The Cryogenic Electron Microscopy Structure of the Cell Adhesion Regulator Metavinculin Reveals an Isoform-Specific Kinked Helix in Its Cytoskeleton Binding Domain
Abstract
1. Introduction
2. Results
2.1. The Metavinculin Polypeptide Chains in the Crystal Structures are Distinct
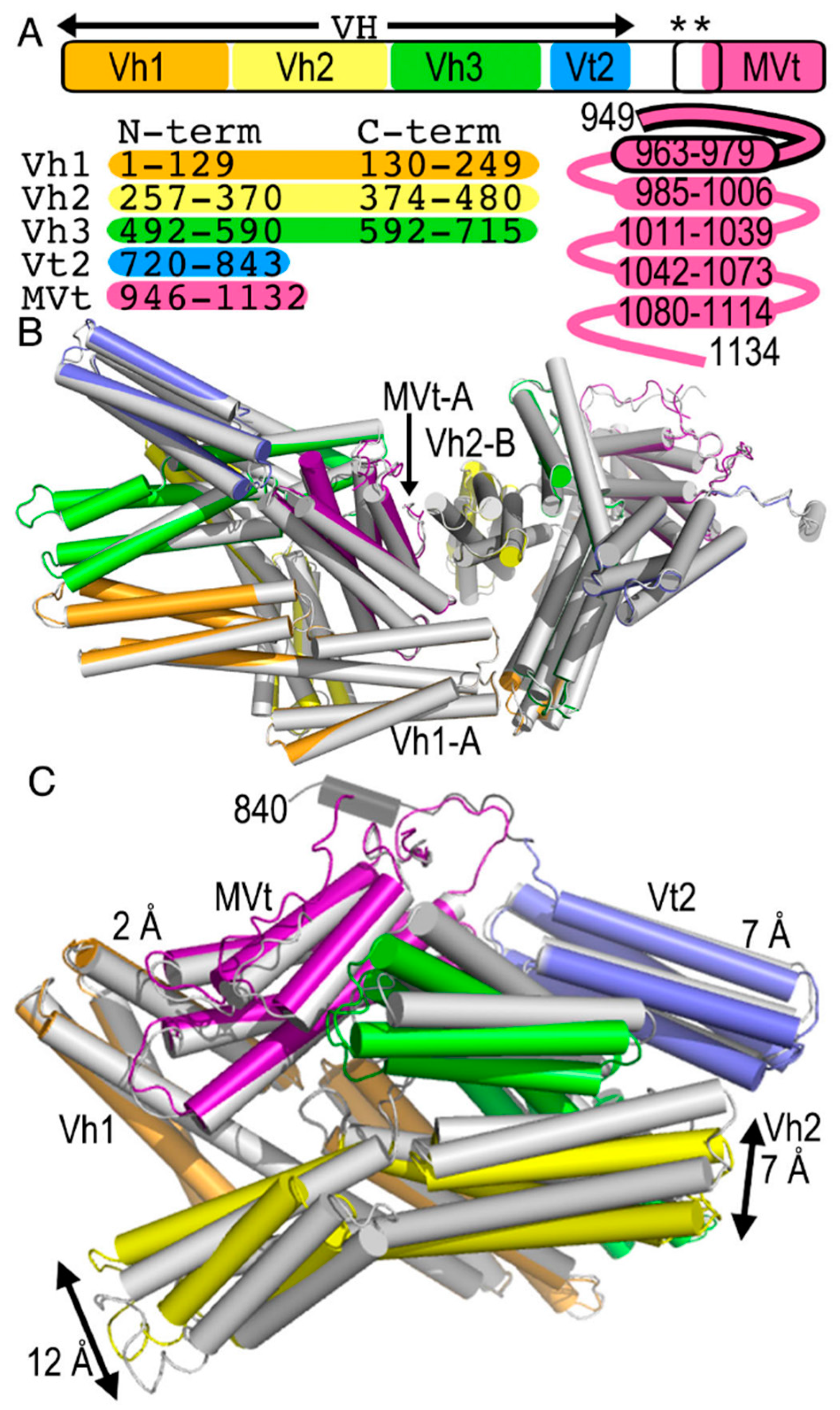
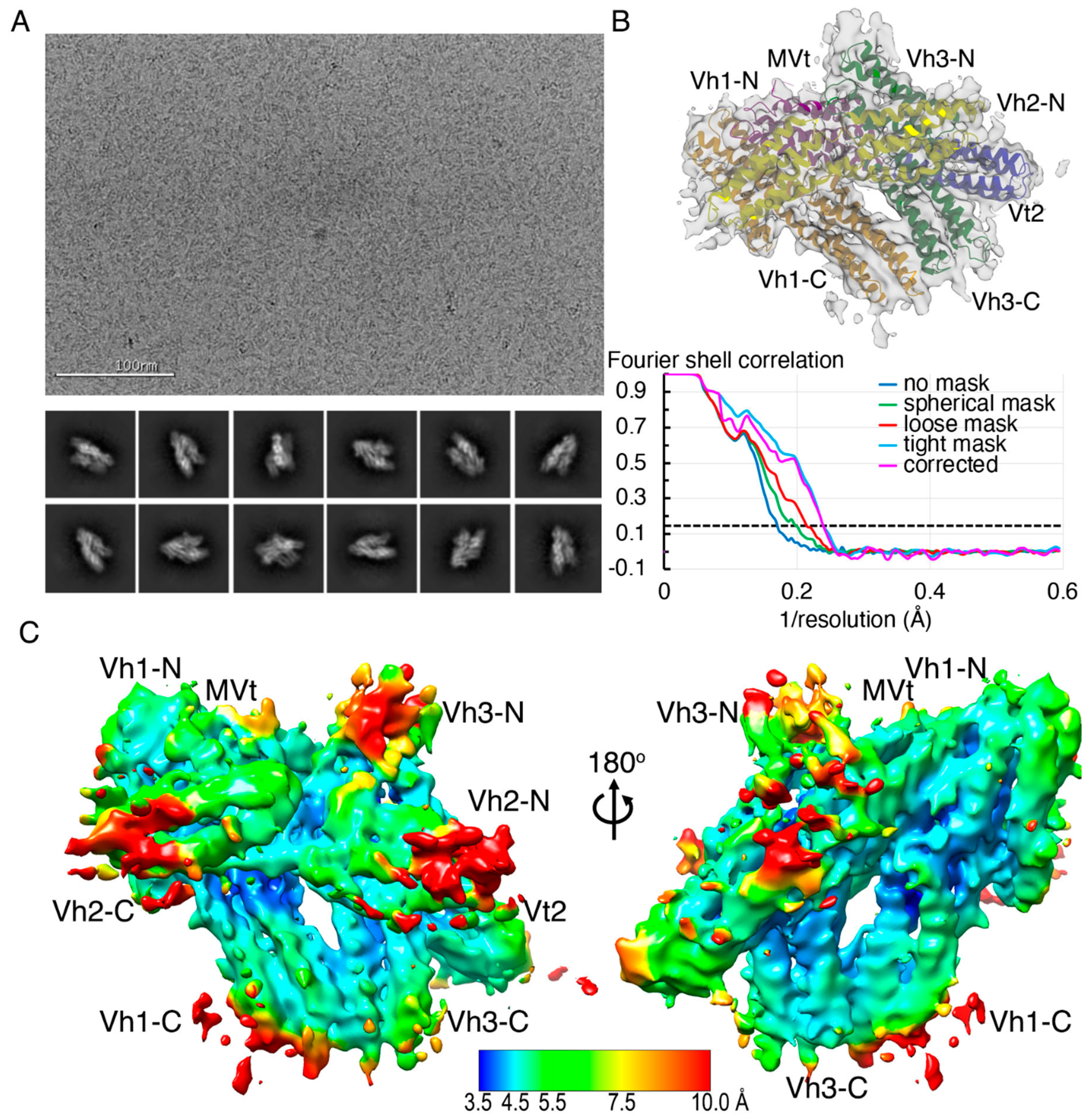
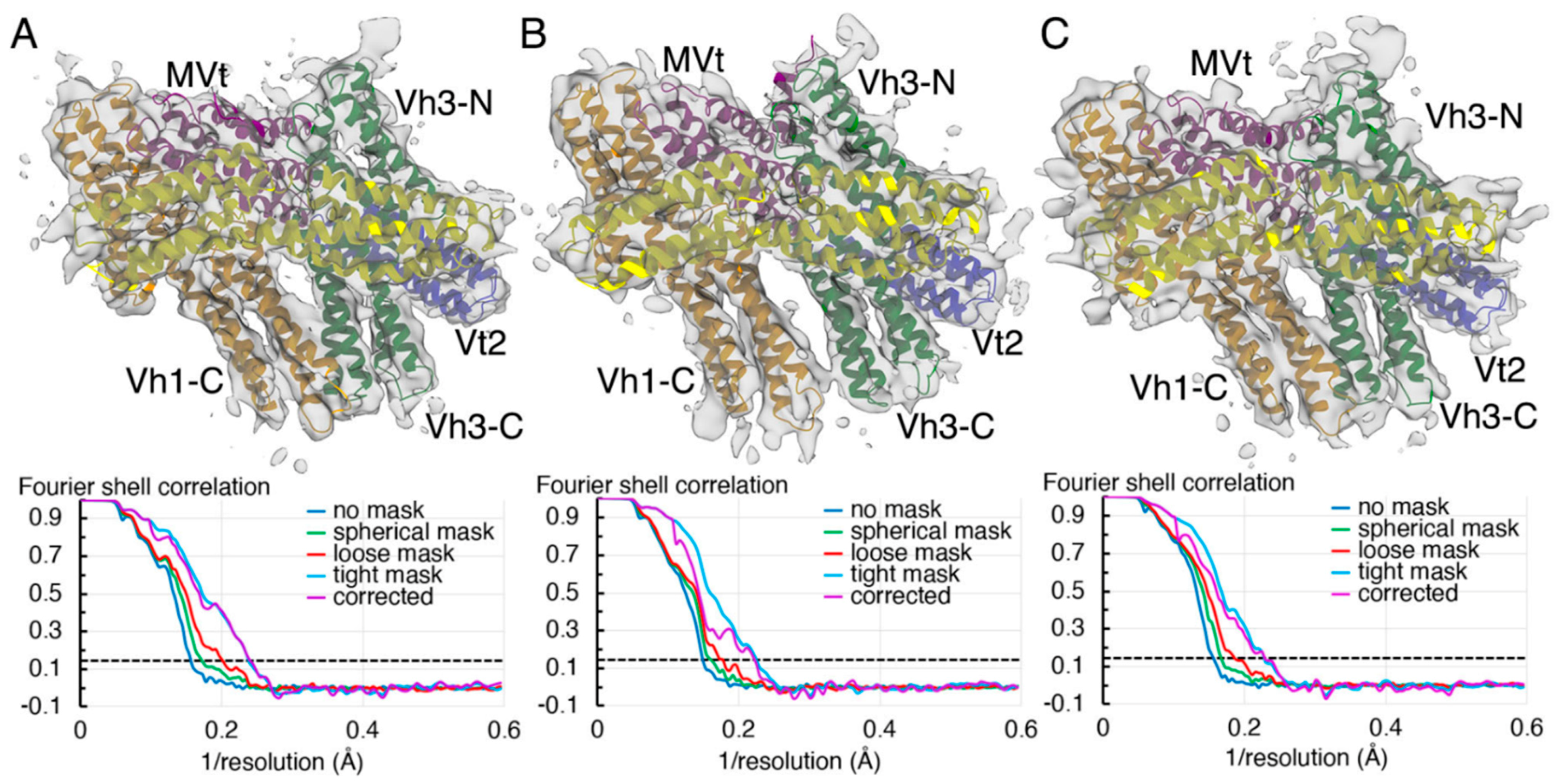
2.2. The Cryogenic Electron Microscopy Metavinculin Structure
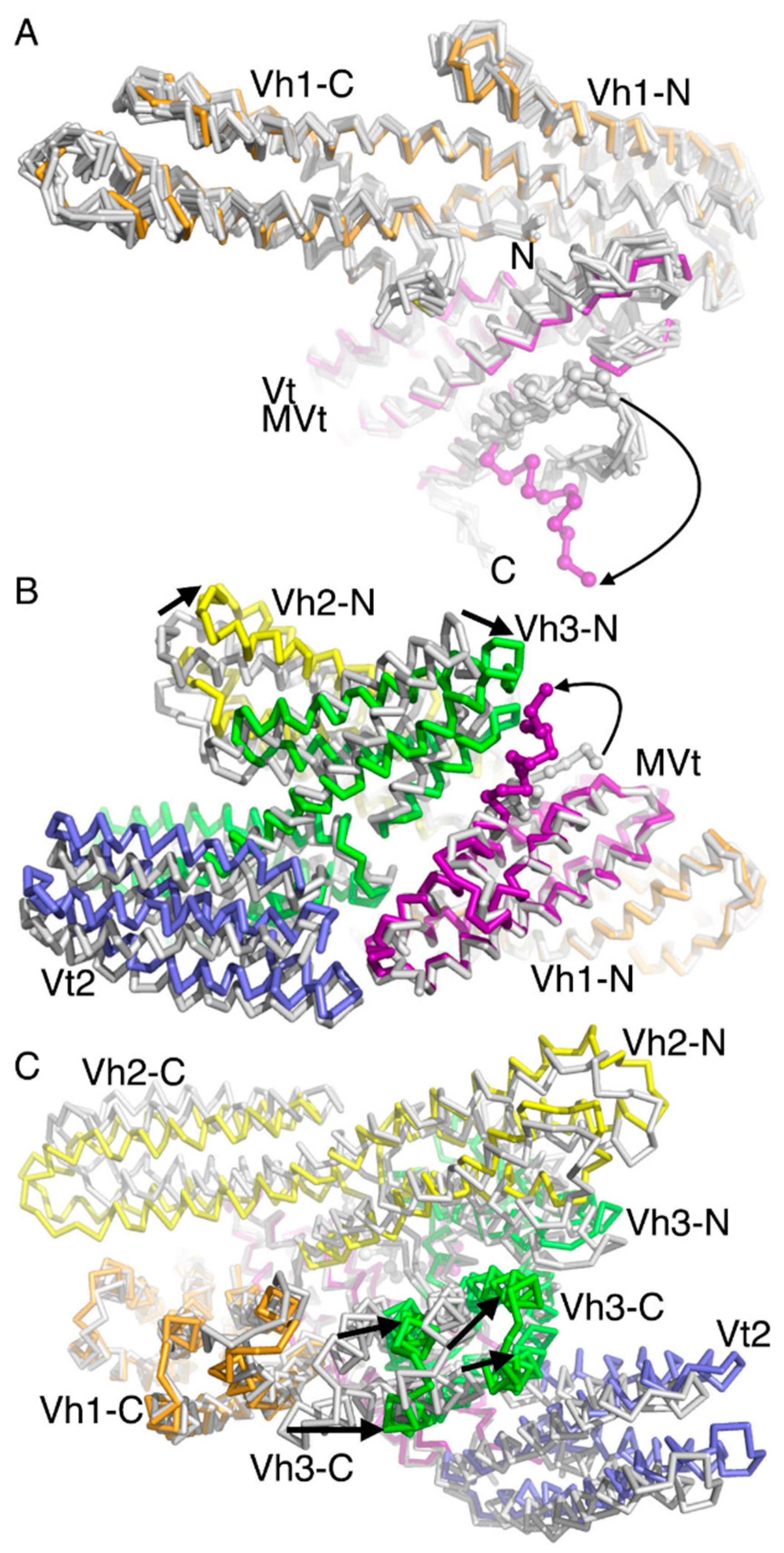
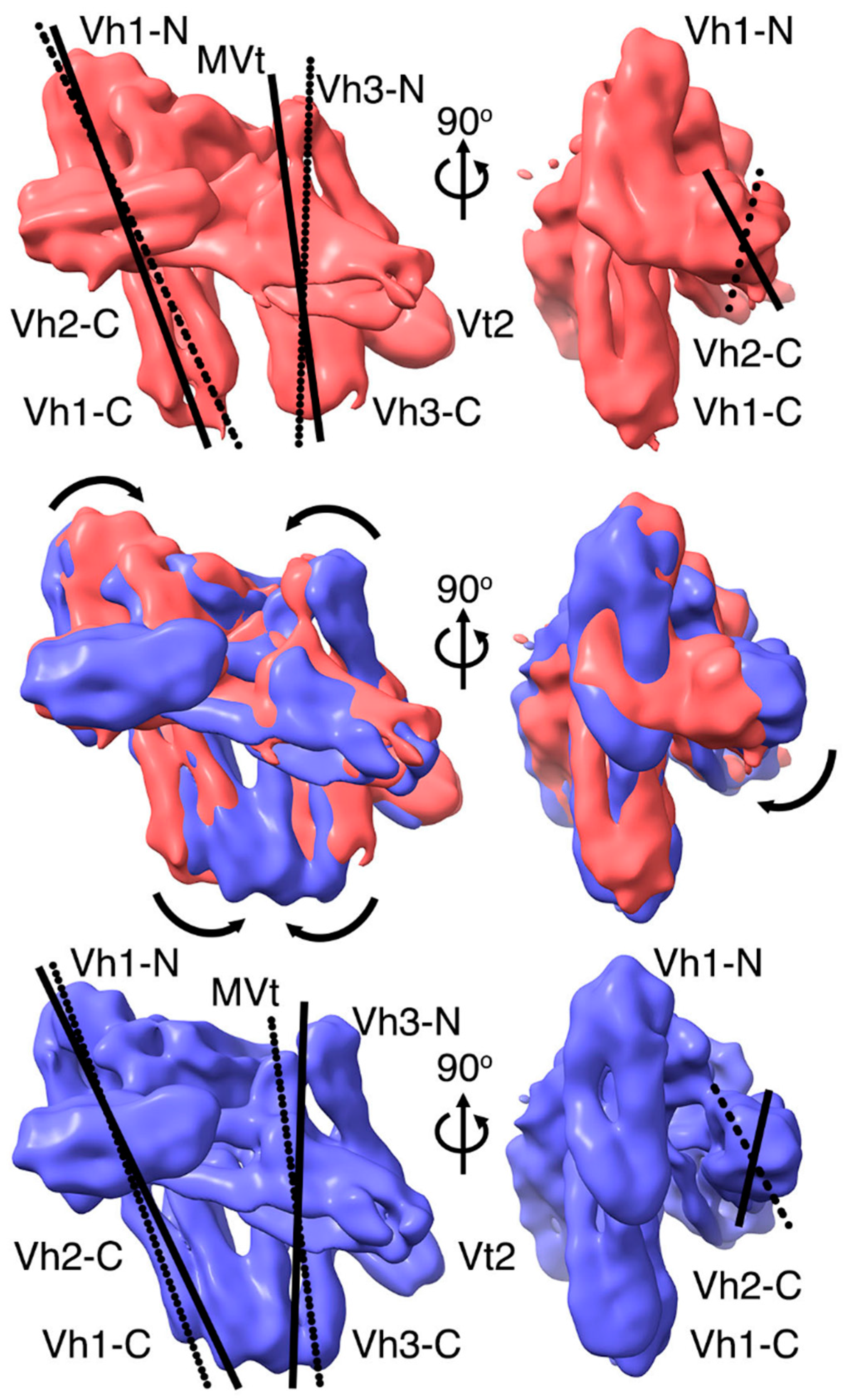
2.3. The Cryogenic Electron Microscopy Metavinculin Conformers
2.4. A Novel Conformer in the F-Actin Binding Domain
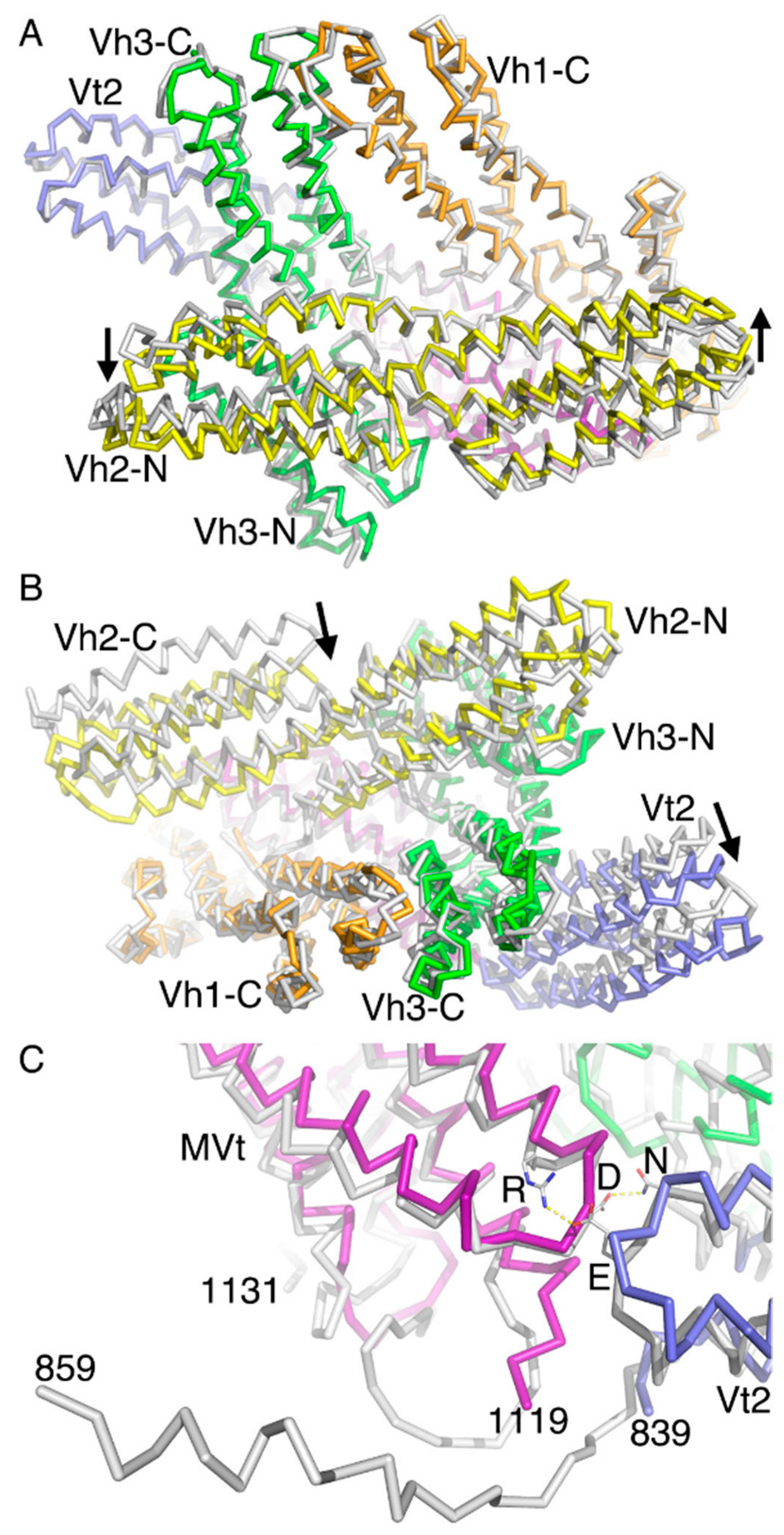
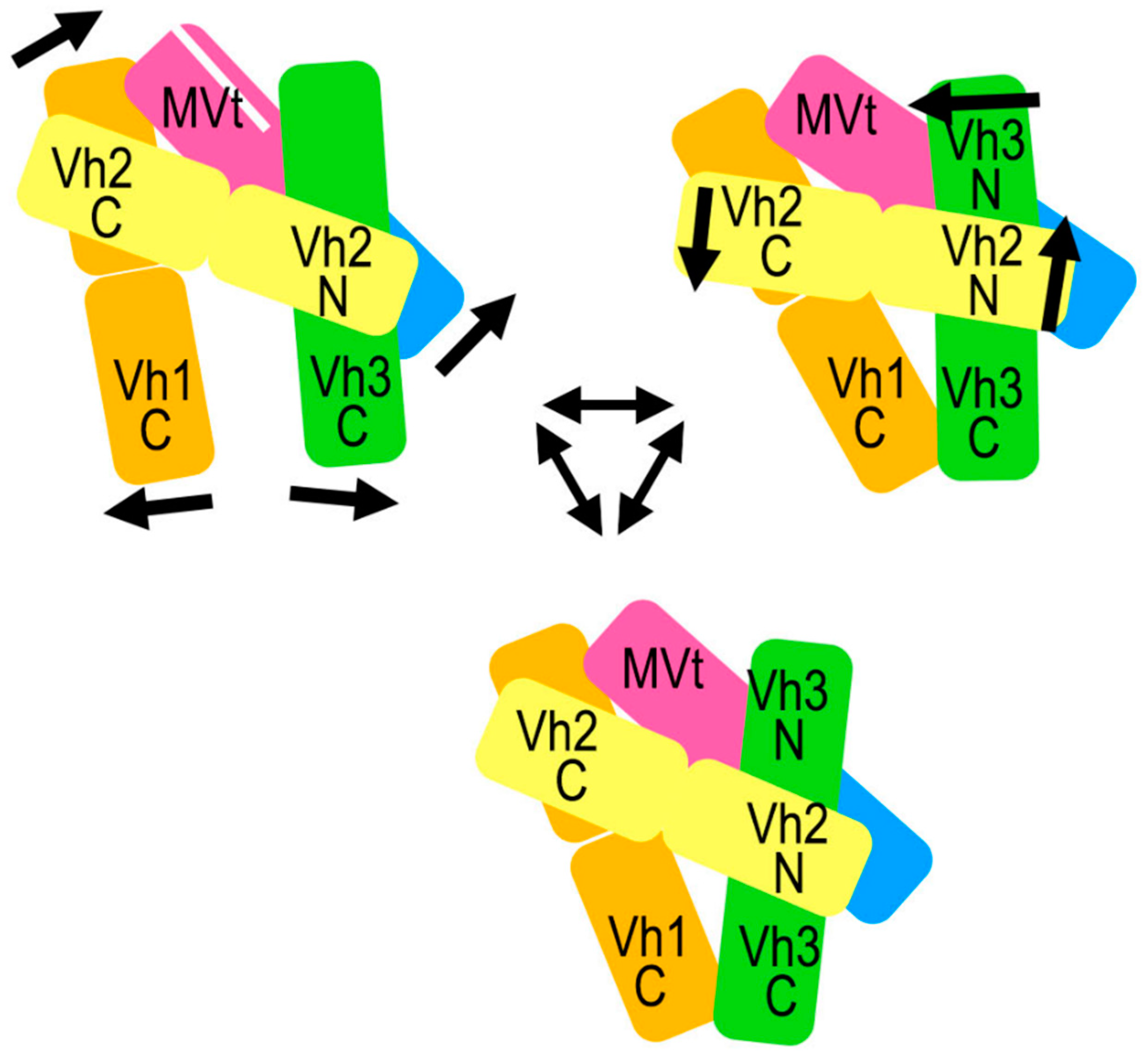
2.5. The Metavinculin Vh2–Vh3 Domain Constellations
2.6. The Two H1’-Parallel Metavinculin Conformations
2.7. The Metavinculin Vh2 C-Terminal Subdomain Constellations
2.8. The Metavinculin Vt2–MVt Domain Constellations
2.9. The Metavinculin C-Terminal F-Actin Binding Domain MVt
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Metavinculin
4.2. Metavinculin Cryogenic Electron Microscopy Data Collection
4.3. Metavinculin Cryogenic Electron Microscopy Data Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kellie, S.; Patel, B.; Mitchell, A.; Critchley, D.R.; Wigglesworth, N.M.; Wyke, J.A. Comparison of the relative importance of tyrosine-specific vinculin phosphorylation and the loss of surface-associated fibronectin in the morphology of cells transformed by Rous sarcoma virus. J. Cell Sci. 1986, 82, 129–142. [Google Scholar] [PubMed]
- Coll, J.L.; Ben-Ze’ev, A.; Ezzell, R.M.; Rodriguez Fernandez, J.L.; Baribault, H.; Oshima, R.G.; Adamson, E.D. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. USA 1995, 92, 9161–9165. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Fernandez, J.L.; Geiger, B.; Salomon, D.; Ben-Ze’ev, A. Overexpression of vinculin suppresses cell motility in BALB/c 3T3 cells. Cell Motil. Cytoskelet. 1992, 22, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Imanaka-Yoshida, K.; Enomoto-Iwamoto, M.; Yoshida, T.; Sakakura, T. Vinculin, Talin, Integrin alpha6beta1 and laminin can serve as components of attachment complex mediating contraction force transmission from cardiomyocytes to extracellular matrix. Cell Motil. Cytoskelet. 1999, 42, 1–11. [Google Scholar] [CrossRef]
- Huveneers, S.; Oldenburg, J.; Spanjaard, E.; Van der Krogt, G.; Grigoriev, I.; Akhmanova, A.; Rehmann, H.; De Rooij, J. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 2012, 196, 641–652. [Google Scholar] [CrossRef]
