A Possible Mechanism of Graphene Oxide to Enhance Thermostability of D-Psicose 3-Epimerase Revealed by Molecular Dynamics Simulations
Abstract
1. Introduction
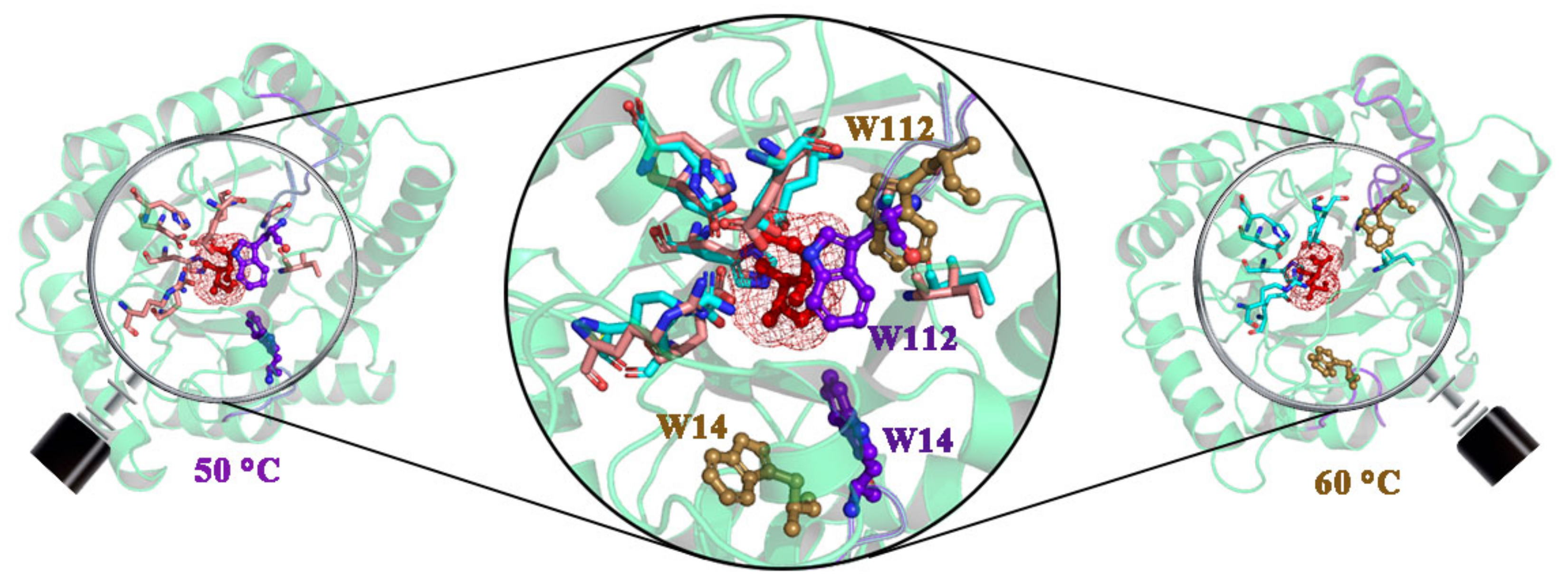
2. Results
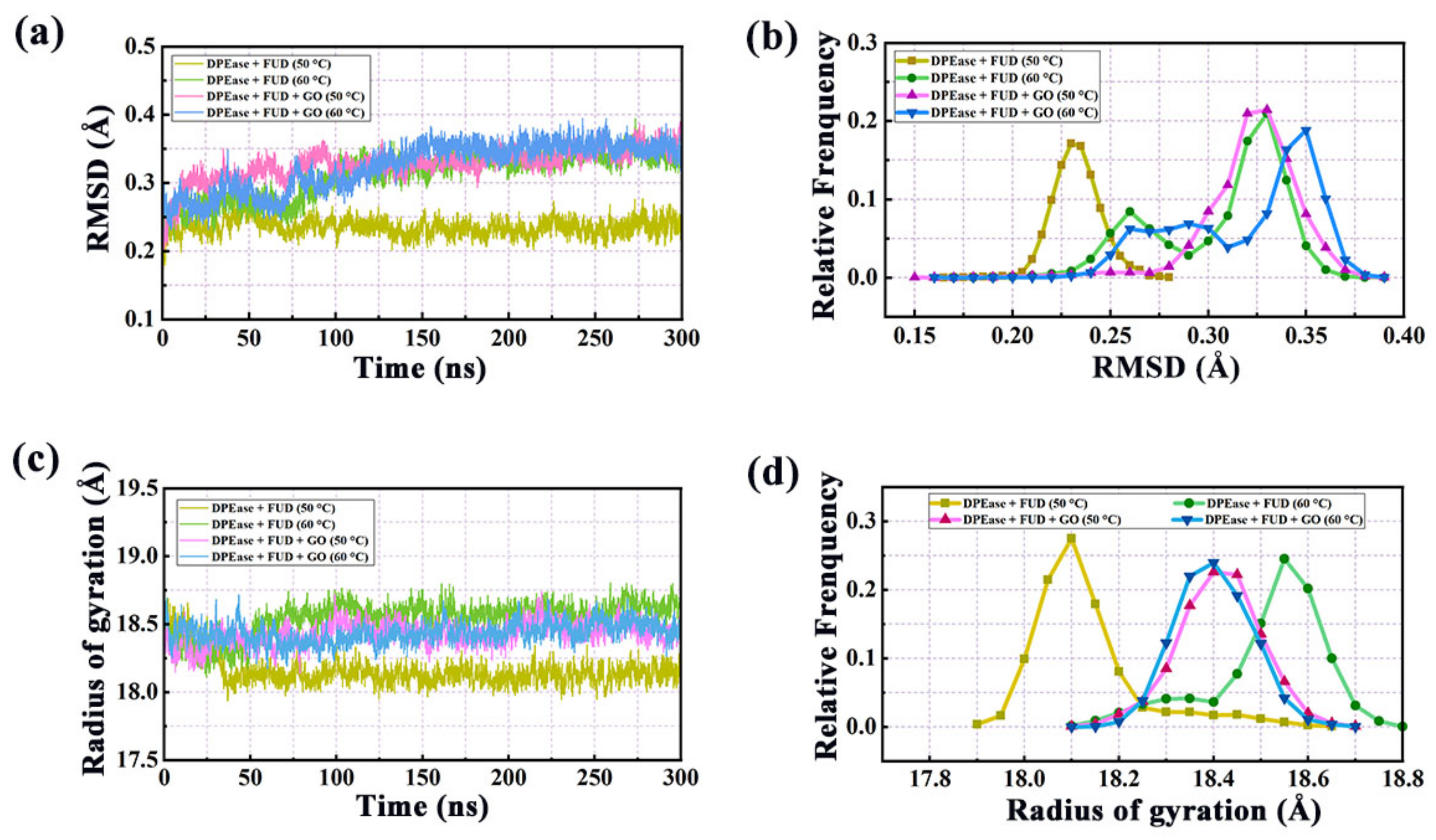
2.1. Structural Stability and Flexibility Analysis
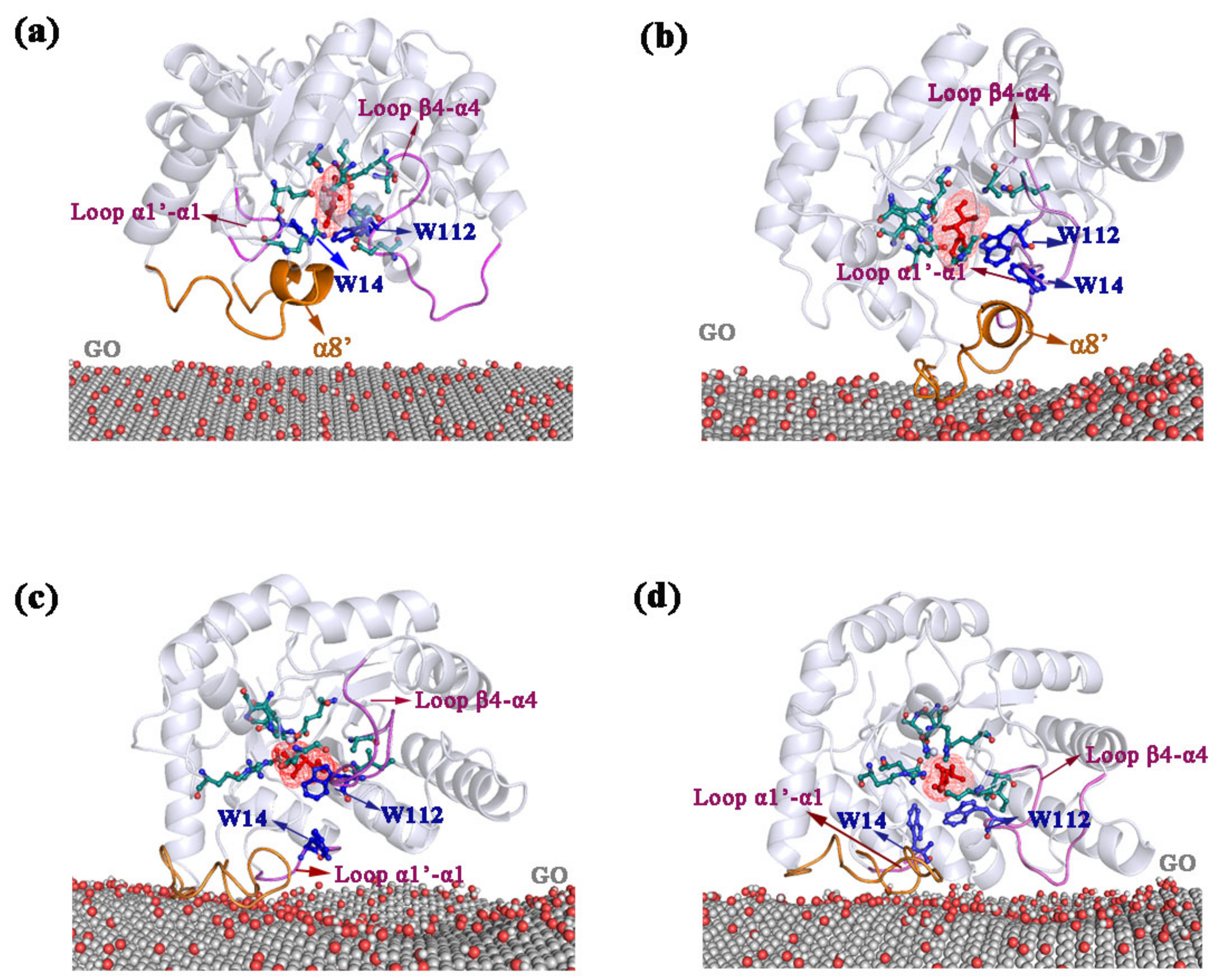
2.2. Adsorption of DPEase onto GO
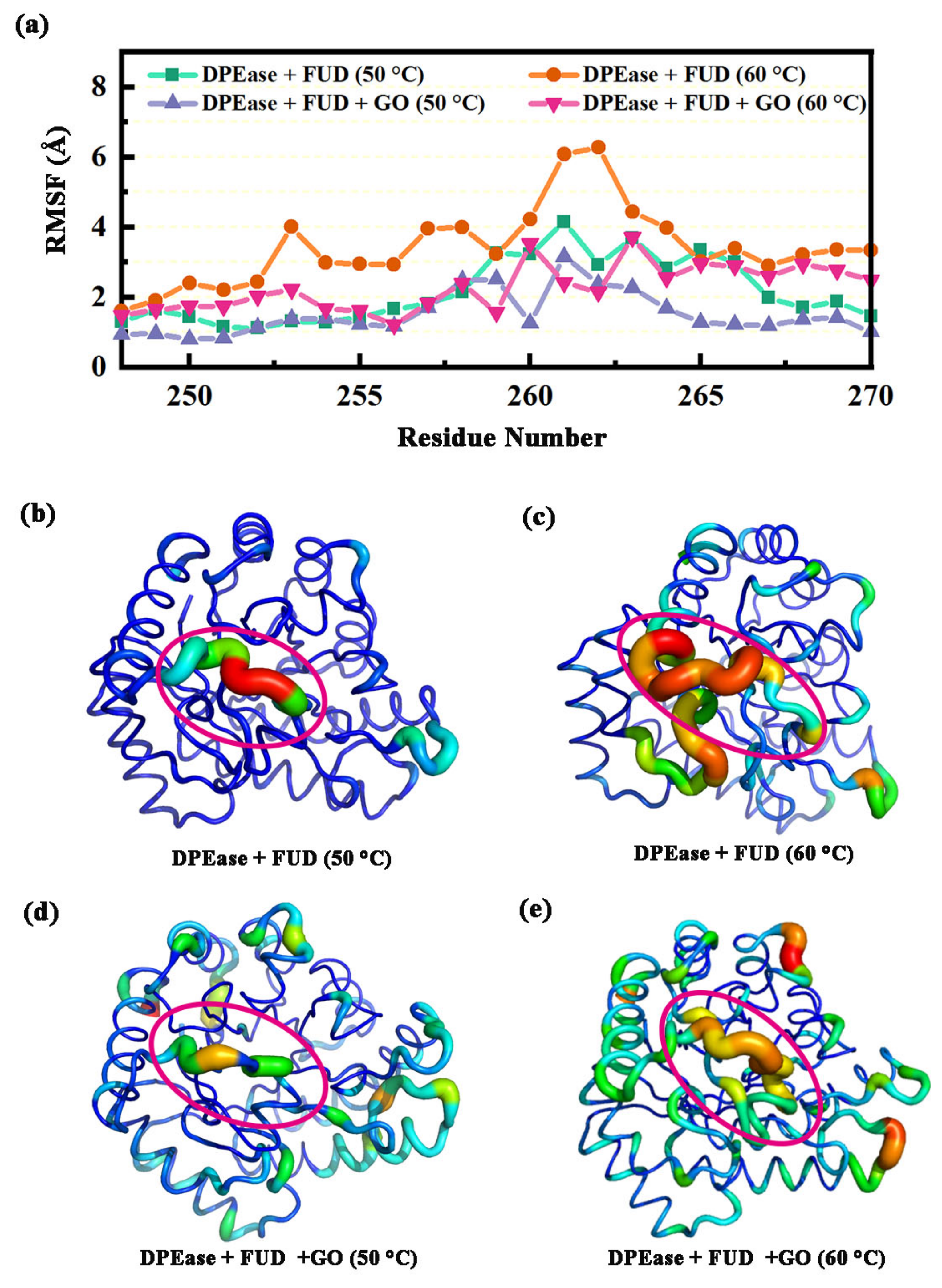
2.3. Dynamics of Regions Adsorbed by GO
2.3.1. Dynamic Analysis of Interaction between Loop α1′-α1 and α8′
2.3.2. Dynamic Analysis of Interaction between Loop β4-α4 and β5-α5
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brena, B.; González-Pombo, P.; Batista-Viera, F. Immobilization of enzymes: A literature survey. Methods Mol. Biol. 2013, 1051, 15–31. [Google Scholar] [PubMed]
- Hermanová, S.; Zarevúcká, M.; Bouša, D.; Pumera, M.; Sofer, Z. Graphene oxide immobilized enzymes show high thermal and solvent stability. Nanoscale 2015, 7, 5852–5858. [Google Scholar] [CrossRef] [PubMed]
- Nawani, N.; Singh, R.; Kaur, J. Immobilization and stability studies of a lipase from thermophilic Bacillus sp: The effect of process parameters on immobilization of enzyme. Electron. J. Biotechnol. 2006, 9, 559–565. [Google Scholar] [CrossRef]
- Cherry, J.R.; Fidantsef, A.L. Directed evolution of industrial enzymes: An update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef]
- Su, R.; Zhang, W.; Zhu, M.; Xu, S.; Ming, Y.; Li, D. Alkaline protease immobilized on graphene oxide: Highly efficient catalysts for the proteolysis of waste-activated sludge. Pol. J. Environ. Stud. 2013, 22, 885–891. [Google Scholar]
- Kao, K.C.; Lin, T.S.; Mou, C.Y. Enhanced activity and stability of lysozyme by immobilization in the matching nanochannels of mesoporous silica nanoparticles. J. Phys. Chem. C 2014, 118, 6734–6743. [Google Scholar] [CrossRef]
