5) | 164 (34.5) | ||
| Tumor characteristics | ||||
| Tumor location, n (%) | ||||
| Cardia and GEJ | 24 (9.4) | -- | ||
| Fundus and corpus | 41 (16.1) | -- | ||
| Antrum and corpus-antrum transition | 157 (61.6) | -- | ||
| Angularis incisura | 7 (2.7) | |||
| Others * | 26 (10.2) | |||
| Grade, n (%) | ||||
| Well-differentiated | 28 (10.8) | -- | ||
| Moderately differentiated | 157 (60.6) | -- | ||
| Poorly differentiated | 63 (24.3) | -- | ||
| Cannot be assessed | 11 (4.2) | -- | ||
| Stage, n (%) | ||||
| I-II | 145 (56.0) | -- | ||
| III-IV | 114 (44.0) | -- | ||
| Synchronous tumors, n (%) | ||||
| Yes | 6 (2.3) | -- | ||
| No | 254 (97.7) | -- | ||
| SNP | Model | Genotype Frequencies | Univariate Analysis | Multivariate Analysis | Age at Diagnosis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases, n (%) | Controls, n (%) | OR | 95% CI | p Value | aOR | 95% CI | p Value | Median (years) | 95% CI | p Value | ||
| PTGS2 | ||||||||||||
| rs689466 | Codominant | |||||||||||
| AA | 121 (61.1) | 322 (68.8) | 1.00 | - | 0.054 | 1.00 | - | 0.021 | 73.00 | 71.25–74.75 | - | |
| AG | 63 (31.8) | 130 (27.8) | 1.29 | 0.89–1.86 | 1.50 | 0.93–2.42 | 73.00 | 68.89–77.11 | 0.21 | |||
| GG | 14 (7.1) | 16 (3.4) | 2.33 | 1.10–4.92 | 3.40 | 1.29–8.97 | 70.00 | 62.49–77.52 | 0.008 | |||
| Dominant | ||||||||||||
| AA | 121 (61.1) | 322 (68.8) | 1.00 | - | 0.056 | 1.00 | - | 0.022 | 73.00 | 71.25–74.75 | - | |
| AG-GG | 77 (38.9) | 146 (31.2) | 1.40 | 0.99–1.98 | 1.69 | 1.08–2.65 | 73.00 | 69.94–76.06 | 0.058 | |||
| Recessive | ||||||||||||
| AA-AG | 184 (92.9) | 452 (96.6) | 1.00 | - | 0.046 | 1.00 | - | 0.027 | 73.00 | 71.48–74.52 | - | |
| GG | 14 (7.1) | 16 (3.4) | 2.15 | 1.03–4.49 | 2.98 | 1.14–7.74 | 70.00 | 62.49–77.52 | 0.011 | |||
| Overdominant | ||||||||||||
| AA-GG | 135 (68.2) | 338 (72.2) | 1.00 | - | 0.30 | 1.00 | - | 0.19 | 73.00 | 71.46–74.54 | - | |
| AG | 63 (31.8) | 130 (27.8) | 1.21 | 0.85–1.74 | 1.37 | 0.86–2.19 | 73.00 | 68.89–77.11 | 0.38 | |||
| Log-additive | - | 1.40 | 1.06–1.86 | 0.021 | 1.66 | 1.15–2.40 | 0.007 | - | ||||
| ABCC4 | ||||||||||||
| rs1678374 | Codominant | |||||||||||
| TT | 90 (40.5) | 161 (33.9) | 1.00 | - | 0.076 | 1.00 | - | 0.063 | 72.00 | 69.48–74.52 | - | |
| TC | 107 (48.2) | 234 (49.3) | 0.82 | 0.58–1.15 | 1.04 | 0.67–1.63 | 72.00 | 69.74–74.26 | 0.47 | |||
| CC | 25 (11.3) | 80 (16.8) | 0.56 | 0.33–0.94 | 0.50 | 0.26–0.97 | 73.00 | 71.00–75.00 | 0.57 | |||
| Dominant | ||||||||||||