- Brown, D.T.; Izard, T. Vinculin-cell membrane interactions. OncoTarget 2015, 6, 34043–34044. [Google Scholar] [CrossRef]
- Izard, T.; Brown, D.T. Mechanisms and Functions of Vinculin Interactions with Phospholipids at Cell Adhesion Sites. J. Biol. Chem. 2016, 291, 2548–2555. [Google Scholar] [CrossRef]
- Johnson, R.P.; Craig, S.W. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 1995, 373, 261–264. [Google Scholar] [CrossRef]
- Bailly, M. Connecting cell adhesion to the actin polymerization machinery: Vinculin as the missing link? Trends Cell Biol. 2003, 13, 163–165. [Google Scholar] [CrossRef]
- Jockusch, B.M.; Isenberg, G. Interaction of α-actinin and vinculin with actin: Opposite effects on filament network formation. Proc. Natl. Acad. Sci. USA 1981, 78, 3005–3009. [Google Scholar] [CrossRef]
- Shiraishi, I.; Simpson, D.G.; Carver, W.; Price, R.; Hirozane, T.; Terracio, L.; Borg, T.K. Vinculin is an essential component for normal myofibrillar arrangement in fetal mouse cardiac myocytes. J. Mol. Cell Cardiol. 1997, 29, 2041–2052. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Baribault, H.; Adamson, E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development 1998, 125, 327–337. [Google Scholar] [PubMed]
- Subauste, M.C.; Pertz, O.; Adamson, E.D.; Turner, C.E.; Junger, S.; Hahn, K.M. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 2004, 165, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Zemljic-Harpf, A.E.; Ponrartana, S.; Avalos, R.T.; Jordan, M.C.; Roos, K.P.; Dalton, N.D.; Phan, V.Q.; Adamson, E.D.; Ross, R.S. Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am. J. Pathol. 2004, 165, 1033–1044. [Google Scholar] [CrossRef]
- Bakolitsa, C.; De Pereda, J.M.; Bagshaw, C.R.; Critchley, D.R.; Liddington, R.C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 1999, 99, 603–613. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Lee, J.H.; Yogesha, S.D.; Izard, T. A helix replacement mechanism directs metavinculin functions. PLoS ONE 2010, 5, e10679. [Google Scholar] [CrossRef]
- Borgon, R.A.; Vonrhein, C.; Bricogne, G.; Bois, P.R.; Izard, T. Crystal structure of human vinculin. Structure 2004, 12, 1189–1197. [Google Scholar] [CrossRef]
- Bakolitsa, C.; Cohen, D.M.; Bankston, L.A.; Bobkov, A.A.; Cadwell, G.W.; Jennings, L.; Critchley, D.R.; Craig, S.W.; Liddington, R.C. Structural basis for vinculin activation at sites of cell adhesion. Nature 2004, 430, 583–586. [Google Scholar] [CrossRef]
