Adaptive Thermogenesis in a Mouse Model Lacking Selenoprotein Biosynthesis in Brown Adipocytes
Abstract
1. Introduction
2. Results
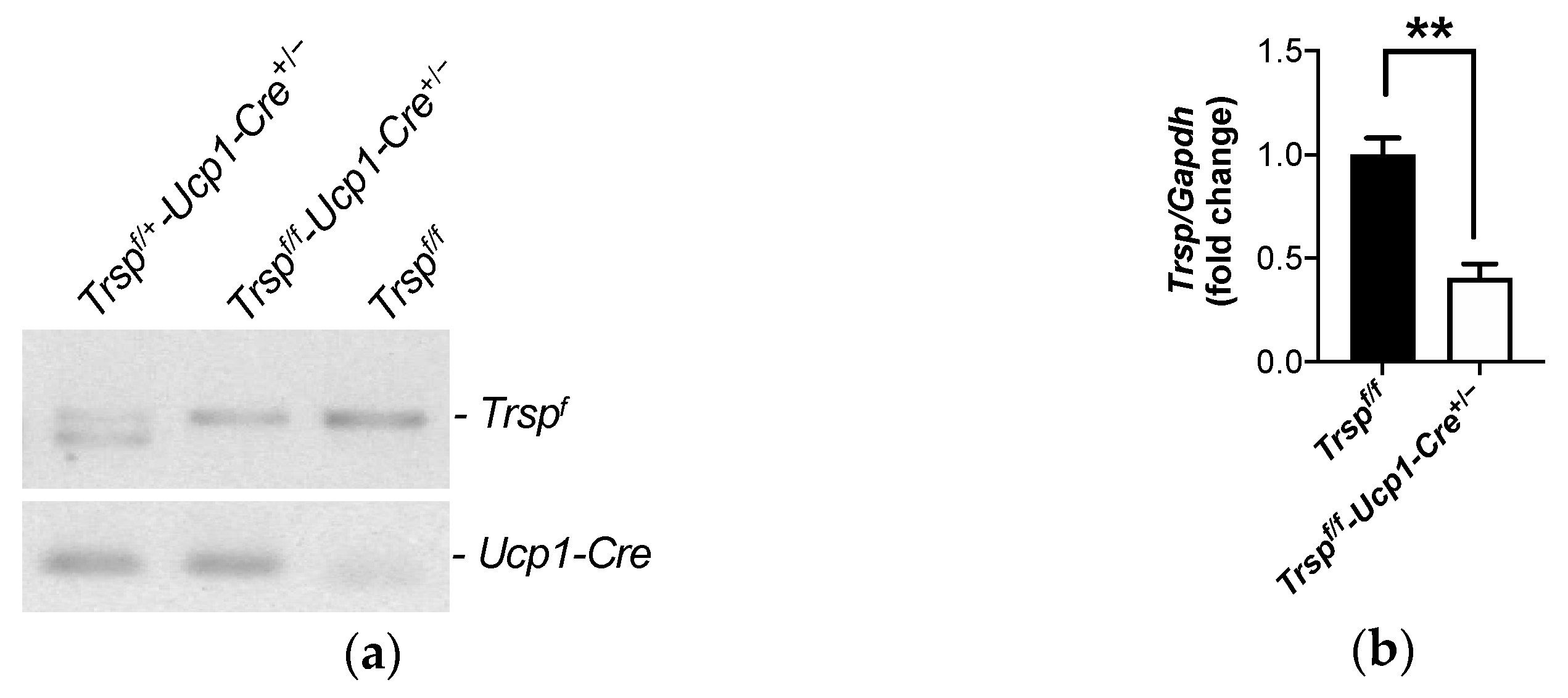
2.1. Generation of the Trspf/f-Ucp1-Cre+/− Mice
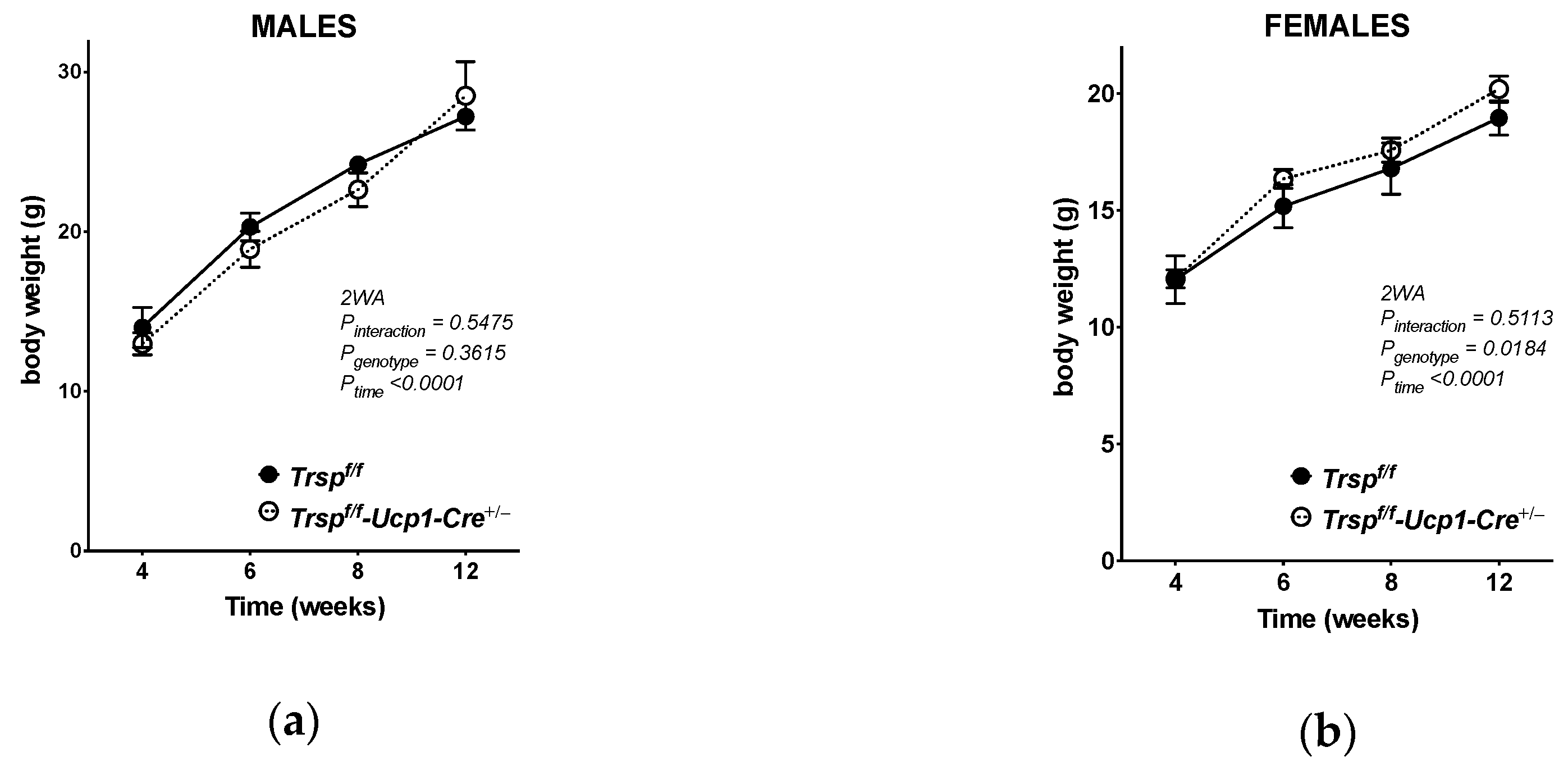
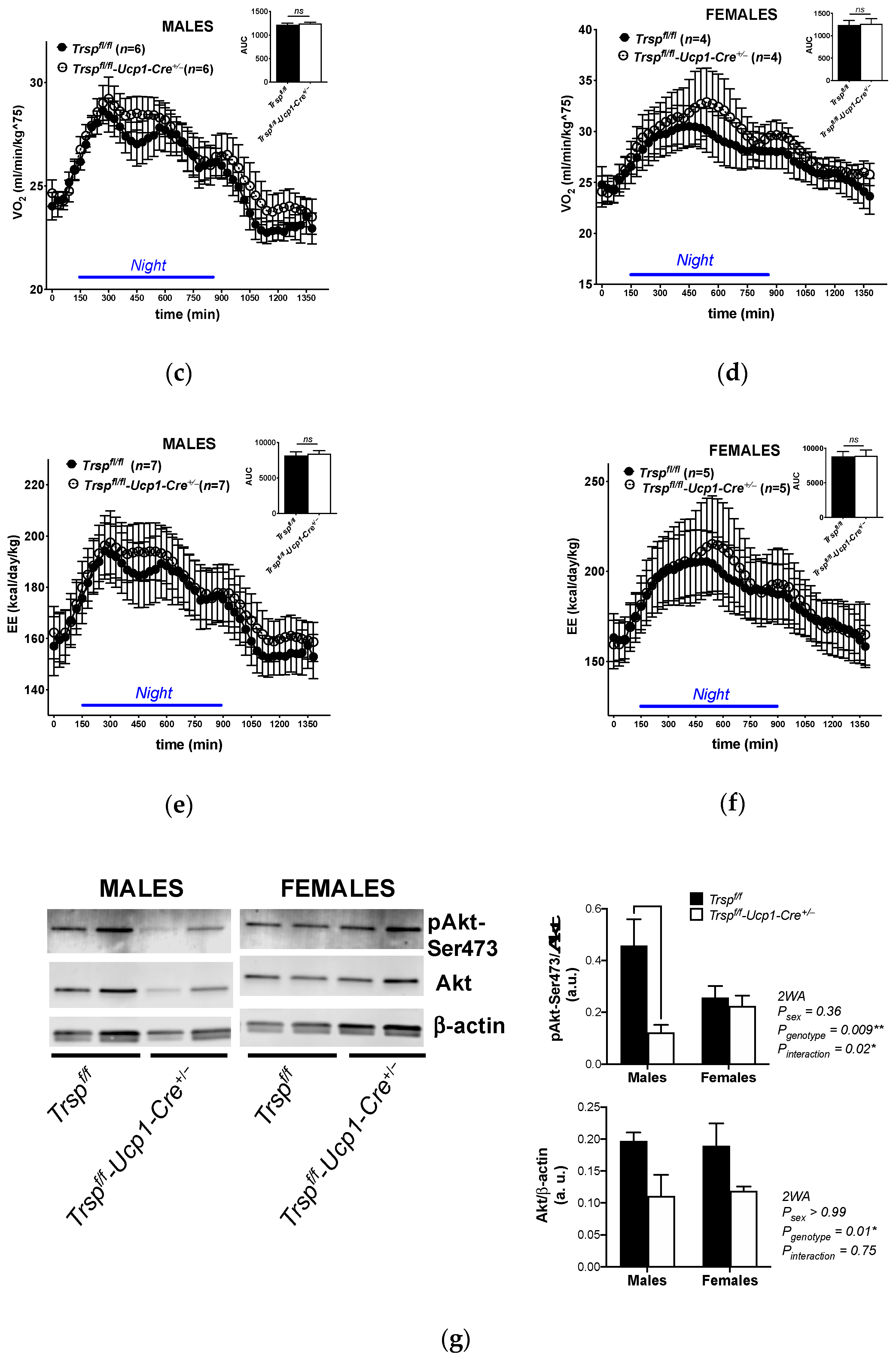
2.2. Trspf/f-Ucp1-Cre+/− Mice Maintain Energy Expenditure
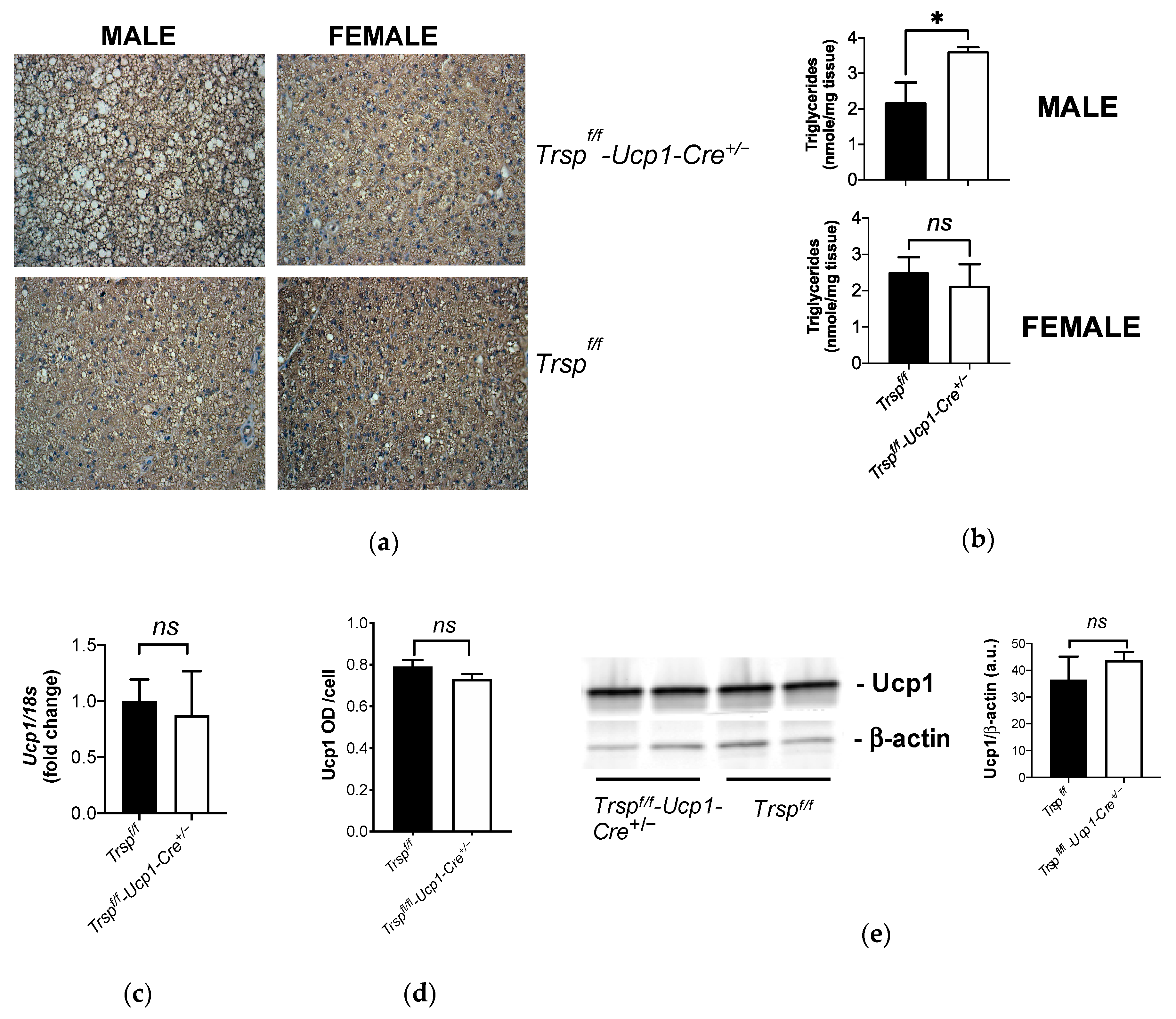
2.3. Mitochondrial Content in BAT of Trspf/f-Ucp1-Cre+/− Mice
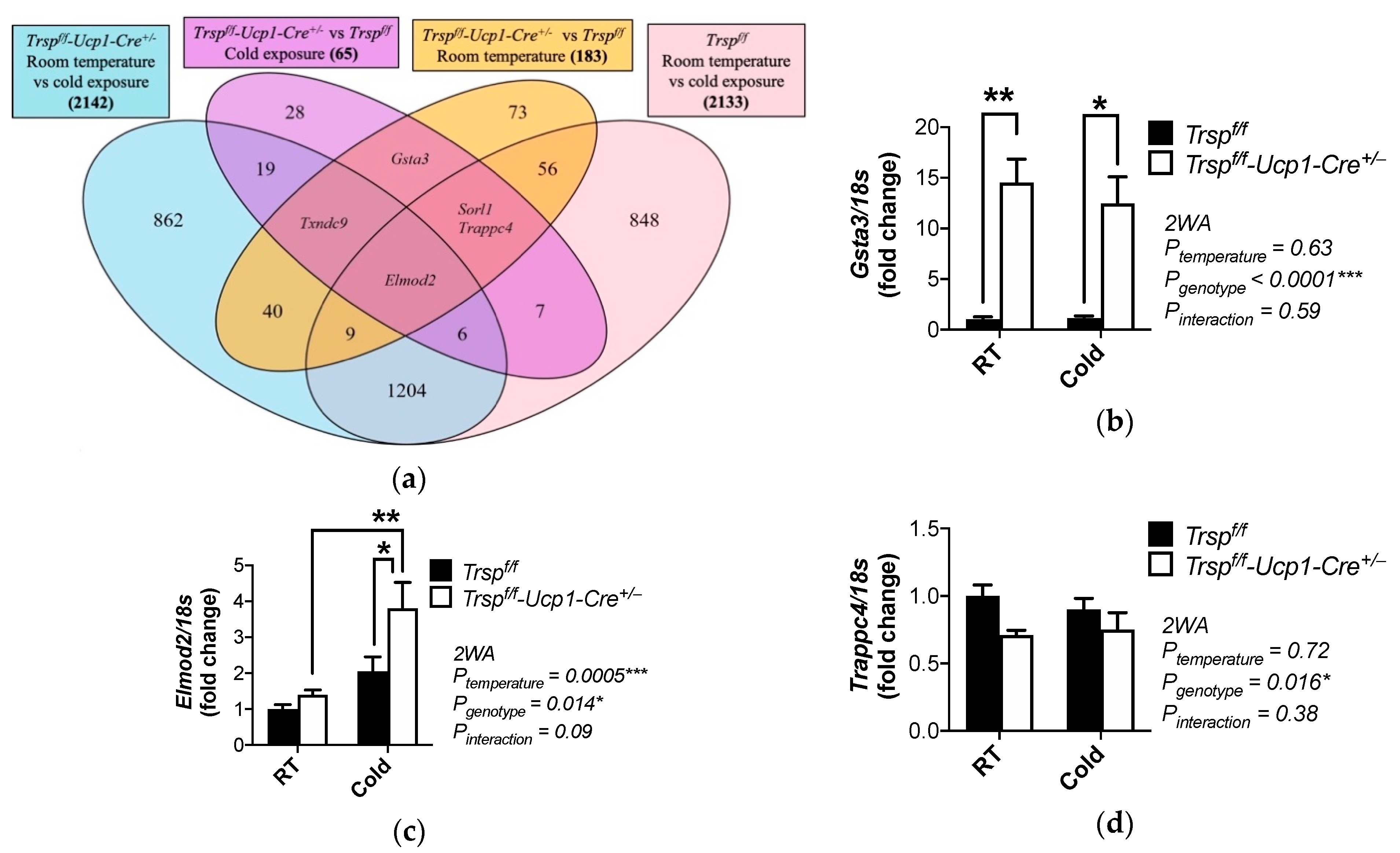
2.4. Changes in Gene Expression and Pathways in BAT of Trspf/f-Ucp1-Cre+/− Mice at Room Temperature
2.5. Body Temperature in the Trspf/f-Ucp1-Cre+/− Mice after Acute Cold Exposure
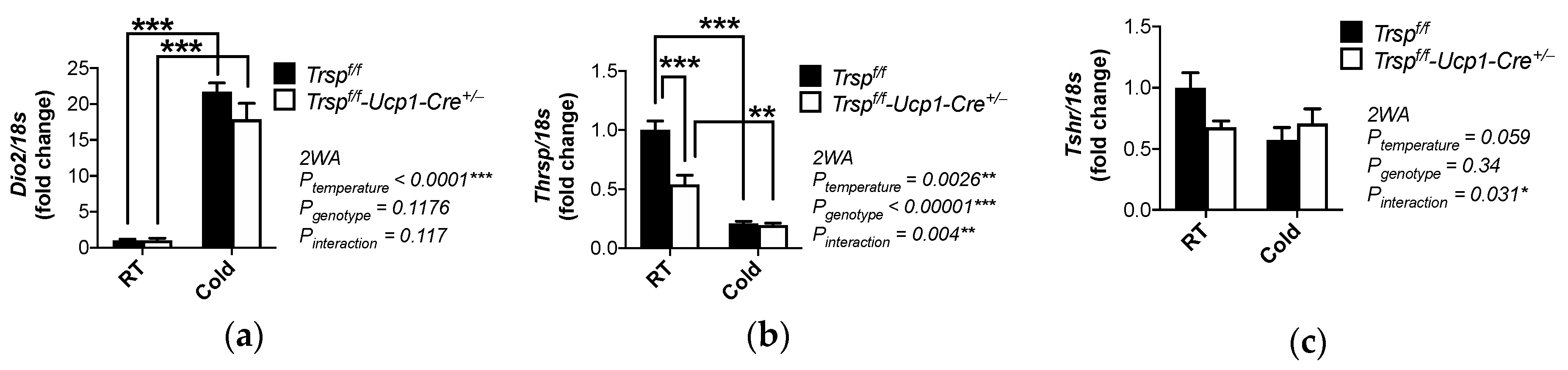
2.6. Changes in Gene Expression and Pathways in BAT of Trspf/f-Ucp1-Cre+/− Mice after Acute Cold Exposure
2.7. Thyroid Function in the Trspf/f-Ucp1-Cre+/− Mice
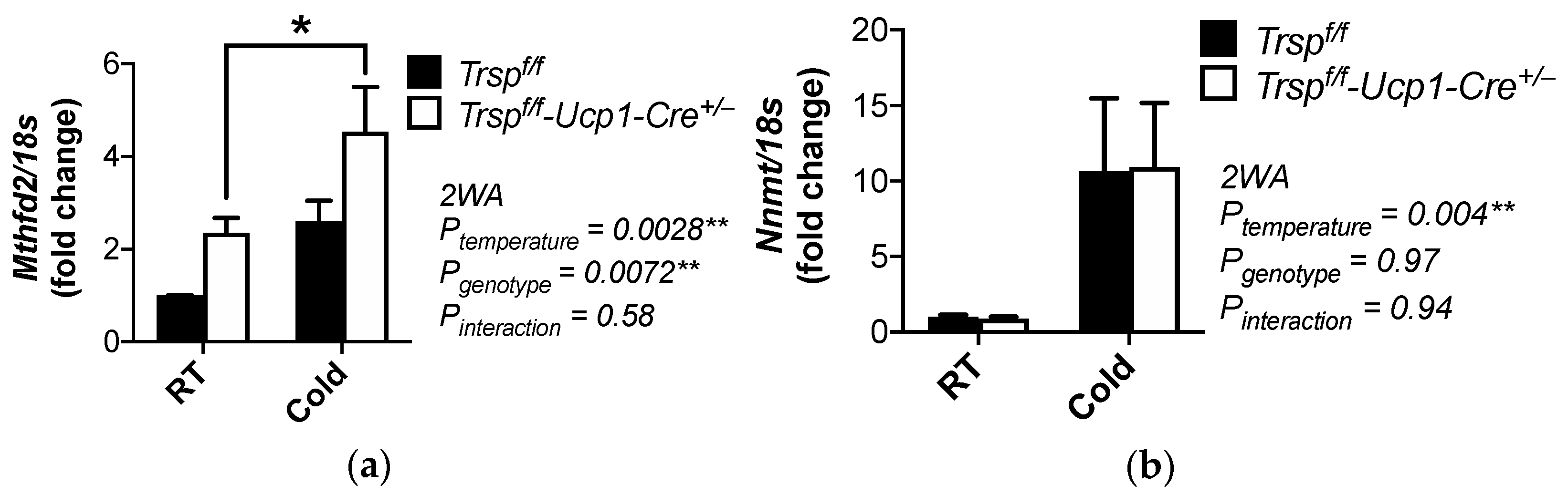
2.8. Methylation Genes in the Trspf/f-Ucp1-Cre+/− Mice
3. Discussion
4. Materials and Methods
4.1. Chemicals and Antibodies
4.2. Animals
4.3. Energy Expenditure Assessment
4.4. Cold Exposure and Core Body Temperature Assessment
4.5. Triglyceride Content
4.6. RNA Extraction
4.7. Microarray Analysis
4.8. Real-Time qPCR
4.9. Immunohistochemistry
4.10. Western Blot
4.11. Thyroid Function
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAT | Brown adipose tissue |
| Dio2 | Iodothyronine deiodinase type 2 |
| qPCR | Quantitative PCR |
| Sec | Selenocysteine |
| T3 | 3,3′,5-triiodothyronine |
| T4 | Thyroxine |
| TSH | Thyroid-stimulating hormone |
| Ucp1 | Uncoupling protein 1 |
References
- Carlisle, A.E.; Lee, N.; Matthew-Onabanjo, A.N.; Spears, M.E.; Park, S.J.; Youkana, D.; Doshi, M.B.; Peppers, A.; Li, R.; Joseph, A.B.; et al. Selenium detoxification is required for cancer-cell survival. Nat. Metab. 2020, 2, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Diamond, A.M. Selenoproteins of the Human Prostate: Unusual Properties and Role in Cancer Etiology. Biol. Trace Elem. Res. 2019, 192, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Kadkol, S.; Diamond, A.M. The Interaction between Dietary Selenium Intake and Genetics in Determining Cancer Risk and Outcome. Nutrients 2020, 12, 2424. [Google Scholar] [CrossRef]
- Duntas, L. Efficacy of selenium treatment in autoimmune thyroiditis demands an intact selenoprotein transport network. Thyroid Off. J. Am. Thyroid Assoc. 2007, 17, 83. [Google Scholar]
- Seiderer, J.; Dambacher, J.; Kuhnlein, B.; Pfennig, S.; Konrad, A.; Torok, H.P.; Haller, D.; Goke, B.; Ochsenkuhn, T.; Lohse, P.; et al. The role of the selenoprotein S (SELS) gene -105G>A promoter polymorphism in inflammatory bowel disease and regulation of SELS gene expression in intestinal inflammation. Tissue Antigens 2007, 70, 238–246. [Google Scholar]
