Generation of a Novel Mesothelin-Targeted Oncolytic Herpes Virus and Implemented Strategies for Manufacturing
Abstract
1. Introduction
2. Results
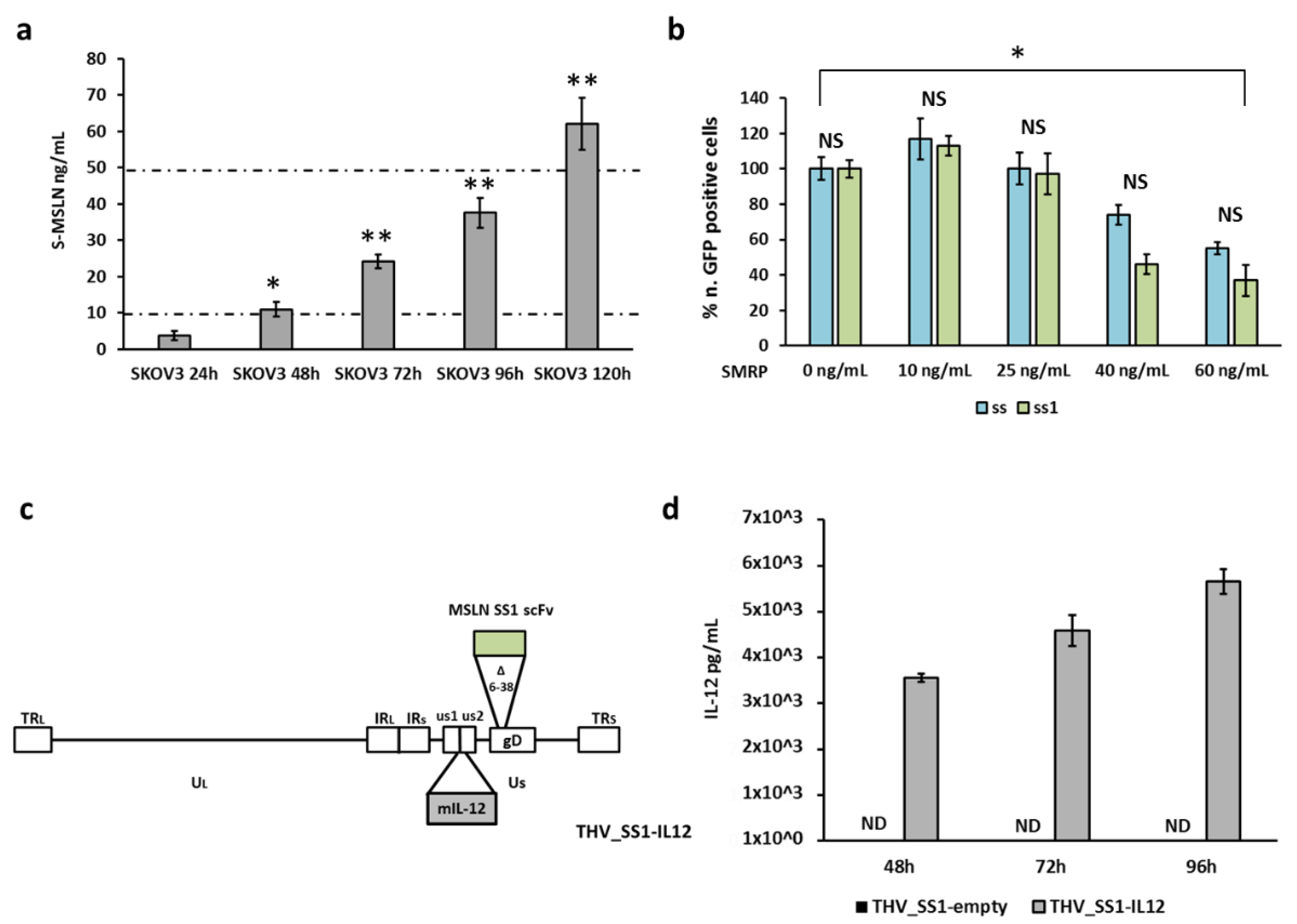
2.1. Generation of Oncolytic Herpes Viruses with a Specific Tropism for Human Mesothelin
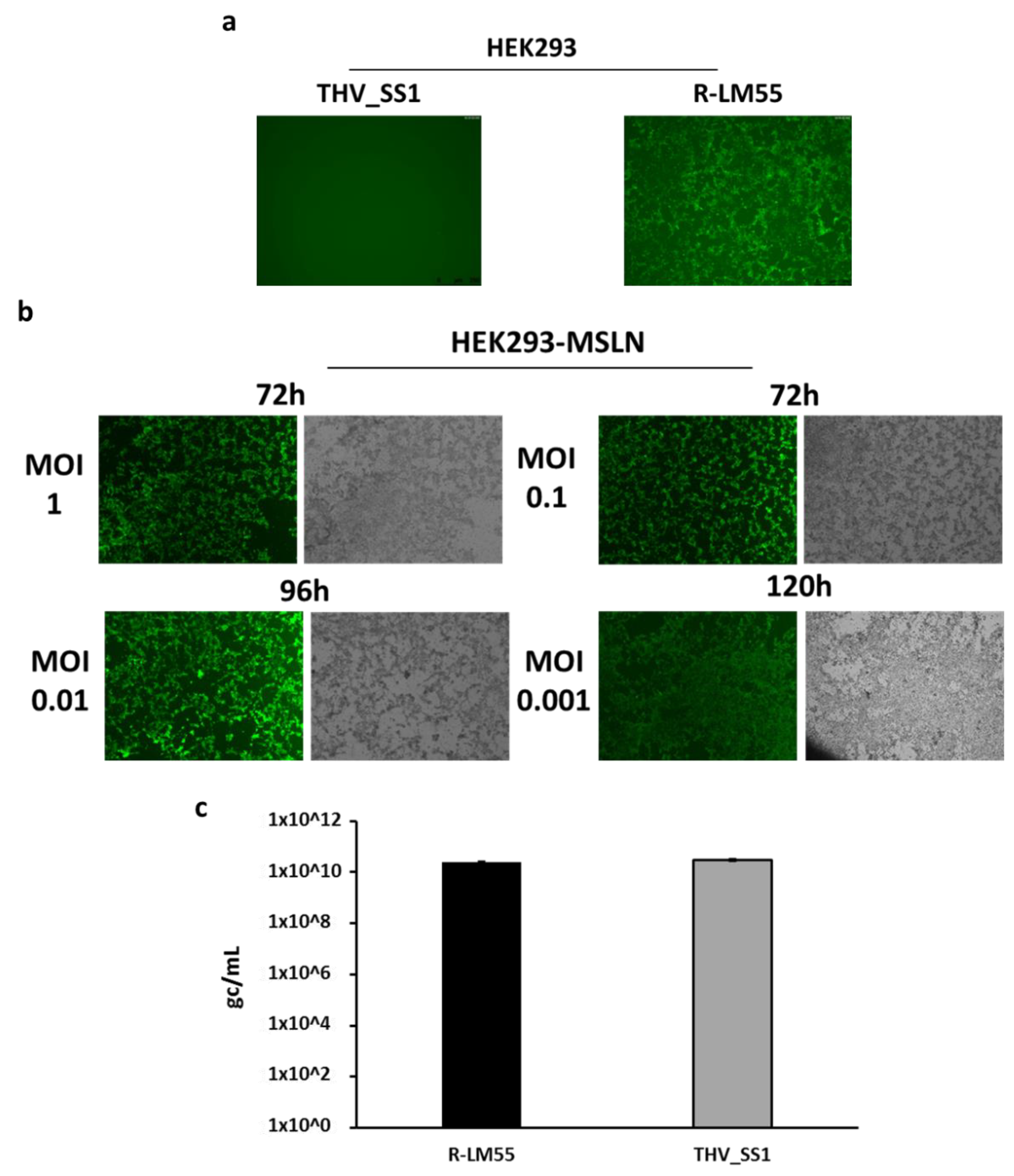
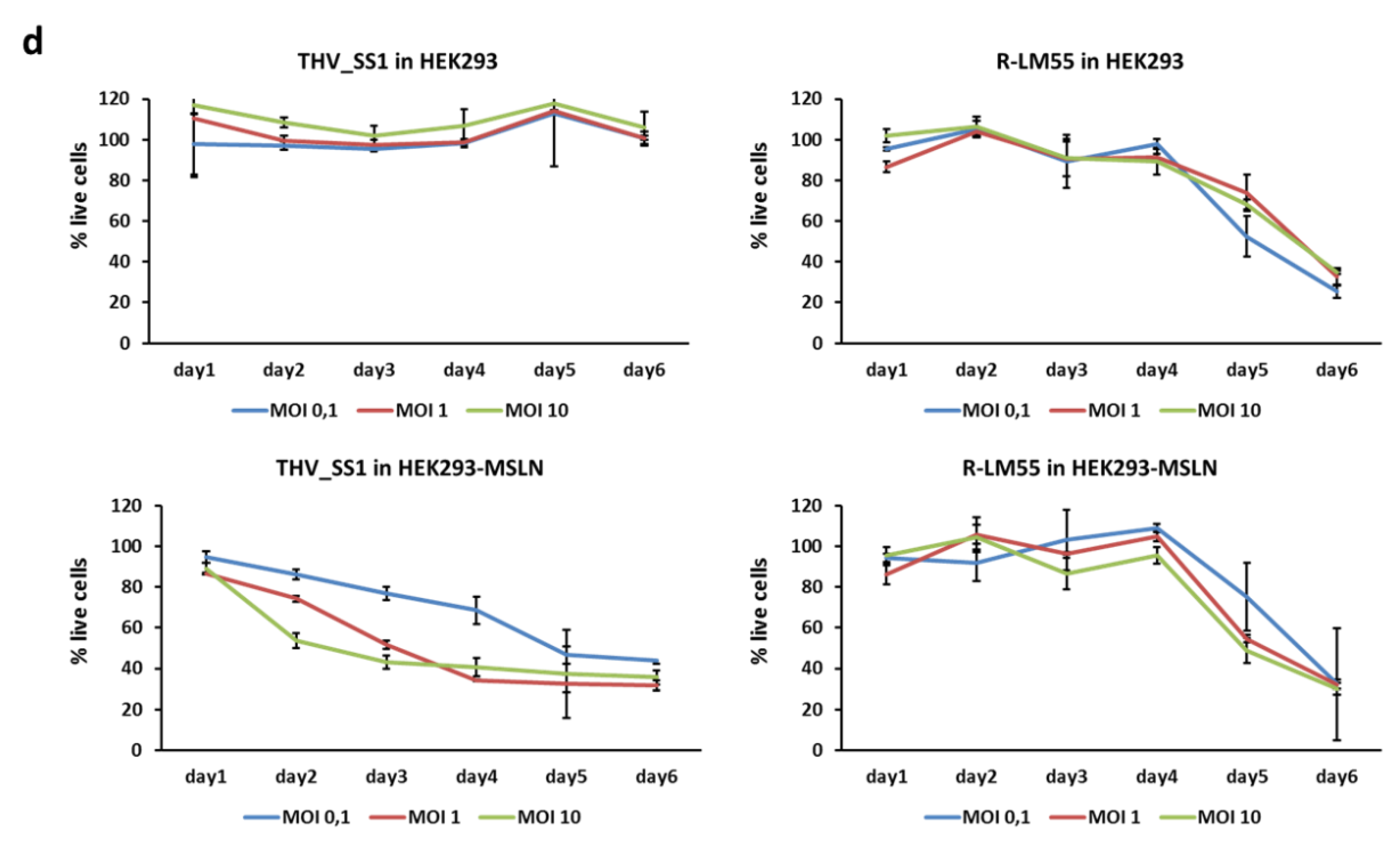
2.2. THV_SS1 Exerts Mesothelin-Dependent Cytotoxicity in Human Cells
2.3. Implementation of a Cell Platform for THV_SS1 Oncolytic Virus Production
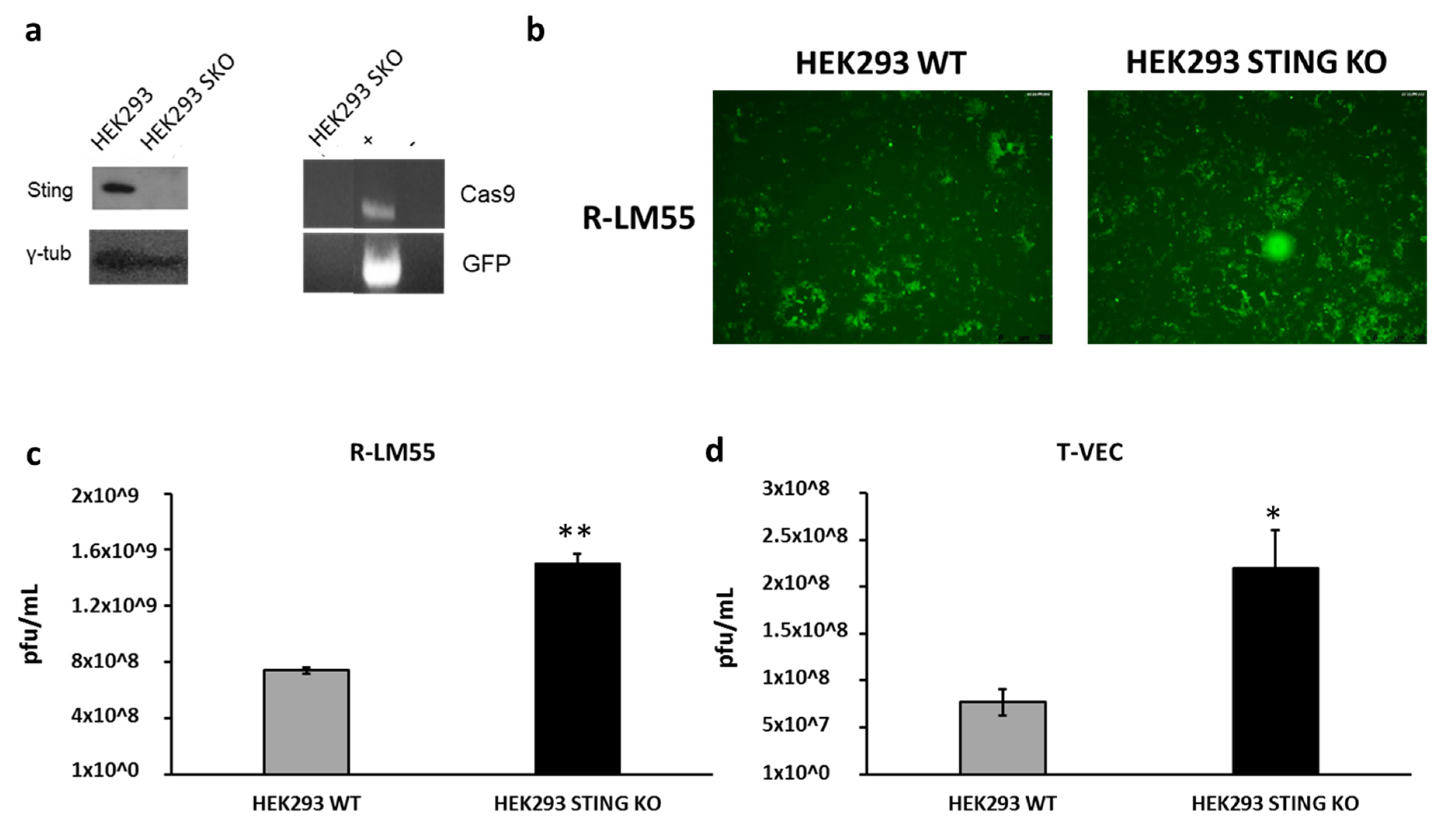
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Manipulation, and Characterization
4.2. Modifications of BAC-HSV-1 Vectors
4.3. Viral Rescue, Production, Titration, and Real Time PCR Analysis
4.4. Tropism of the Recombinant Viruses
4.5. Quantification of Soluble Proteins
4.6. Cytotoxicity Assay
4.7. Figures and Statistical Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MSLN | Mesothelin |
| THV | Targeted Herpes Virus |
| STING | Stimulator of Interferon Genes |
| OV | Oncolytic virus |
| scFv | Single Chain Fragment variable |
| SMRP | Soluble Mesothelin Related Protein |
References
- Sasso, E.; D’Alise, A.M. New viral vector for infectious diseases and cancer. Semin. Immunol. 2020, 50, 101430. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Kohlhapp, F.J. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662, Erratum in: Nat. Rev. Drug Discov. 2016, 15, 660. [Google Scholar] [CrossRef]
- Bommareddy, P.K.; Shettigar, M. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 498–513, Erratum in: Nat. Rev. Immunol. 2018. [Google Scholar] [CrossRef]
- Sivanandam, V.; LaRocca, C.J. Oncolytic Viruses and Immune Checkpoint Inhibition: The Best of Both Worlds. Mol. Ther. Oncolytics 2019, 13, 93–106. [Google Scholar] [CrossRef]
- Twumasi-Boateng, K.; Pettigrew, J.L. Oncolytic viruses as engineering platforms for combination immunotherapy. Nat. Rev. Cancer 2018, 18, 419–432, Erratum in: Nat. Rev. Cancer 2018. [Google Scholar] [CrossRef]
- Van Vloten, J.P.; Workenhe, S.T. Critical Interactions between Immunogenic Cancer Cell Death, Oncolytic Viruses, and the Immune System Define the Rational Design of Combination Immunotherapies. J. Immunol. 2018, 200, 450–458. [Google Scholar] [CrossRef]
- Sasso, E.; Froechlich, G. Replicative conditioning of Herpes simplex type 1 virus by Survivin promoter, combined to ERBB2 retargeting, improves tumour cell-restricted oncolysis. Sci. Rep. 2020, 10, 4307. [Google Scholar] [CrossRef]