- Janssen, M.E.; Liu, H.; Volkmann, N.; Hanein, D. The C-terminal tail domain of metavinculin, vinculin’s splice variant, severs actin filaments. J. Cell Biol. 2012, 197, 585–593. [Google Scholar] [CrossRef]
- Thompson, P.M.; Tolbert, C.E.; Campbell, S.L. Vinculin and metavinculin: Oligomerization and interactions with F-actin. FEBS Lett. 2013, 587, 1220–1229. [Google Scholar] [CrossRef]
- Oztug Durer, Z.A.; McGillivary, R.M.; Kang, H.; Elam, W.A.; Vizcarra, C.L.; Hanein, D.; De La Cruz, E.M.; Reisler, E.; Quinlan, M.E. Metavinculin Tunes the Flexibility and the Architecture of Vinculin-Induced Bundles of Actin Filaments. J. Mol. Biol. 2015, 427, 2782–2798. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.Y.; Thompson, P.M.; Lee, H.T.; Pershad, M.; Campbell, S.L.; Alushin, G.M. The Structural Basis of Actin Organization by Vinculin and Metavinculin. J. Mol. Biol. 2016, 428, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Sarker, M.; Lee, H.T.; Mei, L.; Krokhotin, A.; De Los Reyes, S.E.; Yen, L.; Costantini, L.M.; Griffith, J.; Dokholyan, N.V.; Alushin, G.M.; et al. Cardiomyopathy Mutations in Metavinculin Disrupt Regulation of Vinculin-Induced F-Actin Assemblies. J. Mol. Biol. 2019, 431, 1604–1618. [Google Scholar] [CrossRef] [PubMed]
- Carisey, A.; Tsang, R.; Greiner, A.M.; Nijenhuis, N.; Heath, N.; Nazgiewicz, A.; Kemkemer, R.; Derby, B.; Spatz, J.; Ballestrem, C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013, 23, 271–281. [Google Scholar] [CrossRef]
- Dumbauld, D.W.; Lee, T.T.; Singh, A.; Scrimgeour, J.; Gersbach, C.A.; Zamir, E.A.; Fu, J.; Chen, C.S.; Curtis, J.E.; Craig, S.W.; et al. How vinculin regulates force transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 9788–9793. [Google Scholar] [CrossRef]
- Yao, M.; Qiu, W.; Liu, R.; Efremov, A.K.; Cong, P.; Seddiki, R.; Payre, M.; Lim, C.T.; Ladoux, B.; Mege, R.M.; et al. Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 2014, 5, 4525. [Google Scholar] [CrossRef]
- Hirata, H.; Tatsumi, H.; Lim, C.T.; Sokabe, M. Force-dependent vinculin binding to talin in live cells: A crucial step in anchoring the actin cytoskeleton to focal adhesions. Am. J. Physiol. Cell Physiol. 2014, 306, C607–C620. [Google Scholar] [CrossRef]
- Del Rio, A.; Perez-Jimenez, R.; Liu, R.; Roca-Cusachs, P.; Fernandez, J.M.; Sheetz, M.P. Stretching single talin rod molecules activates vinculin binding. Science 2009, 323, 638–641. [Google Scholar] [CrossRef]
- Carisey, A.; Ballestrem, C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol. 2011, 90, 157–163. [Google Scholar] [CrossRef]
