Prediction Models for Agonists and Antagonists of Molecular Initiation Events for Toxicity Pathways Using an Improved Deep-Learning-Based Quantitative Structure–Activity Relationship System
Abstract
:1. Introduction
2. Results
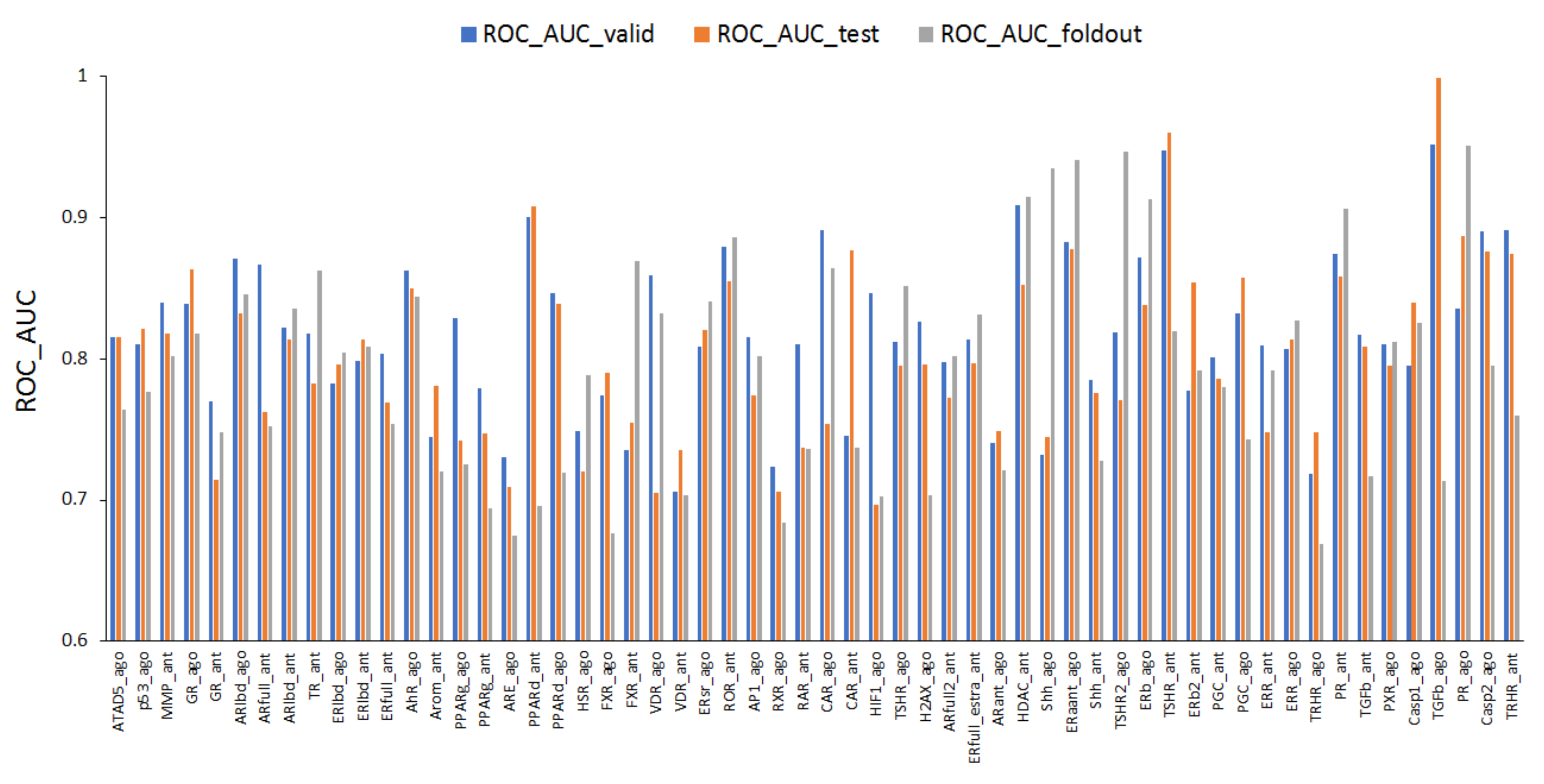
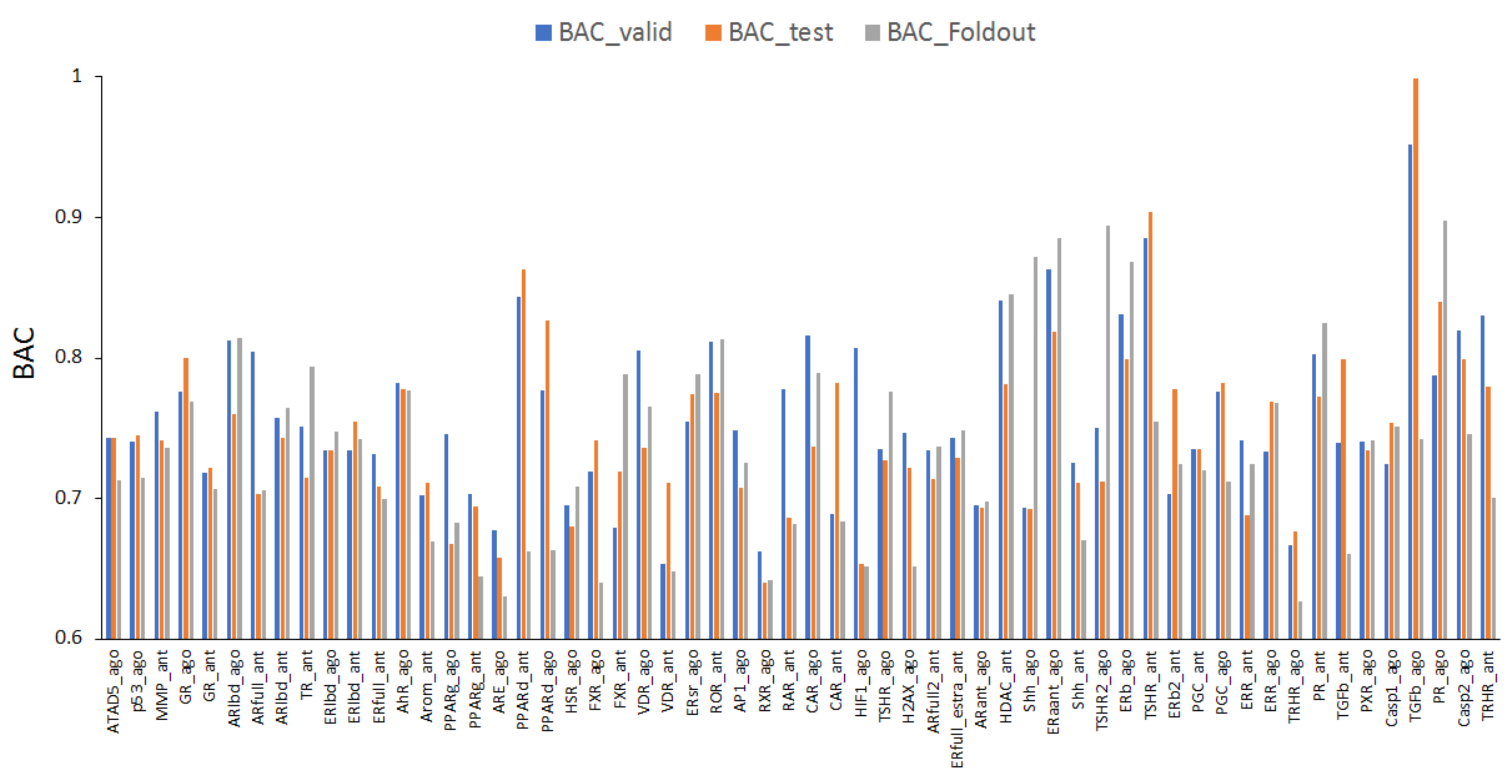
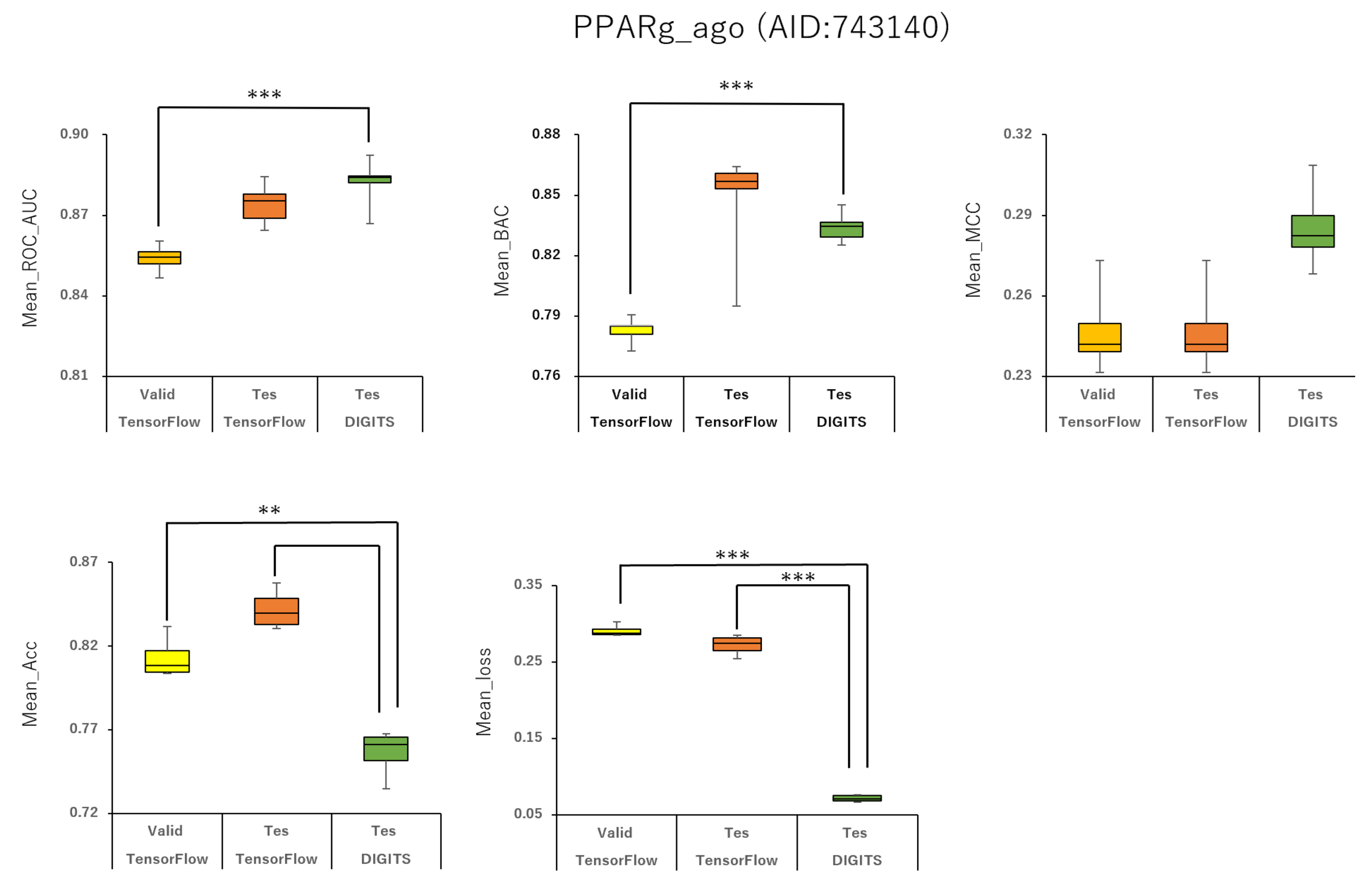
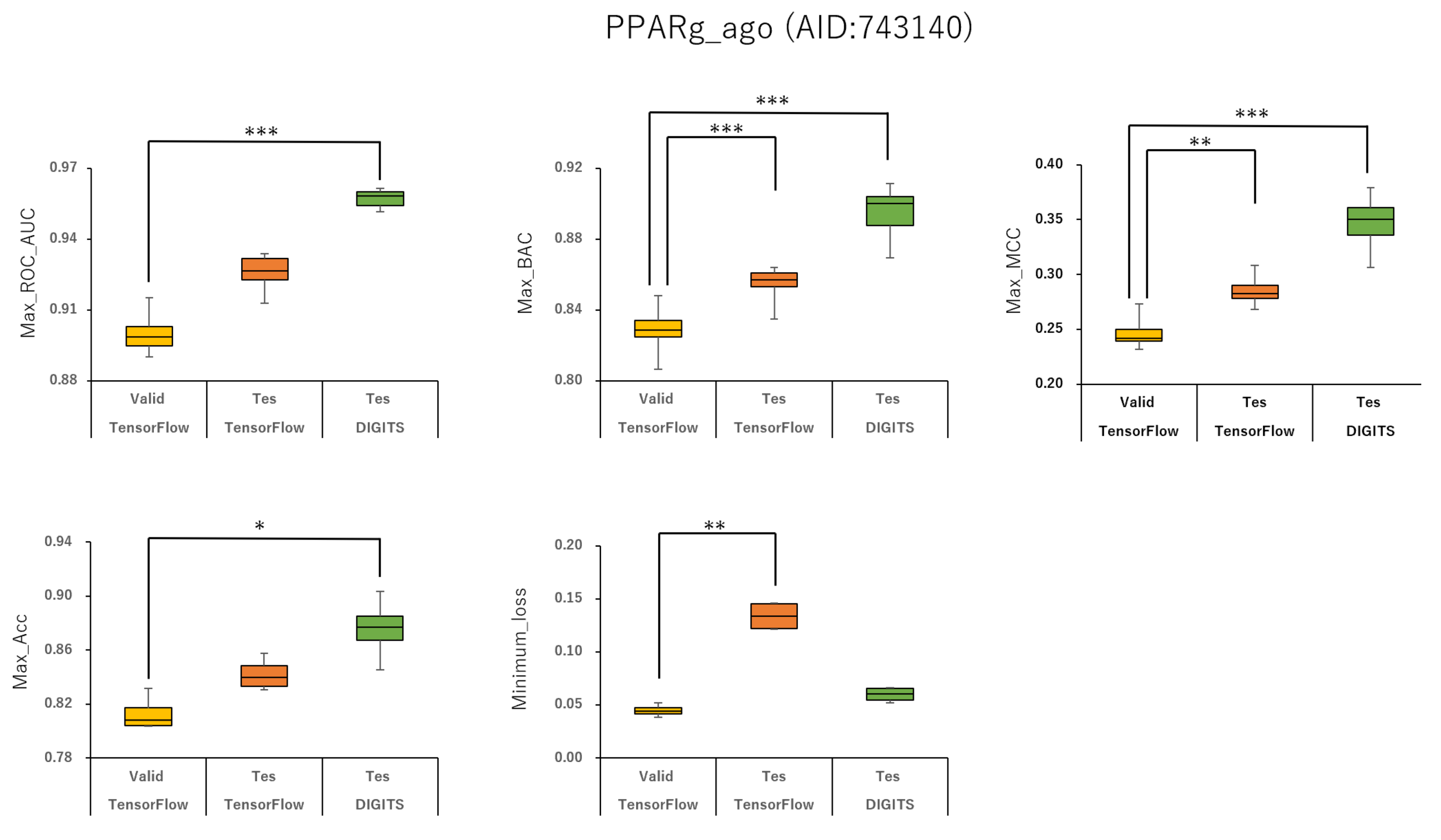
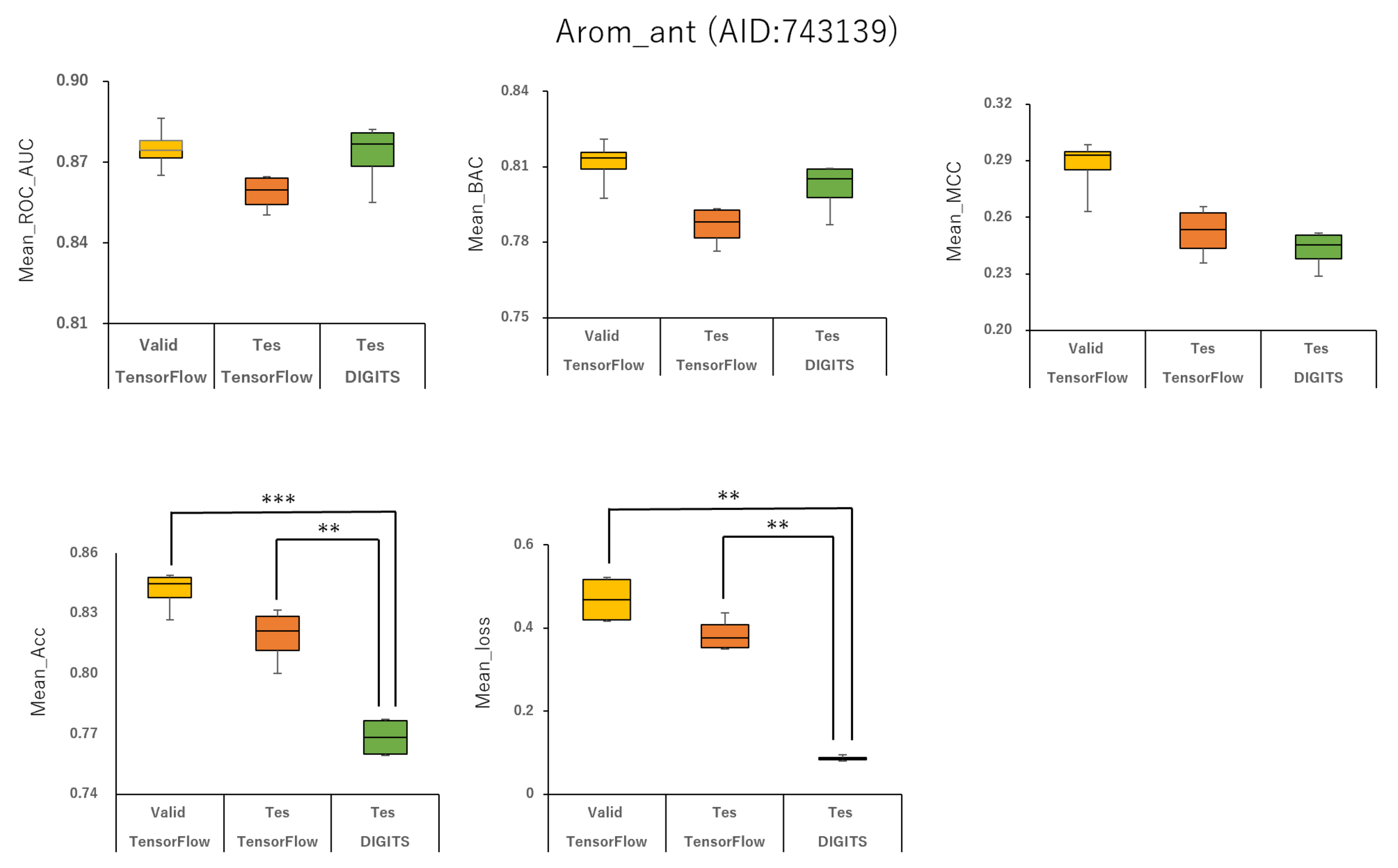
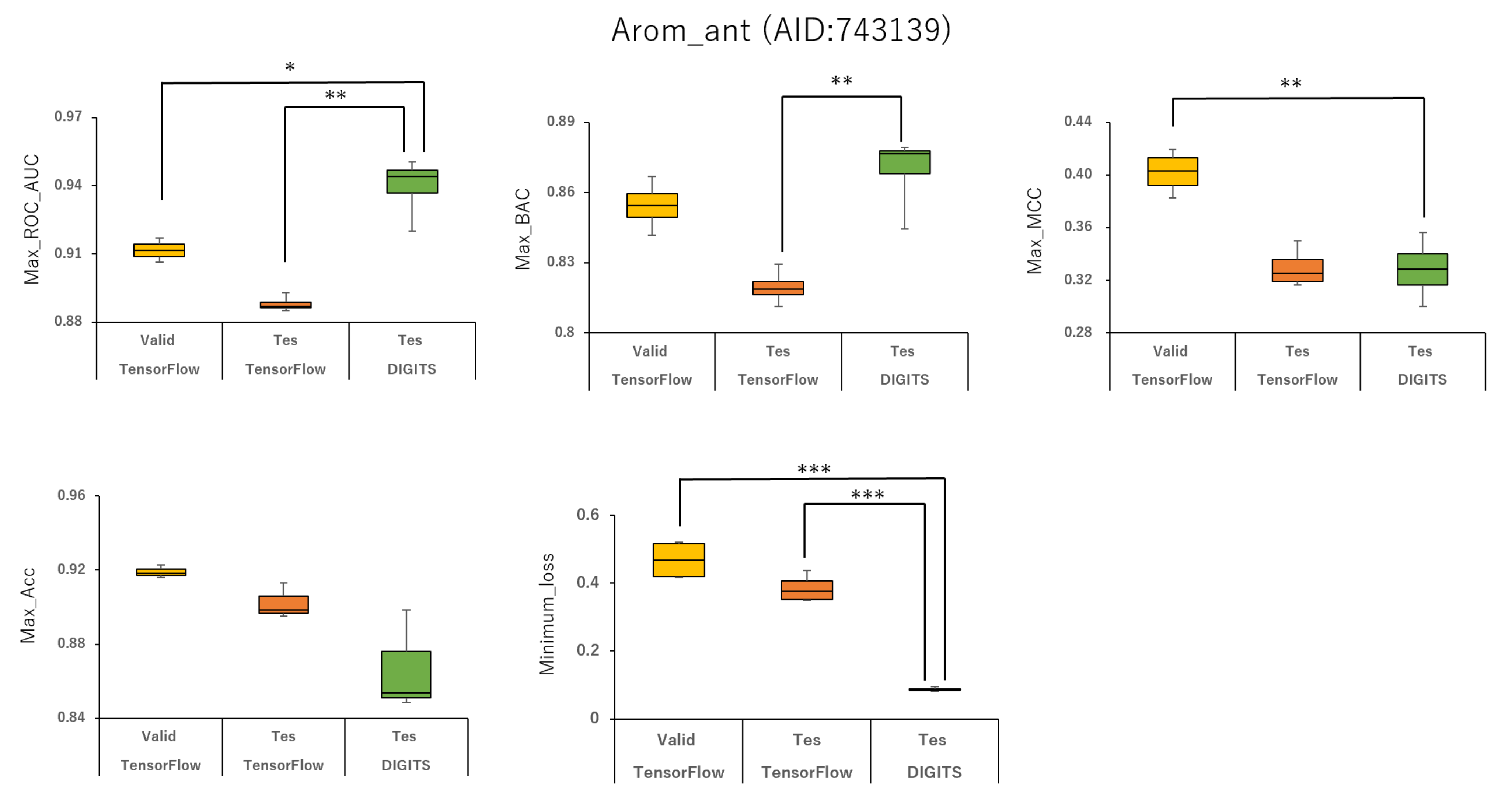
2.1. Prediction Models for 59 MIEs
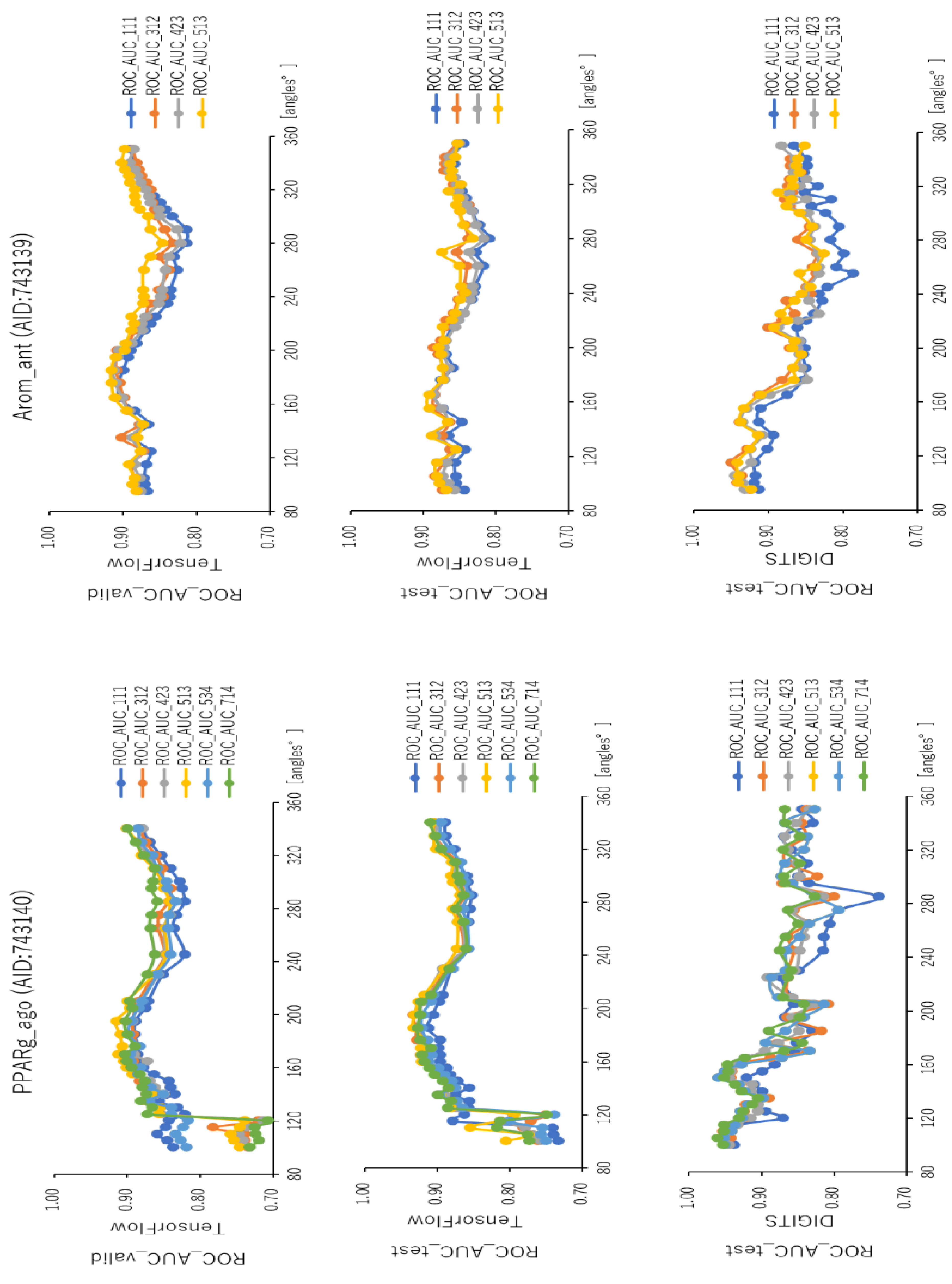
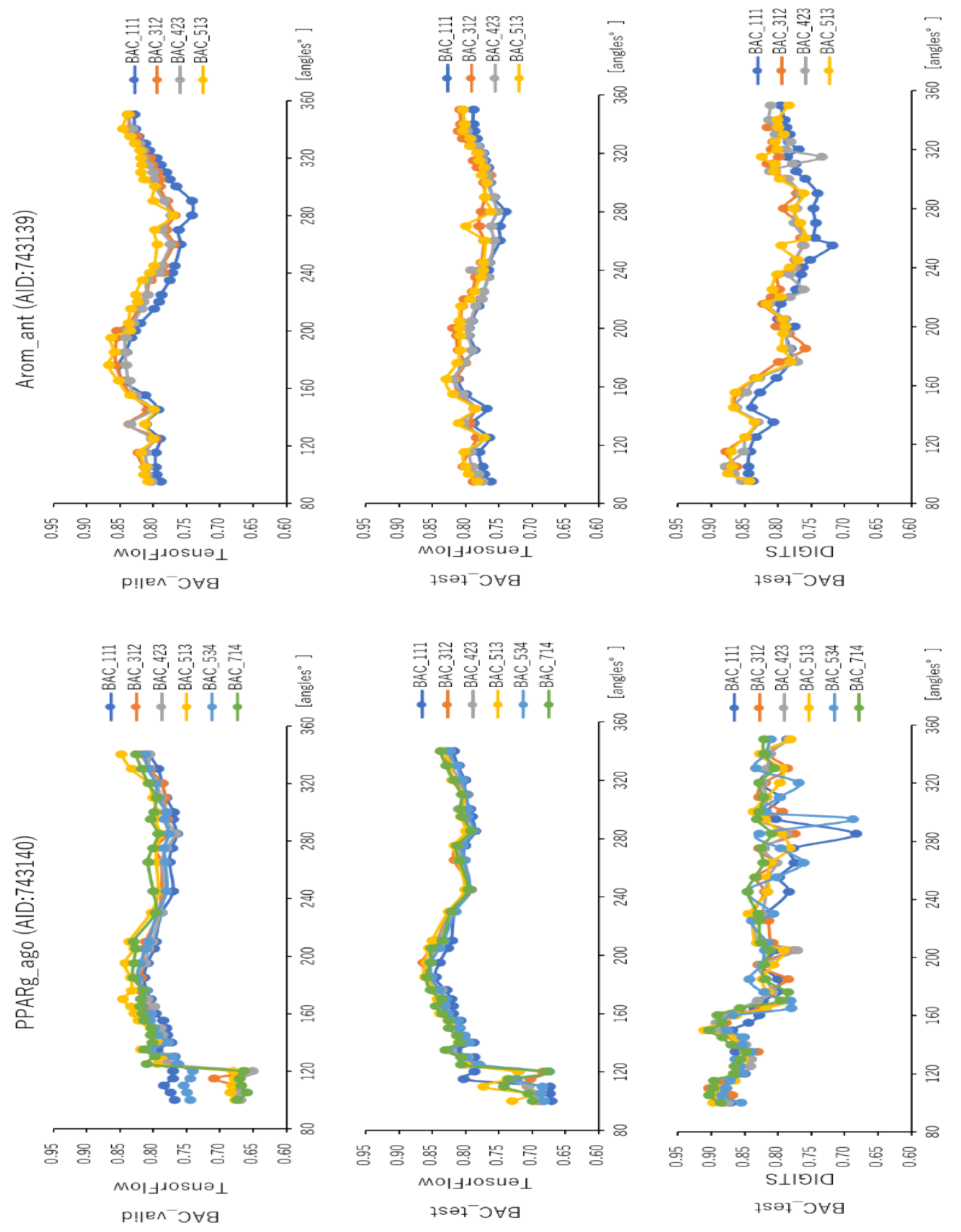
2.2. Angles and Data Split in DeepSnap-DL
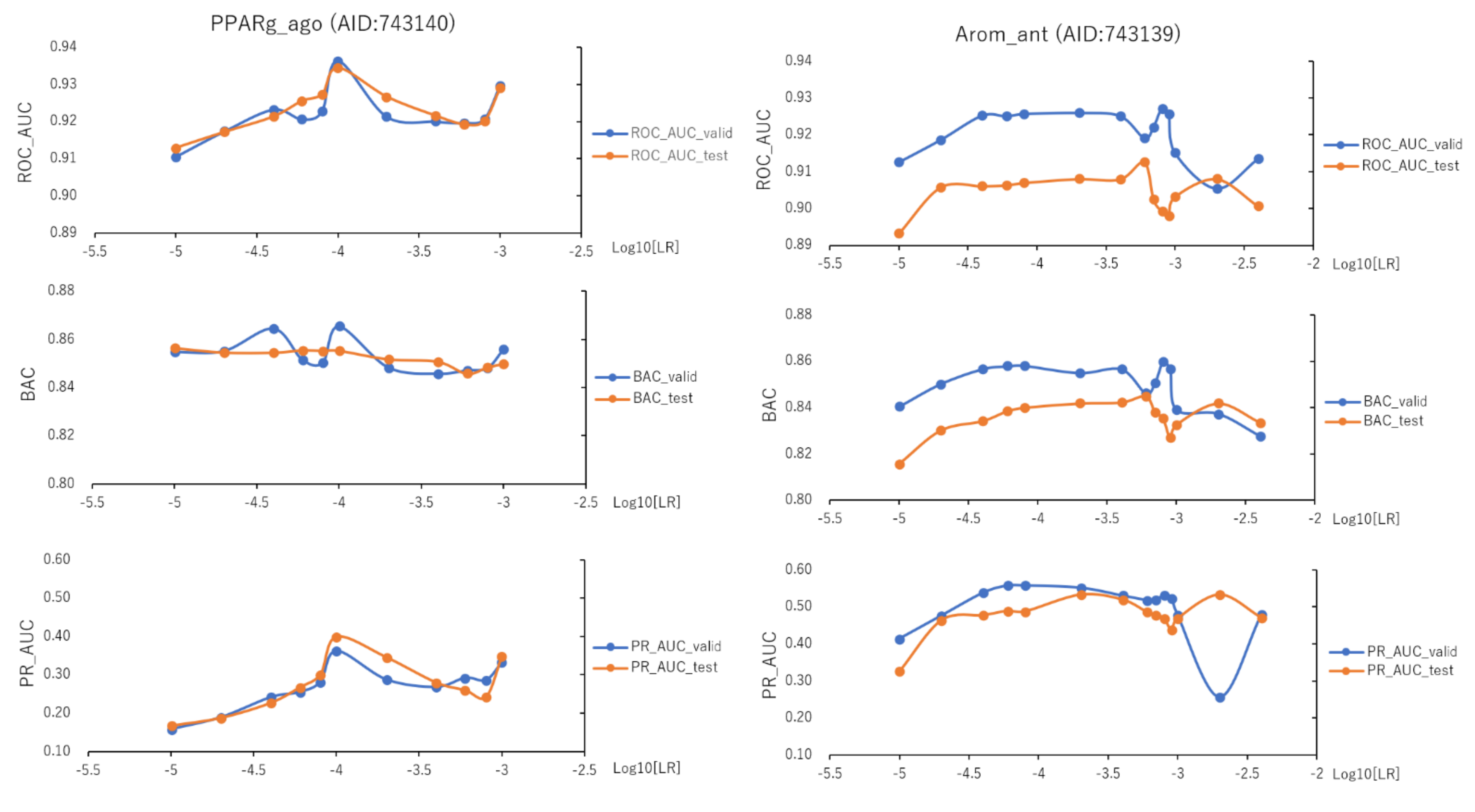