- Misu, H.; Takamura, T.; Takayama, H.; Hayashi, H.; Matsuzawa-Nagata, N.; Kurita, S.; Ishikura, K.; Ando, H.; Takeshita, Y.; Ota, T.; et al. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab. 2010, 12, 483–495. [Google Scholar] [CrossRef]
- Ogawa-Wong, A.N.; Berry, M.J.; Seale, L.A. Selenium and metabolic disorders: An emphasis on type 2 diabetes risk. Nutrients 2016, 8, 80. [Google Scholar] [CrossRef]
- Luongo, C.; Dentice, M.; Salvatore, D. Deiodinases and their intricate role in thyroid hormone homeostasis. Nat. Rev. Endocrinol. 2019, 15, 479–488. [Google Scholar] [CrossRef]
- Negro, R. Selenium and thyroid autoimmunity. Biologics 2008, 2, 265–273. [Google Scholar]
- Carlson, B.A.; Lee, B.J.; Tsuji, P.A.; Copeland, P.R.; Schweizer, U.; Gladyshev, V.N.; Hatfield, D.L. Selenocysteine tRNA([Ser]Sec), the Central Component of Selenoprotein Biosynthesis: Isolation, Identification, Modification, and Sequencing. Methods Mol. Biol. 2018, 1661, 43–60. [Google Scholar] [CrossRef]
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40. [Google Scholar] [PubMed]
- Fixsen, S.M.; Howard, M.T. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J. Mol. Biol. 2010, 399, 385–396. [Google Scholar] [PubMed]
- Gursinsky, T.; Grobe, D.; Schierhorn, A.; Jager, J.; Andreesen, J.R.; Sohling, B. Factors and selenocysteine insertion sequence requirements for the synthesis of selenoproteins from a gram-positive anaerobe in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 1385–1393. [Google Scholar] [PubMed]
- Howard, M.T.; Copeland, P.R. New Directions for Understanding the Codon Redefinition Required for Selenocysteine Incorporation. Biol. Trace Elem. Res. 2019. [Google Scholar] [CrossRef]
- Shetty, S.P.; Shah, R.; Copeland, P.R. Regulation of selenocysteine incorporation into the selenium transport protein, selenoprotein P. J. Biol. Chem. 2014, 289, 25317–25326. [Google Scholar] [CrossRef]
- Simonovic, M.; Puppala, A.K. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2463–2472. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Berry, M.J. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life 2008, 60, 232–235. [Google Scholar]
- Stock, T.; Selzer, M.; Connery, S.; Seyhan, D.; Resch, A.; Rother, M. Disruption and complementation of the selenocysteine biosynthesis pathway reveals a hierarchy of selenoprotein gene expression in the archaeon Methanococcus maripaludis. Mol. Microbiol. 2011, 82, 734–747. [Google Scholar] [CrossRef]
- Bosl, M.R.; Takaku, K.; Oshima, M.; Nishimura, S.; Taketo, M.M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl. Acad. Sci. USA 1997, 94, 5531–5534. [Google Scholar]