- Belkin, A.M.; Koteliansky, V.E. Interaction of iodinated vinculin, metavinculin and α-actinin with cytoskeletal proteins. FEBS Lett. 1987, 220, 291–294. [Google Scholar] [CrossRef]
- Wachsstock, D.H.; Wilkins, J.A.; Lin, S. Specific interaction of vinculin with α-actinin. Biochem. Biophys Res. Commun. 1987, 146, 554–560. [Google Scholar] [CrossRef]
- Bois, P.R.; Borgon, R.A.; Vonrhein, C.; Izard, T. Structural dynamics of α-actinin-vinculin interactions. Mol. Cell. Biol. 2005, 25, 6112–6122. [Google Scholar] [CrossRef]
- Watabe-Uchida, M.; Uchida, N.; Imamura, Y.; Nagafuchi, A.; Fujimoto, K.; Uemura, T.; Vermeulen, S.; Van Roy, F.; Adamson, E.D.; Takeichi, M. α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 1998, 142, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Pokutta, S.; Cadwell, G.W.; Bobkov, A.A.; Bankston, L.A.; Liddington, R.C.; Weis, W.I. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. USA 2012, 109, 8576–8581. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Maiers, J.L.; Choudhury, D.; Craig, S.W.; Demali, K.A. α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 2012, 287, 7728–7737. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, E.S.; Izard, T. α-Catenin unfurls upon binding to vinculin. J. Biol. Chem. 2012, 287, 18492–18499. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Cuff, L.E.; Lawton, C.D.; DeMali, K.A. Vinculin regulates cell-surface E-cadherin expression by binding to β-catenin. J. Cell Sci. 2010, 123, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.B.; Kang, L.; Roe, S.; Borgen, P.I.; Rimm, D.L. Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 1997, 272, 32448–32453. [Google Scholar] [CrossRef]
- Kioka, N.; Sakata, S.; Kawauchi, T.; Amachi, T.; Akiyama, S.K.; Okazaki, K.; Yaen, C.; Yamada, K.M.; Aota, S. Vinexin: A novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 1999, 144, 59–69. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Yao, B.; Shen, W.; Sun, H.; Xu, C.; Wu, J.; Shi, Y. Solution structure of the first SH3 domain of human vinexin and its interaction with vinculin peptides. Biochem. Biophys Res. Commun. 2007, 357, 931–937. [Google Scholar] [CrossRef]
- Brindle, N.P.; Holt, M.R.; Davies, J.E.; Price, C.J.; Critchley, D.R. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 1996, 318, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, M.; Rudiger, M.; Jockusch, B.M.; Walter, U. VASP interaction with vinculin: A recurring theme of interactions with proline-rich motifs. FEBS Lett. 1996, 399, 103–107. [Google Scholar] [CrossRef]