| TT | 90 (40.5) | 161 (33.9) | 1.00 | - | 0.09 | 1.00 | - | 0.55 | 72.00 | 69.48–74.52 | - | |
| TC-CC | 132 (59.5) | 314 (66.1) | 0.75 | 0.54–1.04 | 0.88 | 0.58–1.34 | 72.00 | 70.77–73.23 | 0.66 | |||
| rs1678374 | Recessive | |||||||||||
| TT-TC | 197 (88.7) | 395 (83.2) | 1.00 | - | 0.05 | 1.00 | - | 0.019 | 72.00 | 70.38–73.63 | - | |
| CC | 25 (11.3) | 80 (16.8) | 0.63 | 0.39–1.01 | 0.49 | 0.26–0.91 | 73.00 | 71.00–75.00 | 0.39 | |||
| Overdominant | ||||||||||||
| TT-CC | 115 (51.8) | 241 (50.7) | 1.00 | - | 0.79 | 1.00 | - | 0.28 | 72.00 | 70.46–73.54 | - | |
| TC | 107 (48.2) | 234 (49.3) | 0.96 | 0.70–1.32 | 1.25 | 0.83–1.89 | 72.00 | 69.74–74.26 | 0.31 | |||
| Log-additive | - | 0.77 | 0.60–0.97 | 0.027 | 0.78 | 0.58–1.06 | 0.11 | - | ||||
| rs1678405 | Codominant | |||||||||||
| TT | 108 (50.2) | 196 (41.2) | 1.00 | - | 0.052 | 1.00 | - | 0.09 | 72.00 | 70.40–73.60 | - | |
| TC | 91 (42.3) | 226 (47.5) | 0.73 | 0.52–1.02 | 0.81 | 0.52–1.25 | 72.00 | 69.60–74.40 | 0.58 | |||
| CC | 16 (7.4) | 54 (11.3) | 0.54 | 0.29–0.99 | 0.44 | 0.20–0.95 | 74.00 | 67.60–80.40 | 0.68 | |||
| Dominant | ||||||||||||
| TT | 108 (50.2) | 196 (41.2) | 1.00 | - | 0.027 | 1.00 | - | 0.13 | 72.00 | 70.40–73.60 | - | |
| TC-CC | 107 (49.8) | 280 (58.8) | 0.69 | 0.50–0.96 | 0.73 | 0.48–1.10 | 72.00 | 70.18–73.82 | 0.70 | |||
| Recessive | ||||||||||||
| TT-TC | 199 (92.6) | 422 (88.7) | 1.00 | - | 0.11 | 1.00 | - | 0.049 | 72.00 | 70.58–73.42 | - | |
| CC | 16 (7.4) | 54 (11.3) | 0.63 | 0.35–1.13 | 0.49 | 0.23–1.03 | 74.00 | 67.60–80.40 | 0.58 | |||
| Overdominant | ||||||||||||
| TT-CC | 124 (57.7) | 250 (52.5) | 1.00 | - | 0.21 | 1.00 | - | 0.76 | 72.00 | 70.46–73.54 | - | |
| TC | 91 (42.3) | 226 (47.5) | 0.81 | 0.59–1.12 | 0.94 | 0.62–1.42 | 72.00 | 69.60–74.40 | 0.48 | |||
| Log-additive | - | 0.73 | 0.57–0.94 | 0.015 | 0.72 | 0.52–0.99 | 0.041 | - | ||||
| rs1751031 | Codominant | |||||||||||
| AA | 154 (69.4) | 296 (62.3) | 1.00 | - | 0.10 | 1.00 | - | 0.073 | 72.00 | 70.25–73.75 | - | |
| AG | 59 (26.6) | 164 (34.5) | 0.69 | 0.48 | 0.61 | 0.39–0.95 | 72.00 | 69.72–74.28 | 0.66 | |||
| GG | 9 (4.0) | 15 (302) | 1.15 | 0.49–2.70 | 0.57 | 0.17–1.92 | 78.00 | 69.81–86.19 | 0.29 | |||
| rs1751031 | Dominant | |||||||||||
| AA | 154 (69.4) | 296 (62.3) | 1.00 | - | 0.068 | 1.00 | - | 0.022 | 72.00 | 70.25–73.75 | - | |
| AG-GG | 68 (30.6) | 179 (37.7) | 0.73 | 0.52–1.03 | 0.60 | 0.39–0.94 | 72.00 | 70.08–73.93 | 0.46 | |||
| Recessive | ||||||||||||