2.3. Learning Rate and Batch Size in DeepSnap-DL
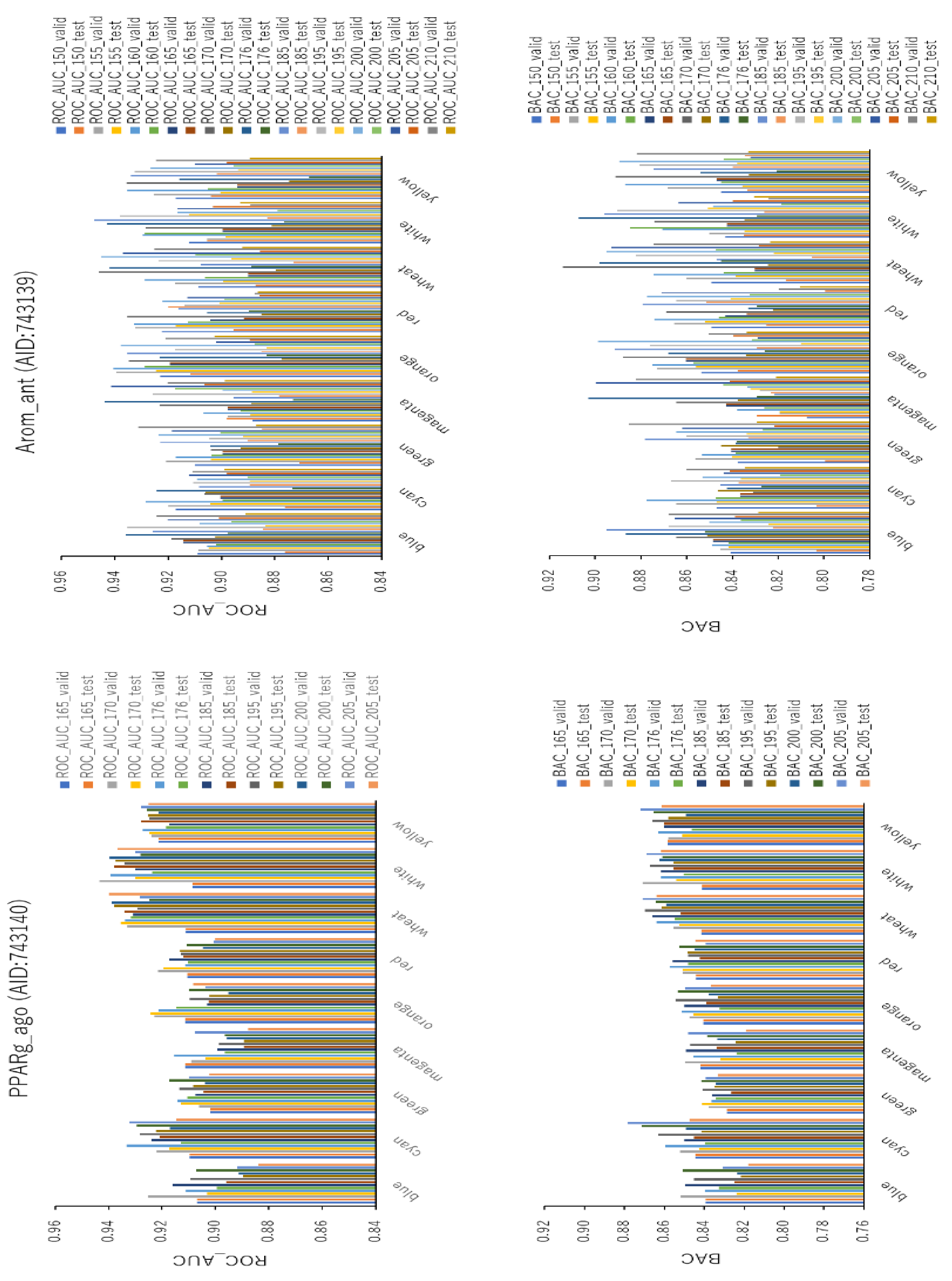
2.4. Background Colors in Images Produced by DeepSnap-DL
3. Discussion
4. Materials and Methods
4.1. Data
4.2. DeepSnap
4.3. Evaluation of Model Quality
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, W.; Chen, J.; Wang, Z.; Xie, H.; Hong, H. Deep learning for predicting toxicity of chemicals: A mini review. J. Environ. Sci. Health. C Environ. Carcinog. Ecotoxicol. Rev. 2018, 36, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Ring, C.; Sipes, N.S.; Hsieh, J.H.; Carberry, C.; Koval, L.E.; Klaren, W.D.; Harris, M.A.; Auerbach, S.S.; Rager, J.E. Predictive