- Li, C.; Huang, M.; Gu, Z.; Hong, Y.; Cheng, L.; Li, Z. Nanosilica sol leads to further increase in polyethylene glycol (PEG) 1000-enhanced thermostability of β-cyclodextrin glycosyltransferase from Bacillus circulans. J. Agric. Food Chem. 2014, 62, 2919–2924. [Google Scholar] [CrossRef]
- Ju, L.; Wang, J.; Gavalas, V.G.; Atwood, D.; Ba Chas, L.G. Alumina−pepsin hybrid nanoparticles with orientation-specific enzyme coupling. Nano Lett. 2015, 3, 55–58. [Google Scholar]
- Li, D.; He, Q.; Cui, Y.; Duan, L.; Li, J. Immobilization of glucose oxidase onto gold nanoparticles with enhanced thermostability. Biochem. Biophys. Res. Commun. 2007, 355, 488–493. [Google Scholar] [CrossRef]
- Xi, L.; Li, S.; Yao, X.; Wei, Y.; Li, J.; Liu, H.; Wu, X. In silico study combining docking and QSAR methods on a series of matrix metalloproteinase 13 inhibitors. Arch. Pharm. 2014, 347, 825–833. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Zhang, Y.; Jin, X.; Liu, H.; Yao, X. Exploring the influence of carbon nanoparticles on the formation of β-sheet-rich oligomers of IAPP₂₂₋₂₈ peptide by molecular dynamics simulation. PLoS ONE 2013, 8, e65579. [Google Scholar]
- Sun, X.; Feng, Z.; Hou, T.; Li, Y. Mechanism of graphene oxide as an enzyme inhibitor from molecular dynamics simulations. ACS Appl. Mater. Interfaces 2014, 6, 7153–7163. [Google Scholar] [CrossRef]
- Ge, C.; Du, J.; Zhao, L.; Wang, L.; Liu, Y.; Li, D.; Yang, Y.; Zhou, R.; Zhao, Y.; Chai, Z.; et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Nati. Acad. Sci. USA 2011, 108, 16968–16973. [Google Scholar] [CrossRef]
- Shih, C.; Lin, S.; Sharma, R.; Strano, M.; Blankschtein, D. Understanding the pH-dependent behavior of graphene oxide aqueous solutions: A comparative experimental and molecular dynamics simulation study. Langmuir 2012, 28, 235–241. [Google Scholar] [CrossRef]
- Lu, N.; Li, Z.; Yang, J. Structure of Graphene Oxide: Thermodynamics versus Kinetics. J. Phys. Chem. C 2011, 115, 11991–11995. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, F.; Yang, H.; Huang, X.; Liu, H.; Zhang, J.; Guo, S. Graphene oxide as a matrix for enzyme immobilization. Langmuir 2010, 26, 6083–6085. [Google Scholar] [CrossRef] [PubMed]