| AA-AG | 213 (96.0) | 460 (96.8) | 1.00 | - | 0.55 | 1.00 | - | 0.52 | 72.00 | 70.61–73.39 | - | |
| GG | 9 (4.0) | 15 (3.2) | 1.30 | 0.56–3.01 | 0.68 | 0.21–2.24 | 78.00 | 69.81–86.19 | 0.31 | |||
| Overdominant | ||||||||||||
| AA-GG | 163 (73.4) | 311 (65.5) | 1.00 | - | 0.034 | 1.00 | - | 0.036 | 72.00 | 70.28-73.72 | - | |
| AG | 59 (26.6) | 164 (34.5) | 0.69 | 0.48-0.98 | 0.62 | 0.40-0.98 | 72.00 | 69.72-74.28 | 0.79 | |||
| Log-additive | - | 0.81 | 0.61-1.09 | 0.17 | 0.65 | 0.44-0.96 | 0.028 | - | ||||
| HPGD | ||||||||||||
| rs2303520 | Codominant | |||||||||||
| GG | 143 (64.4) | 339 (71.4) | 1.00 | - | 0.037 | 1.00 | - | 0.065 | 72.00 | 70.89–73.11 | - | |
| GA | 76 (34.2) | 122 (25.7) | 1.48 | 1.04–2.09 | 1.61 | 1.02–2.54 | 72.00 | 69.12–74.88 | 0.83 | |||
| AA | 3 (1.4) | 14 (3.0) | 0.51 | 0.14–1.79 | 0.51 | 0.11–2.34 | 69.00 | 64.20–73.80 | 0.61 | |||
| Dominant | ||||||||||||
| GG | 143 (64.4) | 339 (71.4) | 1.00 | - | 0.066 | 1.00 | - | 0.086 | 72.00 | 70.89–73.11 | - | |
| GA-AA | 79 (35.6) | 136 (28.6) | 1.38 | 0.98–1.93 | 1.47 | 0.95–2.29 | 72.00 | 69.05–74.96 | 0.92 | |||
| Recessive | ||||||||||||
| GG-GA | 219 (98.7) | 461 (97.0) | 1.00 | - | 0.18 | 1.00 | - | 0.26 | 72.00 | 70.63–73.37 | - | |
| AA | 3 (1.4) | 14 (3.0) | 0.45 | 0.13–1.59 | 0.45 | 0.10–2.04 | 69.00 | 64.20–73.80 | 0.61 | |||
| Overdominant | ||||||||||||
| GG-AA | 146 (65.8) | 353 (74.3) | 1.00 | - | 0.021 | 1.00 | - | 0.031 | 72.00 | 70.87–73.13 | - | |
| GA | 76 (34.2) | 122 (25.7) | 1.51 | 1.07–2.13 | 1.65 | 1.05–2.59 | 72.00 | 69.12–74.88 | 0.81 | |||
| Log-additive | - | 1.21 | 0.90–1.64 | 0.21 | 1.26 | 0.86–1.84 | 0.24 | - | ||||
| SLCO2A1 | ||||||||||||
| rs10935090 | Codominant | |||||||||||
| CC | 162 (73.0) | 378 (79.6) | 1.00 | - | 0.13 | 1.00 | - | 0.026 | 73.00 | 71.81–74.19 | - | |
| CT | 54 (24.3) | 90 (18.9) | 1.40 | 0.95–2.06 | 0.13 | 1.46 | 0.90–2.39 | 0.026 | 70.00 | 67.56–72.44 | 0.034 | |
| TT | 6 (2.7) | 7 (1.5) | 2.00 | 0.66–6.04 | 4.68 | 1.32–16.6 | 62.00 | 59.61–64.39 | <0.001 | |||
| Dominant | ||||||||||||
| CC | 162 (73.0) | 378 (79.6) | 1.00 | - | 0.054 | 1.00 | - | 0.038 | 73.00 | 71.81–74.19 | - | |
| CT-TT | 60 (27.0) | 97 (20.4) | 1.44 | 1.00–2.09 | 1.65 | 1.03–2.63 | 70.00 | 67.93–72.07 | 0.007 | |||
| Recessive | ||||||||||||
| CC-CT | 216 (97.3) | 468 (98.5) | 1.00 | - | 0.28 | 1.00 | - | 0.026 | 72.00 | 70.87–73.14 | - | |
| TT | 6 (2.7) | 7 (1.5) | 1.86 | 0.62–5.59 | 4.30 | 1.22–15.2 | 62.00 | 59.61–64.39 | <0.001 | |||