modeling of biological responses in the rat liver using in vitro Tox21 bioactivity: Benefits from high-throughput toxicokinetics. Comput. Toxicol. 2021, 18, 100166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Norinder, U.; Svensson, F. Deep learning-based conformal prediction of toxicity. J. Chem. Inf. Model. 2021, 61, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kim, H.; Choi, J. In Silico molecular docking and in vivo validation with caenorhabditis elegans to discover molecular initiating events in adverse outcome pathway framework: Case study on endocrine-disrupting chemicals with estrogen and androgen receptors. Int. J. Mol. Sci. 2019, 20, 1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngan, D.K.; Ye, L.; Wu, L.; Xia, M.; Rossoshek, A.; Simeonov, A.; Huang, R. bioactivity signatures of drugs vs. environmental chemicals revealed by tox21 high-throughput screening assays. Front. Big Data 2019, 2, 50. [Google Scholar] [CrossRef] [Green Version]
- Richard, A.M.; Huang, R.; Waidyanatha, S.; Shinn, P.; Collins, B.J.; Thillainadarajah, I.; Grulke, C.M.; Williams, A.J.; Lougee, R.R.; Judson, R.S.; et al. The Tox21 10K compound library: Collaborative chemistry advancing toxicology. Chem. Res. Toxicol. 2021, 34, 189–216. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Wang, S.; Yang, Y.; He, J.; Li, C.; Zhao, Y.H.; Martyniuk, C.J. Identification of active and inactive agonists/antagonists of estrogen receptor based on Tox21 10K compound library: Binomial analysis and structure alert. Ecotoxicol. Environ. Saf. 2021, 214, 112114. [Google Scholar] [CrossRef]
- Dawson, D.E.; Ingle, B.L.; Phillips, K.A.; Nichols, J.W.; Wambaugh, J.F.; Tornero-Velez, R. Designing QSARs for parameters of high-throughput toxicokinetic models using open-source descriptors. Environ. Sci. Technol. 2021, 55, 6505–6517. [Google Scholar] [CrossRef]
- Li, S.; Zhao, J.; Huang, R.; Travers, J.; Klumpp-Thomas, C.; Yu, W.; MacKerell, A.D., Jr.; Sakamuru, S.; Ooka, M.; Xue, F.; et al. Profiling the Tox21 chemical collection for acetylcholinesterase inhibition. Environ. Health Perspect. 2021, 129, 47008. [Google Scholar] [CrossRef]
- Huang, R.; Xia, M.; Nguyen, D.-T.; Zhao, T.; Sakamuru, S.; Zhao, J.; Shahane, S.A.; Rossoshek, A.; Simeonov, A. Tox21Challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Front. Environ. Sci. 2016, 3, 85. [Google Scholar] [CrossRef] [Green Version]
- Picard, S.; Chapdelaine, C.; Cappi, C.; Gardes, L.; Jenn, E.; Lefèvre, B.; Soumarmon, T. Ensuring dataset quality for machine learning certification. arXiv 2020, arXiv:2011.01799. Available online: https://arxiv.org/abs/2011.01799 (accessed on 3 November 2020).