- Sasso, E.; D’Avino, C. Massive parallel screening of phage libraries for the generation of repertoires of human immunomodulatory monoclonal antibodies. MAbs 2018, 10, 1060–1072. [Google Scholar] [CrossRef]
- Rehman, H.; Silk, A.W. Into the clinic: Talimogene laherparepvec (T-VEC), a first-in-class intratumoral oncolytic viral therapy. J. Immunother. Cancer 2016, 4, 53. [Google Scholar] [CrossRef]
- Andtbacka, R.H.; Ross, M. Patterns of Clinical Response with Talimogene Laherparepvec (T-VEC) in Patients with Melanoma Treated in the OPTiM Phase III Clinical Trial. Ann. Surg. Oncol. 2016, 23, 4169–4177. [Google Scholar] [CrossRef]
- Ribas, A.; Dummer, R. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell 2017, 170, 1109–1119, Published Correction Appears in Cell 2018, 174, 1031–1032. [Google Scholar] [CrossRef]
- Woller, N.; Gürlevik, E. Viral Infection of Tumors Overcomes Resistance to PD-1-immunotherapy by Broadening Neoantigenome-directed T-cell Responses. Mol. Ther. 2015, 23, 1630–1640. [Google Scholar] [CrossRef]
- Wang, G.; Kang, X. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat. Commun. 2020, 11, 1395. [Google Scholar] [CrossRef]
- Jayawardena, N.; Poirier, J.T. Virus-Receptor Interactions and Virus Neutralization: Insights for Oncolytic Virus Development. Oncolytic Virother. 2020, 9, 1–15. [Google Scholar] [CrossRef]
- Jhawar, S.R.; Thandoni, A. Oncolytic Viruses—Natural and Genetically Engineered Cancer Immunotherapies. Front. Oncol. 2017, 7, 202. [Google Scholar] [CrossRef]
- Menotti, L.; Avitabile, E. Herpes Simplex Virus Oncolytic Immunovirotherapy: The Blossoming Branch of Multimodal Therapy. Int. J. Mol. Sci. 2020, 21, 8310. [Google Scholar] [CrossRef]
- Menotti, L.; Cerretani, A. Construction of a Fully Retargeted Herpes Simplex Virus 1 Recombinant Capable of Entering Cells Solely via Human Epidermal Growth Factor Receptor 2. J. Virol. 2008, 82, 10153–10161. [Google Scholar] [CrossRef]
- Menotti, L.; Avitabile, E. HSV as A Platform for the Generation of Retargeted, Armed, and Reporter-Expressing Oncolytic Viruses. Viruses 2018, 10, 352. [Google Scholar] [CrossRef]
- Goins, W.F.; Hall, B. Retargeting of herpes simplex virus (HSV) vectors. Curr. Opin. Virol. 2016, 21, 93–101. [Google Scholar] [CrossRef]
- Uchida, H.; Marzulli, M. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 2013, 21, 561–569. [Google Scholar] [CrossRef]
- Zhou, G.; Roizman, B. Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13Ralpha2 receptor of malignant glioma cells. J. Virol. 2005, 79, 5272–5277. [Google Scholar] [CrossRef]