- Carlson, B.A. Selenocysteine tRNA[Ser]Sec mouse models for elucidating roles of selenoproteins in health and development. In Selenium: Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer Science+Business Media, LLC: New York, NY, USA, 2016; pp. 555–566. [Google Scholar]
- Carlson, B.A.; Yoo, M.H.; Tsuji, P.A.; Gladyshev, V.N.; Hatfield, D.L. Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules 2009, 14, 3509–3527. [Google Scholar]
- Oguri, Y.; Kajimura, S. Cellular heterogeneity in brown adipose tissue. J. Clin. Investig. 2020, 130, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Dai, W.; Jang, M.J.; Medrano, L.; Li, Z.; Zhao, H.; Shao, M.; Tan, J.; Li, A.; Ning, T.; et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Investig. 2020, 130, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Ricquier, D. UCP1, the mitochondrial uncoupling protein of brown adipocyte: A personal contribution and a historical perspective. Biochimie 2017, 134, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.E. Full expression of uncoupling protein gene requires the concurrence of norepinephrine and triiodothyronine. Mol. Endocrinol. 1988, 2, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Marsili, A.; Zavacki, A.M.; Harney, J.W.; Larsen, P.R. Physiological role and regulation of iodothyronine deiodinases: A 2011 update. J. Endocrinol. Investig. 2011, 34, 395–407. [Google Scholar] [CrossRef]
- Christoffolete, M.A.; Linardi, C.C.; de Jesus, L.; Ebina, K.N.; Carvalho, S.D.; Ribeiro, M.O.; Rabelo, R.; Curcio, C.; Martins, L.; Kimura, E.T.; et al. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes 2004, 53, 577–584. [Google Scholar] [CrossRef]
- Rehnmark, S.; Bianco, A.C.; Kieffer, J.D.; Silva, J.E. Transcriptional and posttranscriptional mechanisms in uncoupling protein mRNA response to cold. Am. J. Physiol. 1992, 262, E58–E67. [Google Scholar] [CrossRef]
- de Jesus, L.A.; Carvalho, S.D.; Ribeiro, M.O.; Schneider, M.; Kim, S.W.; Harney, J.W.; Larsen, P.R.; Bianco, A.C. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Investig. 2001, 108, 1379–1385. [Google Scholar] [CrossRef]
- Marsili, A.; Aguayo-Mazzucato, C.; Chen, T.; Kumar, A.; Chung, M.; Lunsford, E.P.; Harney, J.W.; Van-Tran, T.; Gianetti, E.; Ramadan, W.; et al. Mice with a targeted deletion of the type 2 deiodinase are insulin resistant and susceptible to diet induced obesity. PLoS ONE 2011, 6, e20832. [Google Scholar] [CrossRef]