- Renggli, C.; Rimanic, L.; Gürel, N.M.; Karlaš, B.; Wu, W.; Zhang, C. A Data quality-driven view of MLOps. arXiv 2021, arXiv:2102.07750. Available online: https://arxiv.org/abs/2102.07750 (accessed on 15 February 2021).
- Basile, A.O.; Yahi, A.; Tatonetti, N.P. Artificial intelligence for drug toxicity and safety. Trends Pharmacol. Sci. 2019, 40, 624–635. [Google Scholar] [CrossRef]
- Huang, D.Z.; Baber, J.C.; Bahmanyar, S.S. The challenges of generalizability in artificial intelligence for ADME/Tox endpoint and activity prediction. Expert Opin. Drug Discov. 2021, in press. [Google Scholar] [CrossRef]
- Karim, A.; Riahi, V.; Mishra, A.; Newton, M.A.H.; Dehzangi, A.; Balle, T.; Sattar, A. Quantitative toxicity prediction via meta ensembling of multitask deep learning models. ACS Omega 2021, 6, 12306–12317. [Google Scholar] [CrossRef]
- Gini, G.; Zanoli, F.; Gamba, A.; Raitano, G.; Benfenati, E. Could deep learning in neural networks improve the QSAR models? SAR QSAR Environ. Res. 2019, 30, 617–642. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Z.; Su, Y.; Zhao, Q.; Li, X.; Ouyang, D. Deep learning for in vitro prediction of pharmaceutical formulations. Acta Pharm. Sin. B 2019, 9, 177–185. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, T.; Liu, C. Prediction of apoptosis protein subcellular localization via heterogeneous features and hierarchical extreme learning machine. SAR QSAR Environ. Res. 2019, 30, 209–228. [Google Scholar] [CrossRef]
- Wenzel, J.; Matter, H.; Schmidt, F. predictive multitask deep neural network models for adme-tox properties: Learning from large data sets. J. Chem. Inf. Model. 2019, 59, 1253–1268. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Yang, Z.; Feng, H.; Tripathi, S.; Dehmer, M. an introductory review of deep learning for prediction models with big data. Front. Artif. Intell. 2020, 3, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Chen, P.; Gu, P.; Wang, B. A deep learning-based chemical system for qsar prediction. IEEE J. Biomed. Health Inform. 2020, 24, 3020–3028. [Google Scholar] [CrossRef] [PubMed]
- Irwin, B.W.J.; Levell, J.R.; Whitehead, T.M.; Segall, M.D.; Conduit, G.J. practical applications of deep learning to impute heterogeneous drug discovery Data. J. Chem. Inf. Model. 2020, 60, 2848–2857. [Google Scholar] [CrossRef]
- Montanari, F.; Kuhnke, L.; Ter Laak, A.; Clevert, D.A. modeling physico-chemical admet endpoints with multitask graph convolutional networks. Molecules 2019, 25, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khemchandani, Y.; O’Hagan, S.; Samanta, S.; Swainston, N.; Roberts, T.J.; Bollegala, D.; Kell, D.B. DeepGraphMolGen, a multi-objective, computational strategy for generating molecules with desirable properties: A graph convolution and reinforcement learning approach. J. Cheminform. 2020, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.; Gini, G. QSAR modeling without descriptors using graph convolutional neural networks: The case of mutagenicity prediction. Mol. Divers. 2021, in press. [Google Scholar] [CrossRef]
- Zhang, S.; Tong, H.; Xu, J.; Maciejewski, R. Graph convolutional networks: A comprehensive review. Comput. Soc. Netw. 2019, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A comprehensive survey on graph neural networks. arXiv 2019, arXiv:1901.00596. Available online: https://arxiv.org/abs/1901.00596 (accessed on 4 December 2019). [CrossRef] [Green Version]
- Wu, Z.; Jiang, D.; Hsieh, C.Y.; Chen, G.; Liao, B.; Cao, D.; Hou, T. Hyperbolic relational graph convolution networks plus: A simple but highly efficient QSAR-modeling method. Brief. Bioinform. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Dong, Y.; Wang, K.; Lee, W.S.; Hooi, B.; Xu, H.; Feng, J. Understanding and Resolving Performance Degradation in Graph Convolutional Networks. arXiv 2020, arXiv:2006.07107. Available online: https://arxiv.org/abs/2006.07107 (accessed on 13 September 2021).
- Chen, M.; Wei, Z.; Huang, Z.; Ding, B.; Li, Y. Simple and Deep Graph Convolutional Networks. arXiv 2020, arXiv:2007.02133. Available online: https://arxiv.org/abs/2007.02133 (accessed on 4 July 2020).
- Das, R.; Boote, B.; Bhattacharya, S.; Maulik, U. Multipath graph convolutional neural networks. arXiv 2021, arXiv:2105.01510. Available online: https://arxiv.org/abs/2105.01510 (accessed on 4 May 2021).
- Chen, H.; Xu, Y.; Huang, F.; Deng, Z.; Huang, W.; Wang, S.; He, P.; Li, Z. Label-aware graph convolutional networks. arXiv 2020, arXiv:1907.04707. Available online: https://arxiv.org/abs/1907.04707 (accessed on 5 September 2020).
- Wang, H.; Leskovec, J. Unifying graph convolutional neural networks and label propagation. arXiv 2020, arXiv:2002.06755. Available online: https://arxiv.org/abs/2002.06755 (accessed on 17 February 2020).