- Whitby, R.L.; Gun’ko, V.M.; Korobeinyk, A.; Busquets, R.; Cundy, A.B.; László, K.; Skubiszewska-Zięba, J.; Leboda, R.; Tombácz, E.; Toth, I.Y.; et al. Driving forces of conformational changes in single-layer graphene oxide. ACS Nano 2012, 6, 3967–3973. [Google Scholar] [CrossRef] [PubMed]
- Luan, V.H.; Tien, H.N.; Le, T.H.; Hien, N.; Hur, S.H. Synthesis of a highly conductive and large surface area graphene oxide hydrogel and its use in a supercapacitor. J. Mater. Chem. A 2013, 1, 208–211. [Google Scholar] [CrossRef]
- Zhu, P.; Sumpter, B.G.; Meunier, V. Electronic, thermal, and structural properties of graphene oxide frameworks. J. Phys. Chem. B 2013, 117, 8276–8281. [Google Scholar] [CrossRef]
- Zhuang, Z.; Peng, Q.; Zhang, B.; Li, Y. Controllable synthesis of Cu2S nanocrystals and their assembly into a superlattice. J. Am. Chem. Soc. 2008, 130, 10482–10483. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, C.; Guo, S.; Zhang, J. Interactions of graphene and graphene oxide with proteins and peptides. Nanotechnol. Rev. 2013, 2, 27–45. [Google Scholar] [CrossRef]
- Jin, L.; Yang, K.; Yao, K.; Zhang, S.; Tao, H.; Lee, S.T.; Liu, Z.; Peng, R. Functionalized graphene oxide in enzyme engineering: A selective modulator for enzyme activity and thermostability. ACS Nano 2012, 6, 4864–4875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zheng, B.; Zhang, J.; Huang, X.; Liu, H.; Guo, S.; Zhang, J. Horseradish peroxidase immobilized on graphene oxide: Physical properties and applications in phenolic compound removal. J. Phys. Chem. C 2010, 114, 8469–8473. [Google Scholar] [CrossRef]
- Pavlidis, I.V.; Vorhaben, T.; Tsoufis, T.; Rudolf, P.; Bornscheuer, U.T.; Gournis, D.; Stamatis, H. Development of effective nanobiocatalytic systems through the immobilization of hydrolases on functionalized carbon-based nanomaterials. Bioresour. Technol. 2012, 115, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Fan, F.; Wang, Y.; Feng, W.; Ji, P. Enzyme immobilization on carboxyl-functionalized graphene oxide for catalysis in organic solvent. Ind. Eng. Chem. Res. 2013, 52, 6343–6348. [Google Scholar] [CrossRef]