- Campadelli-Fiume, G.; Petrovic, B. Retargeting Strategies for Oncolytic Herpes Simplex Viruses. Viruses 2016, 8, 63. [Google Scholar] [CrossRef]
- De Lucia, M.; Cotugno, G. Retargeted and Multi-cytokine-Armed Herpes Virus Is a Potent Cancer Endovaccine for Local and Systemic Anti-tumor Treatment. Mol. Ther. Oncolytics 2020, 19, 253–264. [Google Scholar] [CrossRef]
- Chang, K.; Pastan, I. Isolation and characterization of a monoclonal antibody, K1, reactive with ovarian cancers and normal mesothelium. Int. J. Cancer 1992, 50, 373–381. [Google Scholar] [CrossRef]
- Pastan, I.; Hassan, R. Discovery of mesothelin and exploiting it as a target for immunotherapy. Cancer Res. 2014, 74, 2907–2912. [Google Scholar] [CrossRef]
- Shaw, D.R.; Muminova, Z.E. Mesothelin: A new target for immunotherapy. Clin. Cancer Res. 2004, 10, 8751–8752. [Google Scholar] [CrossRef]
- Lamberts, L.E.; de Groot, D.J. Functional genomic mRNA profiling of a large cancer data base demonstrates mesothelin overexpression in a broad range of tumor types. Oncotarget 2015, 6, 28164–28172. [Google Scholar] [CrossRef][Green Version]
- Inoue, S.; Tsunoda, T. Diffuse mesothelin expression leads to worse prognosis through enhanced cellular proliferation in colorectal cancer. Oncol. Lett. 2020, 19, 1741–1750. [Google Scholar] [CrossRef]
- Chowdhury, P.S.; Viner, J.L. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proc. Natl. Acad. Sci. USA 1998, 95, 669–674. [Google Scholar] [CrossRef]
- Sapede, C.; Gauvrit, A. Aberrant splicing and protease involvement in mesothelin release from epithelioid mesothelioma cells. Cancer Sci. 2008, 99, 590–594. [Google Scholar] [CrossRef]
- Hellstrom, I.; Raycraft, J. Mesothelin variant 1 is released from tumor cells as a diagnostic marker. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Pastan, I.; Zhang, Y. Modulating mesothelin shedding to improve therapy. Oncotarget 2012, 3, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Li, P. Mesothelin as a biomarker for targeted therapy. Biomark. Res. 2019, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Thomas, A. Mesothelin Immunotherapy for Cancer: Ready for Prime Time? J. Clin. Oncol. 2016, 34, 4171–4179. [Google Scholar] [CrossRef]
- Chang, M.C.; Chen, Y.L. Mesothelin-specific cell-based vaccine generates antigen-specific immunity and potent antitumor effects by combining with IL-12 immunomodulator. Gene Ther. 2016, 23, 38–49. [Google Scholar] [CrossRef]
- Arabi, F.; Torabi-Rahvar, M. Antigenic targets of CAR T Cell Therapy. A retrospective view on clinical trials. Exp. Cell Res. 2018, 369, 1–10. [Google Scholar] [CrossRef]