- Jedrychowski, M.P.; Lu, G.Z.; Szpyt, J.; Mariotti, M.; Garrity, R.; Paulo, J.A.; Schweppe, D.K.; Laznik-Bogoslavski, D.; Kazak, L.; Murphy, M.P.; et al. Facultative protein selenation regulates redox sensitivity, adipose tissue thermogenesis, and obesity. Proc. Natl. Acad. Sci. USA 2020, 117, 10789–10796. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, C.; Straub, L.G. Lessons from Cre-Mice and Indicator Mice. In Brown Adipose Tissue; Pfeifer, A., Klingenspor, M., Herzig, S., Eds.; Springer: Cham, Switzerland, 2018; pp. 37–54. [Google Scholar]
- Popov, L.D. Mitochondrial biogenesis: An update. J. Cell Mol. Med. 2020, 24, 4892–4899. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.D.; Lee, S.J.; Jastroch, M.; Kabra, D.; Stemmer, K.; Aichler, M.; Abplanalp, B.; Ananthakrishnan, G.; Bhardwaj, N.; Collins, S.; et al. p62 links beta-adrenergic input to mitochondrial function and thermogenesis. J. Clin. Investig. 2013, 123, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.; Renaut, J.; Cambier, S.; Murk, A.J.; Gutleb, A.C.; Serchi, T. Dataset of liver proteins of eu- and hypothyroid rats affected in abundance by any of three factors: In vivo exposure to hexabromocyclododecane (HBCD), thyroid status, gender differences. Data Brief 2016, 8, 1344–1347. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Murakami, T.; Cheng, J.; Kano, H.; Fukata, M.; Fujimoto, T. ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol. Biol. Cell 2015, 26, 2333–2342. [Google Scholar] [CrossRef]
- Zhao, S.L.; Hong, J.; Xie, Z.Q.; Tang, J.T.; Su, W.Y.; Du, W.; Chen, Y.X.; Lu, R.; Sun, D.F.; Fang, J.Y. TRAPPC4-ERK2 interaction activates ERK1/2, modulates its nuclear localization and regulates proliferation and apoptosis of colorectal cancer cells. PLoS ONE 2011, 6, e23262. [Google Scholar] [CrossRef]
- Kinlaw, W.B.; Church, J.L.; Harmon, J.; Mariash, C.N. Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J. Biol. Chem. 1995, 270, 16615–16618. [Google Scholar] [CrossRef]
- LaFave, L.T.; Augustin, L.B.; Mariash, C.N. S14: Insights from knockout mice. Endocrinology 2006, 147, 4044–4047. [Google Scholar] [CrossRef]
- Chou, W.Y.; Cheng, Y.S.; Ho, C.L.; Liu, S.T.; Liu, P.Y.; Kuo, C.C.; Chang, H.P.; Chen, Y.H.; Chang, G.G.; Huang, S.M. Human spot 14 protein interacts physically and functionally with the thyroid receptor. Biochem. Biophys. Res. Commun. 2007, 357, 133–138. [Google Scholar] [CrossRef]
- Green, N.H.; Galvan, D.L.; Badal, S.S.; Chang, B.H.; LeBleu, V.S.; Long, J.; Jonasch, E.; Danesh, F.R. MTHFD2 links RNA methylation to metabolic reprogramming in renal cell carcinoma. Oncogene 2019, 38, 6211–6225. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, X.; Wang, Y.; Huang, X.; Yang, J.; Zeng, J.; Li, G.; Xie, X.; Zhang, J. Nicotinamide N-methyltransferase inhibits autophagy induced by oxidative stress through suppressing the AMPK pathway in breast cancer cells. Cancer Cell Int. 2020, 20, 191. [Google Scholar] [CrossRef] [PubMed]