- Su, R.; Shi, P.; Zhu, M.; Hong, F.; Li, D. Studies on the properties of graphene oxide-alkaline protease bio-composites. Bioresour. Technol. 2012, 115, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Peng, C.; Li, D.; Zhuo, L.; He, S.; Wang, L.; Huang, Q.; Xu, Q.H.; Fan, C. Metal ion-modulated graphene-DNAzyme interactions: Design of a nanoprobe for fluorescent detection of lead(II) ions with high sensitivity, selectivity and tunable dynamic range. Chem. Commun. 2011, 47, 6278–6280. [Google Scholar] [CrossRef]
- Wang, W.; Donini, O.; Reyes, C.M.; Kollman, P.A. Biomolecular simulations: Recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 211–243. [Google Scholar] [CrossRef]
- Dedania, S.R.; Patel, M.J.; Patel, D.M.; Akhani, R.C.; Patel, D.H. Immobilization on graphene oxide improves the thermal stability and bioconversion efficiency of D-psicose 3-epimerase for rare sugar production. Enzyme Microb. Technol. 2017, 107, 49–56. [Google Scholar] [CrossRef]
- Zhang, L.; Mu, W.; Jiang, B.; Zhang, T. Characterization of D-tagatose-3-epimerase from Rhodobacter sphaeroides that converts D-fructose into D-psicose. Biotechnol. Lett. 2009, 31, 857–862. [Google Scholar] [CrossRef]
- Zhang, W.; Fang, D.; Xing, Q.; Zhou, L.; Jiang, B.; Mu, W. Characterization of a novel metal-dependent D-psicose 3-epimerase from Clostridium scindens 35704. PLoS ONE 2013, 8, e62987. [Google Scholar] [CrossRef]
- Oh, D.K.; Kim, N.H.; Kim, H.J.; Park, C.S.; Kim, S.W.; Ko, M.; Park, B.W.; Min, J. d-Psicose production from d-fructose using an isolated strain, Sinorhizobium sp. World J. Microbiol. Biotechnol. 2007, 23, 559–563. [Google Scholar] [CrossRef]
- Tseng, C.W.; Liao, C.Y.; Sun, Y.; Peng, C.C.; Tzen, J.T.; Guo, R.T.; Liu, J.R. Immobilization of Clostridium cellulolyticum D-psicose 3-epimerase on artificial oil bodies. J. Agric. Food Chem. 2014, 62, 6771–6776. [Google Scholar] [CrossRef]