- Kelly, R.J.; Sharon, E. Mesothelin-targeted agents in clinical trials and in preclinical development. Mol. Cancer Ther. 2012, 11, 517–525. [Google Scholar] [CrossRef]
- Morello, A.; Sadelain, M. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov. 2016, 6, 133–146. [Google Scholar] [CrossRef]
- Lanitis, E.; Poussin, M. Redirected antitumor activity of primary human lymphocytes transduced with a fully human anti-mesothelin chimeric receptor. Mol. Ther. 2012, 20, 633–643. [Google Scholar] [CrossRef]
- Alewine, C.; Ahmad, M. Phase I/II Study of the Mesothelin-targeted Immunotoxin LMB-100 with Nab-Paclitaxel for Patients with Advanced Pancreatic Adenocarcinoma. Clin. Cancer Res. 2020, 26, 828–836. [Google Scholar] [CrossRef]
- Hassan, R.; Ebel, W. Preclinical evaluation of MORAb-009, a chimeric antibody targeting tumor-associated mesothelin. Cancer Immun. 2007, 7, 20. [Google Scholar] [PubMed]
- Lindenberg, L.; Thomas, A. Safety and biodistribution of 111In-amatuximab in patients with mesothelin expressing cancers using single photon emission computed tomography-computed tomography (SPECT-CT) imaging. Oncotarget 2015, 6, 4496–4504. [Google Scholar] [CrossRef] [PubMed]
- Del Bano, J.; Florès-Florès, R. A Bispecific Antibody-Based Approach for Targeting Mesothelin in Triple Negative Breast Cancer. Front. Immunol. 2019, 10, 1593. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Niu, Z. Clinicopathological significance of mesothelin expression in invasive breast cancer. J. Int. Med. Res. 2012, 40, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L. Mesothelin promotes invasion and metastasis in breast cancer cells. J. Int. Med. Res. 2012, 40, 2109–2116. [Google Scholar] [CrossRef]
- Ungerechts, G.; Bossow, S. Moving oncolytic viruses into the clinic: Clinical-grade production, purification, and characterization of diverse oncolytic viruses. Mol. Ther. Methods Clin. Dev. 2016, 3, 16018. [Google Scholar] [CrossRef]
- Froechlich, G.; Caiazza, C. Integrity of the Antiviral STING-mediated DNA Sensing in Tumor Cells Is Required to Sustain the Immunotherapeutic Efficacy of Herpes Simplex Oncolytic Virus. Cancers 2020, 12, 3407. [Google Scholar] [CrossRef]
- Zhang, C.; Shang, G. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678, Erratum in: Nature 2008, 456, 274. [Google Scholar] [CrossRef]
- Ablasser, A.; Goldeck, M. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef]
- Ablasser, A.; Schmid-Burgk, J.L. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 2013, 503, 530–534. [Google Scholar] [CrossRef]