- Bellei, C.; Alattas, H.; Kaaniche, N. Label-GCN: An effective method for adding label propagation to graph convolutional networks. arXiv 2021, arXiv:2104.02153. Available online: https://arxiv.org/abs/2104.02153 (accessed on 5 April 2021).
- Lu, Z.; Pu, H.; Wang, F.; Hu, Z.; Wang, L. The expressive power of neural networks: A view from the width. arXiv 2017, arXiv:1709.02540. Available online: https://arxiv.org/abs/1709.02540 (accessed on 1 November 2017).
- Gupta, T.K.; Raza, K. Optimizing deep feedforward neural network architecture: A tabu search based approach. Neural. Process. Lett. 2020, 51, 2855–2870. [Google Scholar] [CrossRef]
- Uesawa, Y. Quantitative structure-activity relationship analysis using deep learning based on a novel molecular image input technique. Bioorg. Med. Chem. Lett. 2018, 28, 3400–3403. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Uesawa, Y. molecular image-based prediction models of nuclear receptor agonists and antagonists using the deepsnap-deep learning approach with the Tox21 10K library. Molecules 2020, 25, 2764. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Hosaka, T.; Ogaito, A.; Yoshinari, K.; Uesawa, Y. Prediction model of aryl hydrocarbon receptor activation by a novel QSAR approach, deepsnap-deep learning. deep learning. Molecules 2020, 25, 1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaka, Y.; Uesawa, Y. Prediction model with high-performance constitutive androstane receptor (car) using deepsnap-deep learning approach from the Tox21 10K compound library. Int. J. Mol. Sci. 2019, 20, 4855. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaka, Y.; Uesawa, Y. DeepSnap-deep learning approach predicts progesterone receptor antagonist activity with high performance. Front. Bioeng. Biotechnol. 2020, 7, 485. [Google Scholar] [CrossRef] [Green Version]
- Dou, L.; Yang, F.; Xu, L.; Zou, Q. A comprehensive review of the imbalance classification of protein post-translational modifications. Brief. Bioinform. 2021, 22, bbab089. [Google Scholar] [CrossRef] [PubMed]
- Popoola, S.I.; Adebisi, B.; Ande, R.; Hammoudeh, M.; Anoh, K.; Atayero, A.A. smote-drnn: A deep learning algorithm for botnet detection in the internet-of-things networks. Sensors 2021, 21, 2985. [Google Scholar] [CrossRef] [PubMed]
- Haldekar, M.; Ganesan, A.; Oates, T. Identifying spatial relations in images using convolutional neural networks. arXiv 2017, arXiv:1706.04215. Available online: https://arxiv.org/abs/1706.04215 (accessed on 13 June 2017).
- Marcos, D.; Volpi, M.; Tuia, D. Learning rotation invariant convolutional filters for texture classification. arXiv 2016, arXiv:1604.06720. Available online: https://arxiv.org/abs/1604.06720 (accessed on 21 September 2016).
- Marcos, D.; Volpi, M.; Komodakis, N.; Tuia, D. Rotation equivariant vector field networks. arXiv 2017, arXiv:1612.09346. Available online: https://arxiv.org/abs/1612.09346 (accessed on 25 August 2017).
- Marcos, D.; Kellenberger, B.; Lobry, S.; Tuia, D. Scale equivariance in CNNs with vector fields. arXiv 2018, arXiv:1807.11783. Available online: https://arxiv.org/abs/1807.11783 (accessed on 25 August 2017).
- Chidester, B.; Do, M.N.; Ma, J. Rotation Equivariance and Invariance in Convolutional Neural Networks. arXiv 2018, arXiv:1805.12301. Available online: https://arxiv.org/abs/1805.12301 (accessed on 31 May 2018).
- Chidester, B.; Zhou, T.; Do, M.N.; Ma, J. Rotation equivariant and invariant neural networks for microscopy image analysis. Bioinformatics 2019, 35, i530–i537. [Google Scholar] [CrossRef] [PubMed]
- Philbrick, K.A.; Yoshida, K.; Inoue, D.; Akkus, Z.; Kline, T.L.; Weston, A.D.; Korfiatis, P.; Takahashi, N.; Erickson, B.J. What Does Deep Learning See? Insights From a Classifier Trained to Predict Contrast Enhancement Phase From CT Images. Am. J. Roentgenol. 2018, 211, 1184–1193. [Google Scholar] [CrossRef]
- Pubche. Available online: https://pubchem.ncbi.nlm.nih.gov (accessed on 16 September 2004).
- PyMOLWiki. Available online: https://pymolwiki.org/index.php/Color_Values (accessed on 22 January 2011).
- Artificial Intelligence Research. Computing Deviation of Area Under the Precision-recall CURVE (washington.edu). Available online: http://aiweb.cs.washington.edu/ai/mln/auc.html (accessed on 1993).
- Siblini, W.; Frry, J.; He-Guelton, L.; Obl, F.; Wang, Y.-Q. Master your Metrics with Calibration. arXiv 2020, arXiv:1909.02827. Available online: https://arxiv.org/abs/1909.02827 (accessed on 28 April 2020).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaka, Y.; Totoki, S.; Handa, K.; Shiota, T.; Kurosaki, K.; Uesawa, Y. Prediction Models for Agonists and Antagonists of Molecular Initiation Events for Toxicity Pathways Using an Improved Deep-Learning-Based Quantitative Structure–Activity Relationship System. Int. J. Mol. Sci. 2021, 22, 10821. https://doi.org/10.3390/ijms221910821
Matsuzaka Y, Totoki S, Handa K, Shiota T, Kurosaki K, Uesawa Y. Prediction Models for Agonists and Antagonists of Molecular Initiation Events for Toxicity Pathways Using an Improved Deep-Learning-Based Quantitative Structure–Activity Relationship System. International Journal of Molecular Sciences. 2021; 22(19):10821. https://doi.org/10.3390/ijms221910821
Chicago/Turabian StyleMatsuzaka, Yasunari, Shin Totoki, Kentaro Handa, Tetsuyoshi Shiota, Kota Kurosaki, and Yoshihiro Uesawa. 2021. "Prediction Models for Agonists and Antagonists of Molecular Initiation Events for Toxicity Pathways Using an Improved Deep-Learning-Based Quantitative Structure–Activity Relationship System" International Journal of Molecular Sciences 22, no. 19: 10821. https://doi.org/10.3390/ijms221910821
APA StyleMatsuzaka, Y., Totoki, S., Handa, K., Shiota, T., Kurosaki, K., & Uesawa, Y. (2021). Prediction Models for Agonists and Antagonists of Molecular Initiation Events for Toxicity Pathways Using an Improved Deep-Learning-Based Quantitative Structure–Activity Relationship System. International Journal of Molecular Sciences, 22(19), 10821. https://doi.org/10.3390/ijms221910821