- Martinez-deMena, R.; Hernandez, A.; Obregon, M.J. Triiodothyronine is required for the stimulation of type II 5’-deiodinase mRNA in rat brown adipocytes. Am. J. Physiol. Endocrinol. Metab. 2002, 282, E1119–E1127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cinti, S. Between brown and white: Novel aspects of adipocyte differentiation. Ann. Med. 2011, 43, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Ribich, S.; Christoffolete, M.A.; Simovic, G.; Correa-Medina, M.; Patti, M.E.; Bianco, A.C. Absence of thyroid hormone activation during development underlies a permanent defect in adaptive thermogenesis. Endocrinology 2010, 151, 4573–4582. [Google Scholar] [CrossRef] [PubMed]
- Riese, C.; Michaelis, M.; Mentrup, B.; Gotz, F.; Kohrle, J.; Schweizer, U.; Schomburg, L. Selenium-dependent pre- and posttranscriptional mechanisms are responsible for sexual dimorphic expression of selenoproteins in murine tissues. Endocrinology 2006, 147, 5883–5892. [Google Scholar] [CrossRef]
- Seale, L.A.; Ogawa-Wong, A.N.; Berry, M.J. Sexual Dimorphism in Selenium Metabolism and Selenoproteins. Free Radic. Biol. Med. 2018, 127, 198–205. [Google Scholar] [CrossRef]
- Nedergaard, J.; Dicker, A.; Cannon, B. The interaction between thyroid and brown-fat thermogenesis. Central or peripheral effects? Ann. N. Y. Acad. Sci. 1997, 813, 712–717. [Google Scholar] [CrossRef]
- Martinez-deMena, R.; Anedda, A.; Cadenas, S.; Obregon, M.J. TSH effects on thermogenesis in rat brown adipocytes. Mol. Cell Endocrinol. 2015, 404, 151–158. [Google Scholar] [CrossRef]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef]
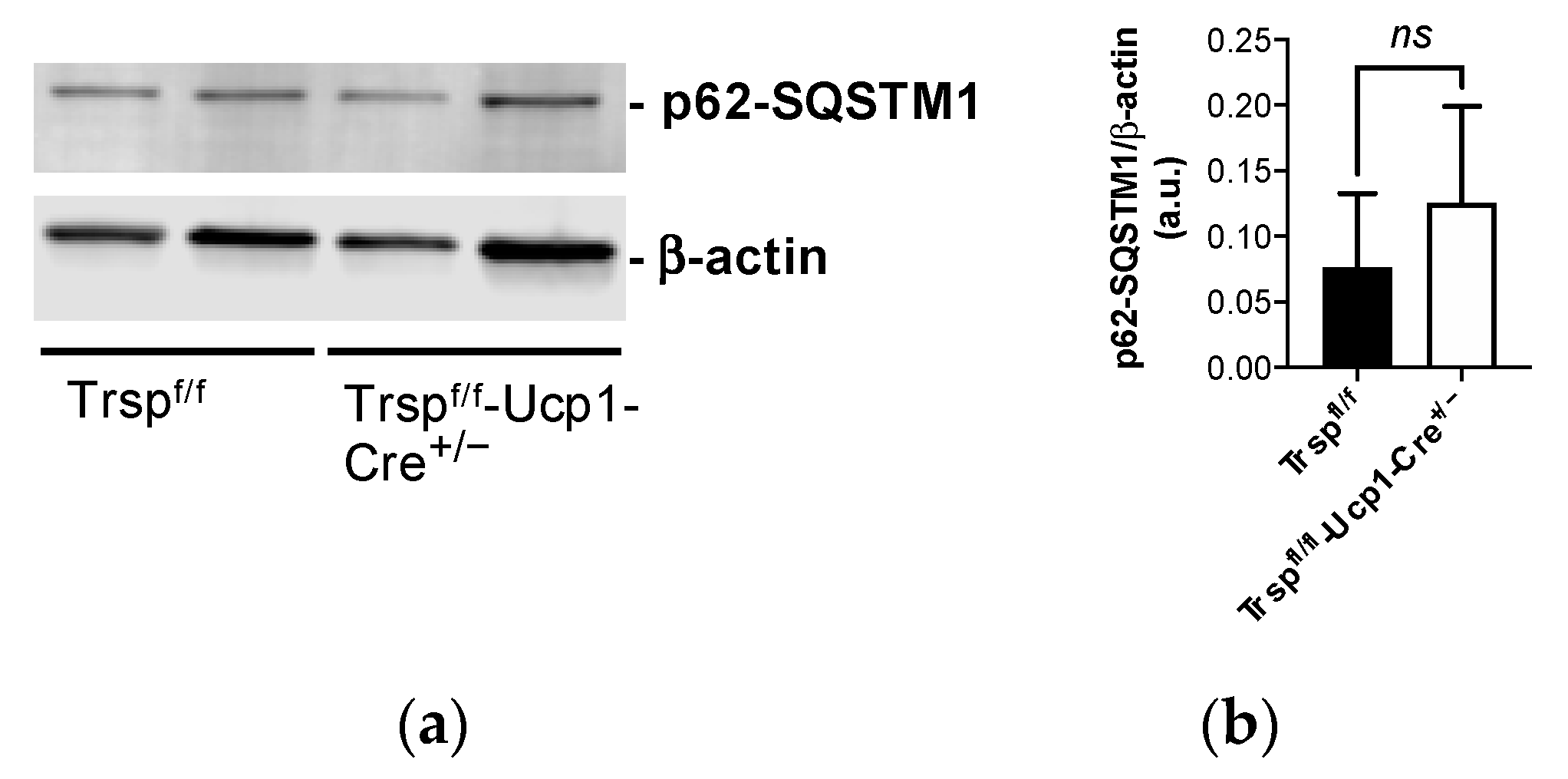
- Ferhat, M.; Funai, K.; Boudina, S. Autophagy in Adipose Tissue Physiology and Pathophysiology. Antioxid. Redox Signal. 2019, 31, 487–501. [Google Scholar] [CrossRef]
- Cairo, M.; Villarroya, J.; Cereijo, R.; Campderros, L.; Giralt, M.; Villarroya, F. Thermogenic activation represses autophagy in brown adipose tissue. Int. J. Obes. 2016, 40, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, N.; Garcia-Macia, M.; Sahu, S.; Athonvarangkul, D.; Liebling, E.; Merlo, P.; Cecconi, F.; Schwartz, G.J.; Singh, R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016, 23, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fujioka, H.; Joshi, D.; Li, Q.; Sangwung, P.; Hsieh, P.; Zhu, J.; Torio, J.; Sweet, D.; Wang, L.; et al. Mitophagy is required for brown adipose tissue mitochondrial homeostasis during cold challenge. Sci. Rep. 2018, 8, 8251. [Google Scholar] [CrossRef] [PubMed]
- Obregon, M.J. Adipose tissues and thyroid hormones. Front. Physiol. 2014, 5, 479. [Google Scholar] [CrossRef] [PubMed]
- Hori, H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014, 5, 144. [Google Scholar] [CrossRef]