- Lam, E.; Stein, S. Adenovirus Detection by the cGAS/STING/TBK1 DNA Sensing Cascade. J. Virol. 2013, 88, 974–981. [Google Scholar] [CrossRef]
- Ni, G.; Ma, Z. cGAS and STING: At the intersection of DNA and RNA virus-sensing networks. PLoS Pathog. 2018, 14. [Google Scholar] [CrossRef]
- Franz, K.M.; Neidermyer, W.J. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E2058–E2067. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Phung, Y. New high affinity monoclonal antibodies recognize non-overlapping epitopes on mesothelin for monitoring and treating mesothelioma. Sci. Rep. 2015, 5, 9928. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, D. Modified CAR T cells targeting membrane-proximal epitope of mesothelin enhances the antitumor function against large solid tumor. Cell Death Dis. 2019, 10, 476, Erratum in: Cell Death Dis. 2020, 11, 235. [Google Scholar] [CrossRef]
- Leshem, Y.; King, E.M. SS1P Immunotoxin Induces Markers of Immunogenic Cell Death and Enhances the Effect of the CTLA-4 Blockade in AE17M Mouse Mesothelioma Tumors. Toxins 2018, 10, 470. [Google Scholar] [CrossRef]
- Hassan, R.; Bullock, S. Phase I study of SS1P, a recombinant anti-mesothelin immunotoxin given as a bolus I.V. infusion to patients with mesothelin-expressing mesothelioma, ovarian, and pancreatic cancers. Clin. Cancer Res. 2007, 13, 5144–5149. [Google Scholar] [CrossRef]
- Baldo, P.; Cecco, S. Amatuximab and novel agents targeting mesothelin for solid tumors. Onco. Targets Ther. 2017, 10, 5337–5353. [Google Scholar] [CrossRef]
- Chowdhury, P.S.; Pastan, I. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat. Biotechnol. 1999, 17, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Karasneh, G.A.; Shukla, D. Herpes simplex virus infects most cell types in vitro: Clues to its success. Virol. J. 2011, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Feng, M. A human single-domain antibody elicits potent antitumor activity by targeting an epitope in mesothelin close to the cancer cell surface. Mol. Cancer Ther. 2013, 12, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ayaru, L. Expansion of anti-mesothelin specific CD4+ and CD8+ T cell responses in patients with pancreatic carcinoma. PLoS ONE 2014, 9, e88133. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xie, L. Plasma Mesothelin as a Novel Diagnostic and Prognostic Biomarker in Colorectal Cancer. J. Cancer 2017, 8, 1355–1361. [Google Scholar] [CrossRef]