- Itoh, H.; Okaya, H.; Khan, A.R.; Tajima, S.; Hayakawa, S.; Izumori, K. Purification and Characterization of D-Tagatose 3-Epimerase from Pseudomonas sp. ST-24. Biosci. Biotechnol. Biochem. 1994, 58, 2168–2171. [Google Scholar] [CrossRef]
- Kim, K.; Kim, H.-J.; Oh, D.; Cha, S.; Rhee, S. Crystal structure of D-psicose 3-epimerase from Agrobacterium tumefaciens and its complex with true substrate D-fructose: A pivotal role of metal in catalysis, an active site for the non-phosphorylated substrate, and its conformational changes. J. Mol. Biol. 2006, 361, 920–931. [Google Scholar] [CrossRef]
- Thompson, A.B.; Calhoun, A.K.; Smagghe, B.J.; Stevens, M.D.; Wotkowicz, M.T.; Hatziioannou, V.M.; Bamdad, C. A gold nanoparticle platform for protein-protein interactions and drug discovery. ACS Appl. Mater. Interfaces 2011, 3, 2979–2987. [Google Scholar] [CrossRef]
- Sahoo, D.; Bhattacharya, P.; Patra, H.K.; Mandal, P.; Chakravorti, S. Gold nanoparticle induced conformational changes in heme protein. J. Nanopart. Res. 2011, 13, 6755–6760. [Google Scholar] [CrossRef]
- Zuo, G.; Huang, Q.; Wei, G.; Zhou, R.; Fang, H. Plugging into proteins: Poisoning protein function by a hydrophobic nanoparticle. ACS Nano 2010, 4, 7508–7514. [Google Scholar] [CrossRef]
- Katoch, J.; Kim, S.N.; Kuang, Z.; Farmer, B.L.; Naik, R.R.; Tatulian, S.A.; Ishigami, M. Structure of a peptide adsorbed on graphene and graphite. Nano Lett. 2012, 12, 2342–2346. [Google Scholar] [CrossRef]
- Zuo, G.; Zhou, X.; Huang, Q.; Fang, H.; Zhou, R. Adsorption of villin headpiece onto graphene, carbon nanotube, and C60: Effect of contacting surface curvatures on binding affinity. J. Phys. Chem. C 2011, 115, 23323–23328. [Google Scholar] [CrossRef]