- Huttelmaier, S.; Mayboroda, O.; Harbeck, B.; Jarchau, T.; Jockusch, B.M.; Rudiger, M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 1998, 8, 479–488. [Google Scholar] [CrossRef]
- DeMali, K.A.; Barlow, C.A.; Burridge, K. Recruitment of the Arp2/3 complex to vinculin: Coupling membrane protrusion to matrix adhesion. J. Cell Biol. 2002, 159, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.W.; Chen, H. Lamellipodia protrusion: Moving interactions of vinculin and Arp2/3. Curr. Biol. 2003, 13, R236–R238. [Google Scholar] [CrossRef]
- Turner, C.E.; Glenney, J.R., Jr.; Burridge, K. Paxillin: A new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990, 111, 1059–1068. [Google Scholar] [CrossRef]
- Wood, C.K.; Turner, C.E.; Jackson, P.; Critchley, D.R. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J. Cell Sci. 1994, 107, 709–717. [Google Scholar]
- Lee, J.H.; Rangarajan, E.S.; Yogesha, S.D.; Izard, T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure 2009, 17, 833–842. [Google Scholar] [CrossRef]
- Lee, J.H.; Rangarajan, E.S.; Vonrhein, C.; Bricogne, G.; Izard, T. The metavinculin tail domain directs constitutive interactions with raver1 and vinculin RNA. J. Mol. Biol. 2012, 422, 697–704. [Google Scholar] [CrossRef]
- Huttelmaier, S.; Illenberger, S.; Grosheva, I.; Rudiger, M.; Singer, R.H.; Jockusch, B.M. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001, 155, 775–786. [Google Scholar] [CrossRef]
- Izard, T.; Tran Van Nhieu, G.; Bois, P.R. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J. Cell Biol. 2006, 175, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Tran Van Nhieu, G.; Izard, T. Vinculin binding in its closed conformation by a helix addition mechanism. EMBO J. 2007, 26, 4588–4596. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, J.H.; Cossart, P.; Gouin, E.; Izard, T. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J. Biol. Chem. 2011, 286, 35096–35103. [Google Scholar] [CrossRef]
- Park, H.; Valencia-Gallardo, C.; Sharff, A.; Tran Van Nhieu, G.; Izard, T. Novel vinculin binding site of the IpaA invasin of Shigella. J. Biol. Chem. 2011, 286, 23214–23221. [Google Scholar] [CrossRef] [PubMed]
- Feramisco, J.R.; Smart, J.E.; Burridge, K.; Helfman, D.M.; Thomas, G.P. Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J. Biol. Chem. 1982, 257, 11024–11031. [Google Scholar] [CrossRef]