- Leoni, V.; Vannini, A. A fully-virulent retargeted oncolytic HSV armed with IL-12 elicits local immunity and vaccine therapy towards distant tumors. PLoS Pathog. 2018, 14, e1007209. [Google Scholar] [CrossRef]
- Hucl, T.; Brody, J.R. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007, 67, 9055–9065. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 420747. [Google Scholar] [CrossRef]
- Cheng, Z.; Dai, T. The interactions between cGAS-STING pathway and pathogens. Signal. Transduct. Target. Ther. 2020, 5, 91. [Google Scholar] [CrossRef]
- Bodda, C.; Reinert, L.S. HSV1 VP1-2 deubiquitinates STING to block type I interferon expression and promote brain infection. J. Exp. Med. 2020, 217, e20191422. [Google Scholar] [CrossRef]
- Georgana, I.; Sumner, R.P. Virulent Poxviruses Inhibit DNA Sensing by Preventing STING Activation. J. Virol. 2018, 92, e02145-17. [Google Scholar] [CrossRef]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Stempel, M.; Chan, B. Coevolution pays off: Herpesviruses have the license to escape the DNA sensing pathway. Med. Microbiol. Immunol. 2019, 208, 495–512. [Google Scholar] [CrossRef]
- Deschamps, T.; Kalamvoki, M. Evasion of the STING DNA-Sensing Pathway by VP11/12 of Herpes Simplex Virus 1. J. Virol. 2017, 91, e00535-17. [Google Scholar] [CrossRef]
- Huang, J.; You, H. Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J. Virol. 2018, 92, e00841-18. [Google Scholar] [CrossRef]
- Yuan, H.; You, J. Herpes Simplex Virus 1 UL36USP Antagonizes Type I Interferon-Mediated Antiviral Innate Immunity. J. Virol. 2018, 92, e01161-18. [Google Scholar] [CrossRef]
- Zheng, C. Evasion of Cytosolic DNA-Stimulated Innate Immune Responses by Herpes Simplex Virus 1. J. Virol. 2018, 92, e00099-17. [Google Scholar] [CrossRef]
- Pan, S.; Liu, X. Herpes Simplex Virus 1 γ134.5 Protein Inhibits STING Activation That Restricts Viral Replication. J. Virol. 2018, 92, e01015-18. [Google Scholar] [CrossRef]
- Margolis, S.R.; Wilson, S.C. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef]
- Kranzusch, P.J.; Wilson, S.C. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2′,3′ cGAMP Signaling. Mol. Cell 2015, 59, 891–903. [Google Scholar] [CrossRef]
- Ma, J.; Tang, W.K. Recognition of mesothelin by the therapeutic antibody MORAb-009: Structural and mechanistic insights. J. Biol. Chem. 2012, 287, 33123–33131. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Vitelli, A.; Folgori, A. Chimpanzee adenoviral vectors as vaccines—Challenges to move the technology into the fast lane. Expert Rev. Vaccines 2017, 16, 1241–1252. [Google Scholar] [CrossRef]