- Diamond, A.M.; Choi, I.S.; Crain, P.F.; Hashizume, T.; Pomerantz, S.C.; Cruz, R.; Steer, C.J.; Hill, K.E.; Burk, R.F.; McCloskey, J.A.; et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem. 1993, 268, 14215–14223. [Google Scholar] [CrossRef]
- Ganichkin, O.M.; Anedchenko, E.A.; Wahl, M.C. Crystal structure analysis reveals functional flexibility in the selenocysteine-specific tRNA from mouse. PLoS ONE 2011, 6, e20032. [Google Scholar] [CrossRef]
- Barroso, M.; Florindo, C.; Kalwa, H.; Silva, Z.; Turanov, A.A.; Carlson, B.A.; de Almeida, I.T.; Blom, H.J.; Gladyshev, V.N.; Hatfield, D.L.; et al. Inhibition of cellular methyltransferases promotes endothelial cell activation by suppressing glutathione peroxidase 1 protein expression. J. Biol. Chem. 2014, 289, 15350–15362. [Google Scholar] [CrossRef]
- Koufaris, C.; Nilsson, R. Protein interaction and functional data indicate MTHFD2 involvement in RNA processing and translation. Cancer Metab. 2018, 6, 12. [Google Scholar] [CrossRef]
- Sun, X.; Maquat, L.E. Nonsense-mediated decay: Assaying for effects on selenoprotein mRNAs. Methods Enzymol. 2002, 347, 49–57. [Google Scholar]
- Seyedali, A.; Berry, M.J. Nonsense-mediated decay factors are involved in the regulation of selenoprotein mRNA levels during selenium deficiency. Rna 2014, 20, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.; Hall, J.A.; Correa-Medina, M.; Ueta, C.; Kang, H.W.; Cohen, D.E.; Bianco, A.C. Disruption of thyroid hormone activation in type 2 deiodinase knockout mice causes obesity with glucose intolerance and liver steatosis only at thermoneutrality. Diabetes 2011, 60, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Kumaraswamy, E.; Carlson, B.A.; Morgan, F.; Miyoshi, K.; Robinson, G.W.; Su, D.; Wang, S.; Southon, E.; Tessarollo, L.; Lee, B.J.; et al. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol. Cell. Biol. 2003, 23, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Oelkrug, R.; Polymeropoulos, E.T.; Jastroch, M. Brown adipose tissue: Physiological function and evolutionary significance. J. Comp. Physiol. B 2015, 185, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Ogawa-Wong, A.N.; Hashimoto, A.C.; Ha, H.; Pitts, M.W.; Seale, L.A.; Berry, M.J. Sexual Dimorphism in the Selenocysteine Lyase Knockout Mouse. Nutrients 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Seale, L.A.; Hashimoto, A.C.; Kurokawa, S.; Gilman, C.L.; Seyedali, A.; Bellinger, F.P.; Raman, A.V.; Berry, M.J. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol. Cell. Biol. 2012, 32, 4141–4154. [Google Scholar] [CrossRef]


| Top Upregulated | |||
|---|---|---|---|
| Gene Symbol | Gene Name | Fold Change | p-Value |