- Byrne, B.J.; Kaczorowski, Y.J.; Coutu, M.D.; Craig, S.W. Chicken vinculin and meta-vinculin are derived from a single gene by alternative splicing of a 207-base pair exon unique to meta-vinculin. J. Biol. Chem. 1992, 267, 12845–12850. [Google Scholar] [CrossRef]
- Belkin, A.M.; Zhidkova, N.I.; Koteliansky, V.E. Localization of talin in skeletal and cardiac muscles. FEBS Lett. 1986, 200, 32–36. [Google Scholar] [CrossRef]
- Turner, C.E.; Burridge, K. Detection of metavinculin in human platelets using a modified talin overlay assay. Eur. J. Cell Biol. 1989, 49, 202–206. [Google Scholar]
- Belkin, A.M.; Ornatsky, O.I.; Glukhova, M.A.; Koteliansky, V.E. Immunolocalization of meta-vinculin in human smooth and cardiac muscles. J. Cell Biol. 1988, 107, 545–553. [Google Scholar] [CrossRef]
- Belkin, A.M.; Ornatsky, O.I.; Kabakov, A.E.; Glukhova, M.A.; Koteliansky, V.E. Diversity of vinculin/meta-vinculin in human tissues and cultivated cells. Expression of muscle specific variants of vinculin in human aorta smooth muscle cells. J. Biol. Chem. 1988, 263, 6631–6635. [Google Scholar] [CrossRef]
- Zemljic-Harpf, A.; Manso, A.M.; Ross, R.S. Vinculin and talin: Focus on the myocardium. J. Investig. Med. 2009, 57, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Zemljic-Harpf, A.E.; Miller, J.C.; Henderson, S.A.; Wright, A.T.; Manso, A.M.; Elsherif, L.; Dalton, N.D.; Thor, A.K.; Perkins, G.A.; McCulloch, A.D.; et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 2007, 27, 7522–7537. [Google Scholar] [CrossRef] [PubMed]
- Vasile, V.C.; Edwards, W.D.; Ommen, S.R.; Ackerman, M.J. Obstructive hypertrophic cardiomyopathy is associated with reduced expression of vinculin in the intercalated disc. Biochem. Biophys Res. Commun. 2006, 349, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Holder, E.; Lowes, B.; Valent, S.; Bies, R.D. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation 1997, 95, 17–20. [Google Scholar] [CrossRef]
- Olson, T.M.; Illenberger, S.; Kishimoto, N.Y.; Huttelmaier, S.; Keating, M.T.; Jockusch, B.M. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation 2002, 105, 431–437. [Google Scholar] [CrossRef]
- Vasile, V.C.; Will, M.L.; Ommen, S.R.; Edwards, W.D.; Olson, T.M.; Ackerman, M.J. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol. Genet. Metab. 2006, 87, 169–174. [Google Scholar] [CrossRef]
- Vasile, V.C.; Ommen, S.R.; Edwards, W.D.; Ackerman, M.J. A missense mutation in a ubiquitously expressed protein, vinculin, confers susceptibility to hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2006, 345, 998–1003. [Google Scholar] [CrossRef]