| Overdominant | ||||||||||||
| CC-TT | 168 (75.7) | 385 (81.0) | 1.00 | - | 0.11 | 1.00 | - | 0.19 | 73.00 | 71.81–74.19 | - | |
| CT | 54 (24.3) | 90 (18.9) | 1.37 | 0.94–2.02 | 1.39 | 0.86–2.27 | 70.00 | 67.56–72.44 | 0.057 | |||
| Log-additive | - | 1.40 | 1.01–1.95 | 0.044 | 1.69 | 1.12–2.53 | 0.012 | - | ||||
| rs11915399 | Codominant | |||||||||||
| CC | 159 (71.6) | 326 (68.6) | 1.00 | - | 0.72 | 1.00 | - | 0.12 | 72.00 | 70.27–73.73 | - | |
| CT | 57 (25.7) | 135 (28.4) | 0.87 | 0.60–1.24 | 0.61 | 0.38–0.99 | 73.00 | 71.47–74.53 | 0.13 | |||
| TT | 6 (2.7) | 14 (3.0) | 0.88 | 0.33–2.33 | 0.75 | 0.22–2.63 | 74.00 | 59.93–88.07 | 0.52 | |||
| Dominant | ||||||||||||
| CC | 159 (71.6) | 326 (68.6) | 1.00 | - | 0.42 | 1.00 | - | 0.043 | 72.00 | 70.27–73.73 | - | |
| CT-TT | 63 (28.4) | 149 (31.4) | 0.87 | 0.61–1.23 | 0.62 | 0.39–0.99 | 73.00 | 71.53–74.47 | 0.11 | |||
| rs11915399 | Recessive | |||||||||||
| CC-CT | 216 (97.3) | 461 (97.0) | 1.00 | - | 0.86 | 1.00 | - | 0.81 | 72.00 | 70.64–73.37 | - | |
| TT | 6 (2.7) | 14 (3.0) | 0.91 | 0.35–2.41 | 0.86 | 0.25–2.96 | 74.00 | 59.93–88.07 | 0.59 | |||
| Overdominant | ||||||||||||
| CC-TT | 165 (74.3) | 340 (71.6) | 1.00 | - | 0.45 | 1.00 | - | 0.045 | 72.00 | 70.19–73.81 | - | |
| CT | 57 (25.7) | 135 (28.4) | 0.87 | 0.61–1.25 | 0.62 | 0.38–1.00 | 73.00 | 71.47–74.53 | 0.16 | |||
| Log-additive | - | 0.89 | 0.65–1.21 | 0.45 | 0.69 | 0.46–1.03 | 0.065 | - | ||||
| rs9821091 | Codominant | |||||||||||
| GG | 87 (39.2) | 179 (37.7) | 1.00 | - | 0.32 | 1.00 | - | 0.045 | 72.00 | 69.77–74.23 | - | |
| GA | 97 (43.7) | 232 (48.8) | 0.86 | 0.61–1.22 | 0.81 | 0.52–1.28 | 73.00 | 71.43–74.57 | 0.11 | |||
| AA | 38 (17.1) | 64 (13.5) | 1.22 | 0.76–1.97 | 1.75 | 0.95–3.20 | 71.00 | 68.17–73.83 | 0.16 | |||
| Dominant | ||||||||||||
| GG | 87 (39.2) | 179 (37.7) | 1.00 | - | 0.70 | 1.00 | - | 0.96 | 72.00 | 69.77–74.23 | - | |
| GA-AA | 135 (60.8) | 296 (62.3) | 0.94 | 0.68–1.30 | 0.99 | 0.65–1.50 | 72.00 | 70.56–73.44 | 0.42 | |||
| Recessive | ||||||||||||
| GG-GA | 184 (82.9) | 411 (86.5) | 1.00 | - | 0.21 | 1.00 | - | 0.02 | 72.00 | 70.76–73.24 | - | |
| AA | 38 (17.1) | 64 (16.5) | 1.33 | 0.86–1.12 | 1.95 | 1.12–3.40 | 71.00 | 68.17–73.83 | 0.017 | |||
| Overdominant | ||||||||||||
| GG-AA | 125 (56.3) | 243 (51.2) | 1.00 | - | 0.20 | 1.00 | - | 0.085 | 71.00 | 69.06–72.94 | - | |
| GA | 97 (43.7) | 232 (48.8) | 0.81 | 0.59–1.12 | 0.70 | 0.46–1.05 | 73.00 | 71.43–74.57 | 0.018 | |||