- Kim, H.J.; Hyun, E.K.; Kim, Y.S.; Lee, Y.J.; Oh, D.K. Characterization of an Agrobacterium tumefaciens D-psicose 3-epimerase that converts D-fructose to D-psicose. Appl. Environ. Microbiol. 2006, 72, 981–985. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Jiang, B.; Mu, W. Biochemical characterization of a D-psicose 3-epimerase from Treponema primitia ZAS-1 and its application on enzymatic production of D-psicose. J. Sci. Food Agric. 2016, 96, 49–56. [Google Scholar] [CrossRef]
- Zhang, W.; Fang, D.; Zhang, T.; Zhou, L.; Jiang, B.; Mu, W. Characterization of a metal-dependent D-psicose 3-epimerase from a novel strain, Desmospora sp. 8437. J. Agric. Food Chem. 2013, 61, 11468–11476. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Men, Y.; Bai, W.; Li, X.; Zhang, L.; Sun, Y.; Ma, Y. Overexpression of D-psicose 3-epimerase from Ruminococcus sp. in Escherichia coli and its potential application in D-psicose production. Biotechnol. Lett. 2012, 34, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.C.; Kim, H.J.; Oh, D.K. A stable immobilized d -psicose 3-epimerase for the production of d -psicose in the presence of borate. Process Biochem. 2009, 44, 822–828. [Google Scholar] [CrossRef]
- Srivastava, G.; Singh, K.; Talat, M.; Srivastava, O.N.; Kayastha, A.M. Functionalized graphene sheets as immobilization matrix for fenugreek β-amylase: Enzyme kinetics and stability studies. PLoS ONE 2014, 9, e113408. [Google Scholar] [CrossRef][Green Version]
- Chang, M.Y.; Juang, R.S. Use of chitosan–clay composite as immobilization support for improved activity and stability of β-glucosidase. Biochem. Eng. J. 2007, 35, 93–98. [Google Scholar] [CrossRef]
- Calvaresi, M.; Hoefinger, S.; Zerbetto, F. Probing the structure of lysozyme-carbon-nanotube hybrids with molecular dynamics. Chemistry 2012, 18, 4308–4313. [Google Scholar] [CrossRef] [PubMed]
- Vaitheeswaran, S.; Garcia, A.E. Protein stability at a carbon nanotube interface. J. Chem. Phys. 2011, 134, 125101. [Google Scholar] [CrossRef]