- Kanoldt, V.; Kluger, C.; Barz, C.; Schweizer, A.L.; Ramanujam, D.; Windgasse, L.; Engelhardt, S.; Chrostek-Grashoff, A.; Grashoff, C. Metavinculin modulates force transduction in cell adhesion sites. Nat. Commun. 2020, 11, 6403. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. Cryosparc: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Mei, L.; Espinosa de Los Reyes, S.; Reynolds, M.J.; Leicher, R.; Liu, S.; Alushin, G.M. Molecular mechanism for direct actin force-sensing by alpha-catenin. Elife 2020, 9, e62514. [Google Scholar] [CrossRef]
- Panjani, A.; Fleet, D.J. 3D Variability analysis: Directly resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM images. BioRxiv 2020. [Google Scholar] [CrossRef]
- Izard, T.; Evans, G.; Borgon, R.A.; Rush, C.L.; Bricogne, G.; Bois, P.R. Vinculin activation by talin through helical bundle conversion. Nature 2004, 427, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.P.; Pokutta, S.; Torres, M.; Swift, M.F.; Hanein, D.; Volkmann, N.; Weis, W.I. Structural basis of alphaE-catenin-F-actin catch bond behavior. Elife 2020, 9, e60878. [Google Scholar] [CrossRef]
- Chorev, D.S.; Volberg, T.; Livne, A.; Eisenstein, M.; Martins, B.; Kam, Z.; Jockusch, B.M.; Medalia, O.; Sharon, M.; Geiger, B. Conformational states during vinculin unlocking differentially regulate focal adhesion properties. Sci. Rep. 2018, 8, 2693. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.; Pulokas, J.; Fellmann, D.; Cheng, A.; Guerra, F.; Quispe, J.; Stagg, S.; Potter, C.S.; Carragher, B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005, 151, 41–60. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef]
- Cardone, G.; Heymann, J.B.; Steven, A.C. One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 2013, 184, 226–236. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Adams, P.D.; Urzhumtsev, A. Bulk-solvent and overall scaling revisited: Faster calculations, improved results. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 625–634. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein. Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
| (A) Metavinculin electron microscopy data collection and model reconstruction | ||||
| Conformer | Total | 4.15 Å | 4.5 Å | 4.27 Å |
| Microscope | EF-Krios | EF-Krios | EF-Krios | EF-Krios |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Detector | Gatan K3 | Gatan K3 | Gatan K3 | Gatan K3 |
| Magnification (nominal) | 105,000 | 105,000 | 105,000 | 105,000 |
| Dose rate (e−/Å2/sec) | 42.45 | 42.45 | 42.45 | 42.45 |