| Log-additive | - | 1.05 | 0.83–1.32 | 0.70 | 1.19 | 0.89–1.61 | 0.24 | - | ||||
| Gene/Haplotype | % Cases | % Controls | aOR | 95% CI | p Value |
|---|---|---|---|---|---|
| ABCC4£ | |||||
| T-T-A | 49.09 | 42.16 | 1.00 | Reference | - |
| C-C-A | 13.84 | 16.87 | 0.67 | 0.42–1.09 | 0.11 |
| C-T-A | 13.12 | 12.46 | 0.94 | 0.52–1.69 | 0.83 |
| C-C-G | 7.07 | 9.68 | 0.47 | 0.23–0.93 | 0.032 |
| T-T-G | 7.42 | 7.81 | 0.52 | 0.26–1.06 | 0.074 |
| T-C-A | 6.61 | 8.09 | 0.70 | 0.35–1.43 | 0.33 |
| C-T-G | 1.33 | 2.49 | 0.56 | 0.11–2.74 | 0.47 |
| SLCO2A1¥ | |||||
| C-C-G | 45.37 | 45.31 | 1.00 | Reference | - |
| C-C-A | 26.04 | 27.74 | 0.89 | 0.60–1.32 | 0.57 |
| C-T-G | 9.23 | 11.12 | 0.58 | 0.32–1.07 | 0.084 |
| T-C-A | 6.61 | 4.98 | 2.78 | 1.41–5.48 | 0.0034 |
| T-C-G | 6.44 | 0.48 | 0.91 | 0.44–1.87 | 0.80 |
| CV Accuracy | CV Consistency | aOR | 95% CI | p Value | |
|---|---|---|---|---|---|
| Best model | |||||
| rs689466 | 0.621 | 10/10 | 2.743 | 1.967–3.826 | <0.0001 |
| age, rs1678374 | 0.687 | 5/10 | 4.953 | 3.434–7.143 | <0.0001 |
| age, rs689466, rs1678374 | 0.807 | 8/10 | 17.581 | 11.672–26.482 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, C.; Pereira, C.; Farinha, M.; Medeiros, R.; Dinis-Ribeiro, M. Genetic Variations in Prostaglandin E2 Pathway Identified as Susceptibility Biomarkers for Gastric Cancer in an Intermediate Risk European Country. Int. J. Mol. Sci. 2021, 22, 648. https://doi.org/10.3390/ijms22020648
Lopes C, Pereira C, Farinha M, Medeiros R, Dinis-Ribeiro M. Genetic Variations in Prostaglandin E2 Pathway Identified as Susceptibility Biomarkers for Gastric Cancer in an Intermediate Risk European Country. International Journal of Molecular Sciences. 2021; 22(2):648. https://doi.org/10.3390/ijms22020648
Chicago/Turabian StyleLopes, Catarina, Carina Pereira, Mónica Farinha, Rui Medeiros, and Mário Dinis-Ribeiro. 2021. "Genetic Variations in Prostaglandin E2 Pathway Identified as Susceptibility Biomarkers for Gastric Cancer in an Intermediate Risk European Country" International Journal of Molecular Sciences 22, no. 2: 648. https://doi.org/10.3390/ijms22020648
APA StyleLopes, C., Pereira, C., Farinha, M., Medeiros, R., & Dinis-Ribeiro, M. (2021). Genetic Variations in Prostaglandin E2 Pathway Identified as Susceptibility Biomarkers for Gastric Cancer in an Intermediate Risk European Country. International Journal of Molecular Sciences, 22(2), 648. https://doi.org/10.3390/ijms22020648