| Plekhm2 | pleckstrin homology and RUN domain containing M2 | 4.59 | 0.0361 |
| Sntb2 | syntrophin beta 2 | 4.02 | 0.0008 |
| Phf20 | PHD finger protein 20 | 3.78 | 0.0271 |
| Vps4a | vacuolar protein sorting 4 homolog A | 3.33 | 0.0296 |
| Fads3 | fatty acid desaturase 3 | 3.3 | 0.0119 |
| S100pbp | S100P binding protein | 3.26 | 0.0005 |
| Lcp1 | lymphocyte cytosolic protein 1 | 3.17 | 0.0453 |
| Fxyd5 | FXYD domain containing ion transport regulator 5 | 3.07 | 0.0334 |
| Elmod2 | ELMO domain containing 2 | 2.79 | 0.003 |
| Kpna2 | karyopherin subunit alpha 2 | 2.72 | 0.0061 |
| Top Downregulated | |||
| Gene Symbol | Gene Name | Fold Change | p-Value |
| Tmsb4x | thymosin, beta 4, X chromosome | −4.24 | 0.0172 |
| Trappc4 | trafficking protein particle complex 4 | −3.63 | 0.0183 |
| Hscb | HscB mitochondrial iron-sulfur cluster cochaperone | −3.12 | 0.0196 |
| Ccdc85a | coiled-coil domain containing 85A | −2.98 | 0.003 |
| Skil | SKI-like proto-oncogene | −2.87 | 0.0297 |
| Sorl1 | sortilin related receptor 1 | −2.8 | 0.0299 |
| Gk | glycerol kinase | −2.66 | 0.0094 |
| Nbea | neurobeachin | −2.46 | 0.0228 |
| Tmx4 | thioredoxin related transmembrane protein 4 | −2.43 | 0.0234 |
| Per2 | period circadian regulator 2 | −2.42 | 0.0481 |
| Canonical Pathways | −log(p-Value) | Molecules |
|---|---|---|
| Nicotine Degradation II | 1.85 | FMO2,FMO5 |
| Antigen Presentation Pathway | 1.78 | HLA-DMB,MR1 |
| Glycerol Degradation I | 1.59 | Gk |
| LPS/IL-1 Mediated Inhibition of RXR Function | 1.58 | GSTA3,FMO2,SCARB1,FMO5 |
| Myo-inositol Biosynthesis | 1.47 | IMPAD1 |
| Top Upregulated | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Name | Fold Change | p-Value | |
| Nnmt | nicotinamide N-methyltransferase | 24.07 | 0.0012 | |
| Elmod2 | ELMO domain containing 2 | 4.1 | 0.0018 | |
| Gsta3 | glutathione S-transferase alpha 3 | 4.05 | 0.0038 | |
| Laptm5 | lysosomal protein transmembrane 5 | 3.15 | 0.0029 | |
| Lyz | lysozyme | 3.15 | 0.0087 | |
| Ear2 | eosinophil-associated, ribonuclease A family, member 2 | 2.74 | 0.0013 | |
| Mthfd2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 | 2.69 | 0.0126 | |
| Rab3a | RAB3A, member RAS oncogene family | 2.6 | 0.0131 | |
| Sorl1 | sortilin related receptor 1 | 2.44 | 0.0467 | |
| Ctss | cathepsin S | 2.41 | 0.0376 | |
| Top Downregulated | ||||
| Gene Symbol | Gene Name | Fold Change | p-Value | |
| Mbp | myelin basic protein | −3.17 | 0.0408 | |
| Gucd1 | guanylyl cyclase domain containing 1 | −2.86 | 0.0375 | |
| Rnf149 | ring finger protein 149 | −2.56 | 0.0272 | |
| Rnf125 | ring finger protein 125 | −2.54 | 0.0276 | |
| Thrsp | thyroid hormone responsive (Spot14) | −2.47 | 0.0044 | |
| Tshr | thyroid stimulating hormone receptor | −2.46 | 0.0266 | |