- Sun, X.; Feng, Z.; Zhang, L.; Hou, T.; Li, Y. The selective interaction between silica nanoparticles and enzymes from molecular dynamics simulations. PLoS ONE 2014, 9, e107696. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Chou, S.S.; Dravid, V.P. Graphene oxide as an enzyme inhibitor: Modulation of activity of α-chymotrypsin. J. Am. Chem. Soc. 2011, 133, 17524–17527. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.A.; Chou, M.Y. Oxidation functional groups on graphene: Structural and electronic properties. Phys. Rev. 2010, 82, 125401–125403. [Google Scholar] [CrossRef]
- Bogetti, A.T.; Mostofian, B.; Dickson, A.; Pratt, A.J.; Saglam, A.S.; Harrison, P.O.; Adelman, J.L.; Dudek, M.; Torrillo, P.A.; DeGrave, A.J.; et al. A suite of tutorials for the WESTPA rare-events sampling software [Article v1.0]. Living J. Comput. Mol. Sci. 2019, 1, 10607. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Domański, J.; Stansfeld, P.J.; Sansom, M.S.; Beckstein, O. Lipidbook: A public repository for force-field parameters used in membrane simulations. J. Membr. Biol. 2010, 236, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Tepper, H.L.; Voth, G.A. Flexible simple point-charge water model with improved liquid-state properties. J. Chem. Phys. 2006, 124, 024503. [Google Scholar] [CrossRef] [PubMed]
- Eslami, H.; Mojahedi, F.; Moghadasi, J. Molecular dynamics simulation with weak coupling to heat and material baths. J. Chem. Phys. 2010, 133, 084105. [Google Scholar] [CrossRef]
- Martonák, R.; Laio, A.; Parrinello, M. Predicting crystal structures: The Parrinello-Rahman method revisited. Phys. Rev. Lett. 2003, 90, 075503. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]





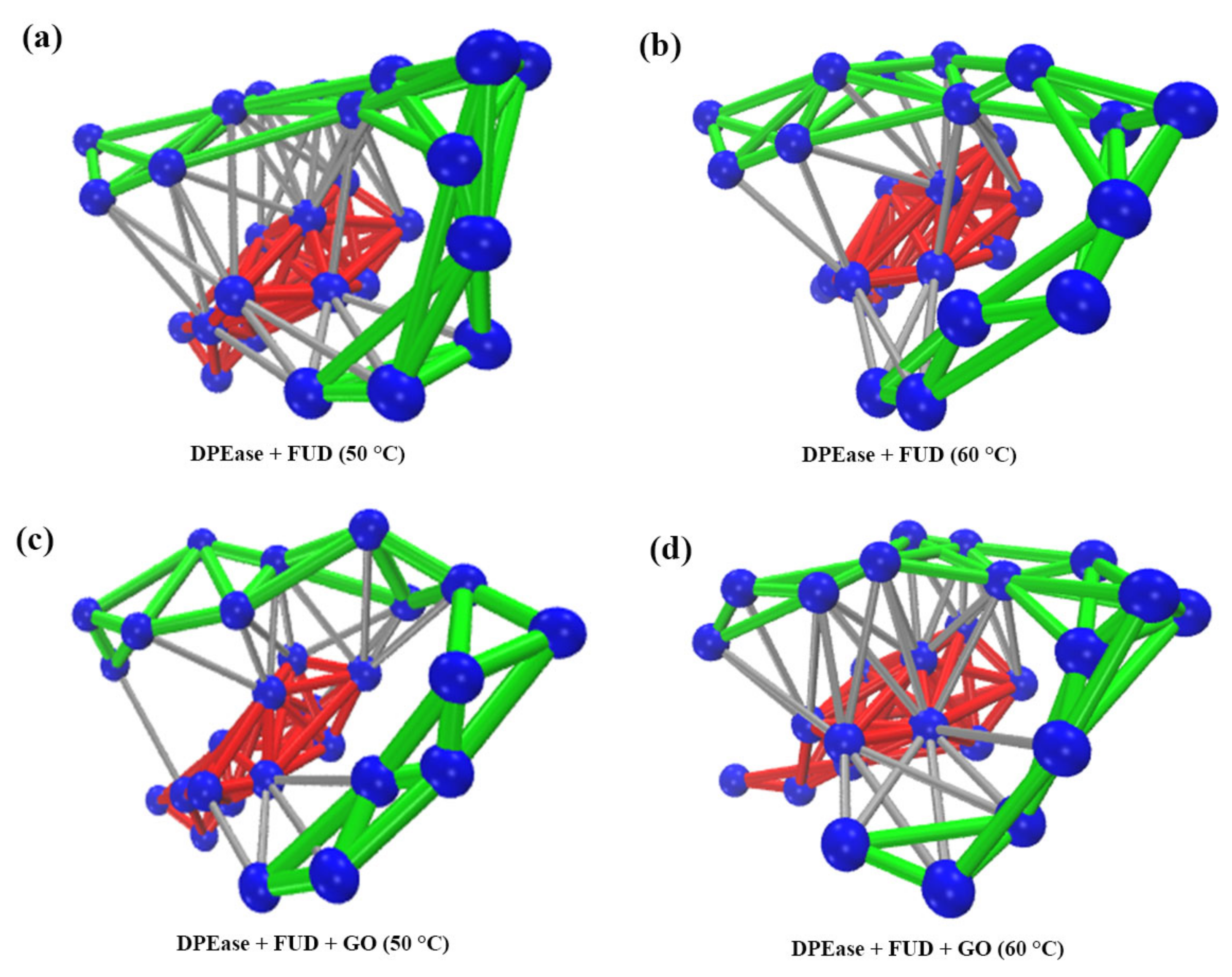
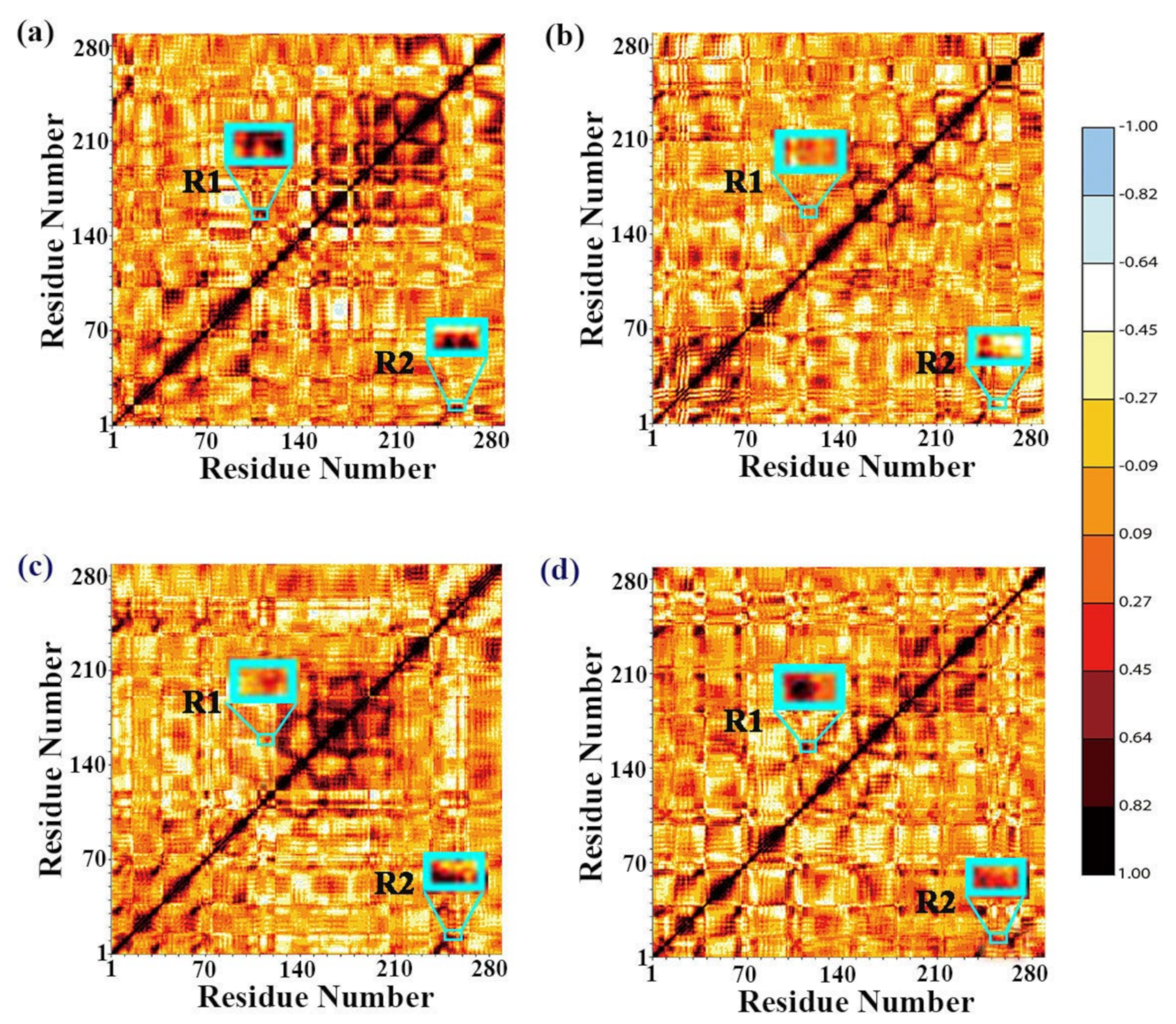
| Regions | DPEase + FUD | DPEase + FUD + GO | ||
|---|---|---|---|---|
| 50 °C (Model 1) | 60 °C (Model 2) | 50 °C (Model 3) | 60 °C (Model 4) | |
| loop α1′-α1 | 0.64 ± 0.17 | 1.89 ± 0.29 | 1.34 ± 0.24 | 1.47 ± 0.18 |
| loop β4-α4 | 0.85 ± 0.35 | 1.55 ± 0.33 | 1.66 ± 0.34 | 1.59 ± 0.24 |
| loop β5-α5 | 0.50 ± 0.077 | 0.52 ± 0.09 | 0.59 ± 0.09 | 0.60 ± 0.08 |
| α8′ | 0.52 ± 0.22 | 1.71 ± 0.55 | 1.64 ± 0.53 | 1.85 ± 0.58 |