| Frames per exposure | 40 | 40 | 40 | 40 |
| Total exposure (e−/Å2) | 67.92 | 67.92 | 67.92 | 67.92 |
| Pixel size (Å/pix) | 0.825 | 0.825 | 0.825 | 0.825 |
| Defocus (mm) | 1.4 | 1.4 | 1.4 | 1.4 |
| Micrograph collected | 3011 | 3011 | 3011 | 3011 |
| Initial particles (no.) | 1,303,504 | 1,303,504 | 1,303,504 | 1,303,504 |
| Symmetry | C1 | C1 | C1 | C1 |
| Resolution (Å) | 4.17 | 4.15 | 4.5 | 4.27 |
| Particles used (no.) | 1,303,504 | 458,305 | 413,507 | 366,802 |
| (B) Metavinculin model refinement and statistics of our three distinct conformers | ||||
| Conformer | Total | 4.15 Å | 4.5 Å | 4.27 Å |
| Atoms | 7568 | 7577 | 7577 | 7577 |
| Protein Residues | 994 | 995 | 995 | 995 |
| Bonds (root means squares deviations) | ||||
| Length (Å) | 0.007 | 0.006 | 0.006 | 0.007 |
| Angles (°) | 1.016 | 0.955 | 1.030 | 1.032 |
| Molprobity score | 2.00 | 2.12 | 2.17 | 2.02 |
| Clash score | 12.79 | 12.91 | 17.86 | 14.28 |
| Ramachandran plot | ||||
| Favored (%) | 94.34 | 95.66 | 93.54 | 94.85 |
| Allowed (%) | 5.66 | 4.34 | 6.46 | 5.15 |
| Outliers (%) | 0.00 | 0.00 | 0.00 | 0.00 |
| Rotamer outliers (%) | 0.25 | 0.37 | 0.12 | 0.12 |
| Cβ outliers (%) | 0.00 | 0.00 | 0.00 | 0.00 |
| Mean temperature factors | 158.11 | 217.30 | 217.93 | 210.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangarajan, E.S.; Izard, T. The Cryogenic Electron Microscopy Structure of the Cell Adhesion Regulator Metavinculin Reveals an Isoform-Specific Kinked Helix in Its Cytoskeleton Binding Domain. Int. J. Mol. Sci. 2021, 22, 645. https://doi.org/10.3390/ijms22020645
Rangarajan ES, Izard T. The Cryogenic Electron Microscopy Structure of the Cell Adhesion Regulator Metavinculin Reveals an Isoform-Specific Kinked Helix in Its Cytoskeleton Binding Domain. International Journal of Molecular Sciences. 2021; 22(2):645. https://doi.org/10.3390/ijms22020645
Chicago/Turabian StyleRangarajan, Erumbi S., and Tina Izard. 2021. "The Cryogenic Electron Microscopy Structure of the Cell Adhesion Regulator Metavinculin Reveals an Isoform-Specific Kinked Helix in Its Cytoskeleton Binding Domain" International Journal of Molecular Sciences 22, no. 2: 645. https://doi.org/10.3390/ijms22020645
APA StyleRangarajan, E. S., & Izard, T. (2021). The Cryogenic Electron Microscopy Structure of the Cell Adhesion Regulator Metavinculin Reveals an Isoform-Specific Kinked Helix in Its Cytoskeleton Binding Domain. International Journal of Molecular Sciences, 22(2), 645. https://doi.org/10.3390/ijms22020645