- Hall, B.L.; Leronni, D. Generation of an Oncolytic Herpes Simplex Viral Vector Completely Retargeted to the GDNF Receptor GFRα1 for Specific Infection of Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 8815. [Google Scholar] [CrossRef]
- Ramasamy, M.N.; Minassian, A.M. Oxford COVID Vaccine Trial Group. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2020, 396, 1979–1993. [Google Scholar] [CrossRef]
- Zhu, F.C.; Li, Y.H. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J. Oxford COVID Vaccine Trial Group. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478, Erratum in: Lancet 2020, 396, 466. [Google Scholar] [CrossRef]
- Sasso, E.; Vitale, M. Binding of carbonic anhydrase IX to 45S rDNA genes is prevented by exportin-1 in hypoxic cells. Biomed. Res. Int. 2015, 2015, 674920. [Google Scholar] [CrossRef]
- Rusciano, G.; Sasso, E. Revealing membrane alteration in cellsoverexpressing CA IX and EGFR by Surface-Enhanced Raman Scattering. Sci Rep. 2019, 9, 1832, Published Correction Appears in Sci Rep. 2019, 9, 9001. [Google Scholar] [CrossRef]
- Liu, X.; Chan, A. Multiple proteases are involved in mesothelin shedding by cancer cells. Commun. Biol. 2020, 1, 728. [Google Scholar] [CrossRef]
- Sasso, E.; Paciello, R. One-Step Recovery of scFv Clones from High-Throughput Sequencing-Based Screening of Phage Display Libraries Challenged to Cells Expressing Native Claudin-1. Biomed. Res. Int. 2015, 703213. [Google Scholar] [CrossRef] [PubMed]
- Paciello, R.; Urbanowicz, R.A. Novel human anti-claudin 1 mAbs inhibit hepatitis C virus infection and may synergize with anti-SRB1 mAb. J. Gen. Virol. 2016, 97, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Sasso, E.; Latino, D. A long non-coding SINEUP RNA boosts semi-stable production of fully human monoclonal antibodies in HEK293E cells. MAbs 2018, 10, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Takahashi, H. An NMR-based approach reveals the core structure of the functional domain of SINEUP lncRNAs. Nucleic Acids Res. 2020, 48. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J.M. The software that powers scientific illustration. Nature 2020, 582, 137–138. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence |
|---|---|
| CAS9_Fwd | 5′-gctctttgatgccctcttcg-3′ |
| CAS9_Rev | 5′-gctgaccctgacactgtttg-3′ |
| GFP_Fwd | 5′-cacgacttcttcaagtccgc-3′ |
| GFP_Rev | 5′-ggtgttctgctggtagtggt-3′ |
| hSting gRNA1_Fwd | 5′-caccggtgacccctgggacacggga-3′ |
| hSting gRNA1_Rev | 5′-aaactcccgtgtcccaggggtcacc-3′ |
| hSting gRNA2_Fwd | 5′-caccggctgggactgctgttaaac-3′ |