| Insig1 | insulin induced gene 1 | −2.43 | 0.0413 | |
| Art3 | ADP-ribosyltransferase 3 | −2.39 | 0.0255 | |
| Il18 | interleukin 18 | −2.36 | 0.0035 | |
| Akap12 | A-kinase anchoring protein 12 | −2.35 | 0.0085 | |
| Canonical Pathways | −log(p-Value) | Molecules |
|---|---|---|
| autophagy | 2.13 | WDFY3,CTSS |
| Histidine Degradation III | 1.94 | MTHFD2 |
| Tetrahydrofolate Salvage from 5,10-methenyltetrahydrofolate | 1.81 | MTHFD2 |
| Folate Transformations I | 1.72 | MTHFD2 |
| LXR/RXR Activation | 1.58 | IL18,LYZ |
| Room Temperature | Cold Exposure | Two-Way ANOVA | |||||
|---|---|---|---|---|---|---|---|
| Trspf/f | Trspf/f-Ucp1-Cre+/− | Trspf/f | Trspf/f-Ucp1-Cre+/− | Pg | Pt | Pi | |
| TSH (pg/mL) | |||||||
| Males | 1895.2 ± 86.0 | 2036 ± 46.05 a | 1312 ± 112.9 | 1161 ± 211.7 b | 0.861 | <0.0001 | 0.031 |
| Females | 710.3 ± 75.4 | 541.1 ± 282.8 | 363.7 ± 107.2 | 503.8 ± 149.4 | 0.043 | 0.868 | 0.094 |
| T4 (μg/dl) | |||||||
| Males | 3.768 ± 1.18 | 4.06 ± 1.01 | 3.447 ± 0.46 | 3.16 ± 0.61 | 0.994 | 0.154 | 0.487 |
| Females | 3.661 ± 0.65 | 3.421 ± 0.42 | 3.082 ± 0.58 | 3.009 ± 0.19 | 0.488 | 0.038 | 0.712 |
| T3 (ng/mL) | |||||||
| Males | 1.02 ±0.19 | 0.79 ± 0.34 | 1.08 ± 0.24 | 0.87 ± 0.21 | 0.01 | 0.42 | 0.9 |
| Females | 0.62 ± 0.24 | 0.58 ± 0.1 | 0.86 ± 0.22 | 0.75 ± 0.04 | 0.36 | 0.01 | 0.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seale, L.A.; Ogawa-Wong, A.N.; Watanabe, L.M.; Khadka, V.S.; Menor, M.; Torres, D.J.; Carlson, B.A.; Hatfield, D.L.; Berry, M.J. Adaptive Thermogenesis in a Mouse Model Lacking Selenoprotein Biosynthesis in Brown Adipocytes. Int. J. Mol. Sci. 2021, 22, 611. https://doi.org/10.3390/ijms22020611
Seale LA, Ogawa-Wong AN, Watanabe LM, Khadka VS, Menor M, Torres DJ, Carlson BA, Hatfield DL, Berry MJ. Adaptive Thermogenesis in a Mouse Model Lacking Selenoprotein Biosynthesis in Brown Adipocytes. International Journal of Molecular Sciences. 2021; 22(2):611. https://doi.org/10.3390/ijms22020611
Chicago/Turabian StyleSeale, Lucia A., Ashley N. Ogawa-Wong, Ligia M. Watanabe, Vedbar S. Khadka, Mark Menor, Daniel J. Torres, Bradley A. Carlson, Dolph L. Hatfield, and Marla J. Berry. 2021. "Adaptive Thermogenesis in a Mouse Model Lacking Selenoprotein Biosynthesis in Brown Adipocytes" International Journal of Molecular Sciences 22, no. 2: 611. https://doi.org/10.3390/ijms22020611
APA StyleSeale, L. A., Ogawa-Wong, A. N., Watanabe, L. M., Khadka, V. S., Menor, M., Torres, D. J., Carlson, B. A., Hatfield, D. L., & Berry, M. J. (2021). Adaptive Thermogenesis in a Mouse Model Lacking Selenoprotein Biosynthesis in Brown Adipocytes. International Journal of Molecular Sciences, 22(2), 611. https://doi.org/10.3390/ijms22020611