| hSting gRNA2_Rev | 5′-aaacgtttaacagcagtcccagcc-3′ |
| Step I gD_Fwd | 5′-agtgggcctccatggggtccgcggcaaatatgccttggcgacccctatttgtttatttttct-3′ |
| Step I gD_Rev | 5′-tggggggctggaacgggtccggtaggcccgcctggatgtgttatttgttaactgttaattgtc-3′ |
| Step II gD_SD1_Fwd | 5′-catggggtccgcggcaaatatgccttggcgcaggtgcagctggtgcagtc-3′ |
| Step II gD_SS/SS1_Fwd | 5′-agtgggcctccatggggtccgcggcaaatatgccttggcgcaggttcagctgcagcagtc-3′ |
| Step II gD_SD1/SS/SS1_Rev | 5′-tggggggctggaacgggtccggtaggcccgcctggatgtgagatcctccgcttccgctgc-3′ |
| Step I US1/US2_Fwd | 5′-cgtttgtcccagcgtcttaatggcgggaagacccctatttgtttattttt-3′ |
| Step I US1/US2_Rev | 5′-ccatgtacgcgtggtctgtttctctccgccttatttgttaactgttaatt-3′ |
| Step II US1/US2_mIL12_Fwd | 5′-cgtttgtcccagcgtcttaatggcgggaagacattgattattgactagtt-3′ |
| Step II US1/US2_mIL12_Rev | 5′-ccatgtacgcgtggtctgtttctctccgccgccatagagcccaccgcatc-3′ |
| Taqman DNApol_Fwd | 5′-catcaccgacccggagagggac-3′ |
| Taqman DNApol_Rev | 5′-gggccaggcgcttgttggtgta-3′ |
| Taqman Probe | FAM-ccgccgaactgagcagacacccgcgc-Tamra |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Froechlich, G.; Gentile, C.; Infante, L.; Caiazza, C.; Pagano, P.; Scatigna, S.; Cotugno, G.; D’Alise, A.M.; Lahm, A.; Scarselli, E.; et al. Generation of a Novel Mesothelin-Targeted Oncolytic Herpes Virus and Implemented Strategies for Manufacturing. Int. J. Mol. Sci. 2021, 22, 477. https://doi.org/10.3390/ijms22020477
Froechlich G, Gentile C, Infante L, Caiazza C, Pagano P, Scatigna S, Cotugno G, D’Alise AM, Lahm A, Scarselli E, et al. Generation of a Novel Mesothelin-Targeted Oncolytic Herpes Virus and Implemented Strategies for Manufacturing. International Journal of Molecular Sciences. 2021; 22(2):477. https://doi.org/10.3390/ijms22020477
Chicago/Turabian StyleFroechlich, Guendalina, Chiara Gentile, Luigia Infante, Carmen Caiazza, Pasqualina Pagano, Sarah Scatigna, Gabriella Cotugno, Anna Morena D’Alise, Armin Lahm, Elisa Scarselli, and et al. 2021. "Generation of a Novel Mesothelin-Targeted Oncolytic Herpes Virus and Implemented Strategies for Manufacturing" International Journal of Molecular Sciences 22, no. 2: 477. https://doi.org/10.3390/ijms22020477
APA StyleFroechlich, G., Gentile, C., Infante, L., Caiazza, C., Pagano, P., Scatigna, S., Cotugno, G., D’Alise, A. M., Lahm, A., Scarselli, E., Nicosia, A., Mallardo, M., Sasso, E., & Zambrano, N. (2021). Generation of a Novel Mesothelin-Targeted Oncolytic Herpes Virus and Implemented Strategies for Manufacturing. International Journal of Molecular Sciences, 22(2), 477. https://doi.org/10.3390/ijms22020477