| α5 | 0.74 ± 0.17 | 0.88 ± 0.30 | 1.01 ± 0.20 | 0.84 ± 0.17 |
| System | Donor | Receptor | Occupancy (%) | Donor | Receptor | Occupancy (%) |
|---|---|---|---|---|---|---|
| DPEase + FUD (50 °C) | FUD:O1 | E150:OE2 | 67.06 | FUD:O6 | E34:OE2 | 24.35 |
| FUD:O4 | E244:OE2 | 64.52 | R215:NH2 | FUD:O1 | 19.13 | |
| FUD:O1 | E156:OE2 | 46.00 | FUD:O6 | E34:OE1 | 54.90 | |
| FUD:O1 | E156:OE1 | 45.16 | H186:NE2 | FUD:O1 | 34.88 | |
| FUD:O5 | E244:OE2 | 33.98 | FUD:O3 | E244:OE2 | 14.76 | |
| DPEase + FUD (60 °C) | FUD:O6 | E34:OE1 | 40.77 | FUD:O3 | E244:OE2 | 32.21 |
| E34:OE1 | FUD:O6 | 40.77 | Y7:OH | FUD:O6 | 30.88 | |
| FUD:O6 | E34:OE2 | 39.91 | H186:NE2 | FUD:O1 | 29.28 | |
| FUD:O1 | E150:OE2 | 39.34 | FUD:O4 | E244:OE1 | 24.12 | |
| FUD:O4 | E244:OE2 | 39.14 | FUD:O1 | E156:OE2 | 10.12 | |
| DPEase + FUD + GO (50 °C) | FUD:O4 | E244:OE2 | 60.99 | FUD:O1 | E156:CD | 10.83 |
| FUD:O3 | E244:OE2 | 62.43 | FUD:O5 | Y6:OH | 11.41 | |
| FUD:O6 | E34:OE2 | 52.02 | FUD:O6 | E34:OE1 | 29.27 | |
| FUD:O1 | E156:OE1 | 17.54 | R215:NH2 | FUD:O1 | 6.12 | |
| Y6:OH | FUD:O6 | 11.29 | R215:NH1 | FUD:O1 | 2.55 | |
| DPEase + FUD + GO (60 °C) | FUD:O4 | E244:OE2 | 85.88 | FUD:O1 | E156:OE1 | 33.61 |
| FUD:O3 | E244:OE2 | 61.38 | Y6:OH | FUD:O3 | 21.65 | |
| FUD:O6 | E34:OE2 | 47.20 | R215:NH2 | FUD:O1 | 21.47 | |
| FUD:O1 | E156:OE2 | 38.68 | Y7:OH | FUD:O6 | 14.27 | |
| FUD:O6 | E34:OE1 | 37.99 | H186:NE2 | FUD:O2 | 15.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Lu, Z.; Wang, M.; Chen, S.; Han, L.; Han, W. A Possible Mechanism of Graphene Oxide to Enhance Thermostability of D-Psicose 3-Epimerase Revealed by Molecular Dynamics Simulations. Int. J. Mol. Sci. 2021, 22, 10813. https://doi.org/10.3390/ijms221910813
Li C, Lu Z, Wang M, Chen S, Han L, Han W. A Possible Mechanism of Graphene Oxide to Enhance Thermostability of D-Psicose 3-Epimerase Revealed by Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2021; 22(19):10813. https://doi.org/10.3390/ijms221910813
Chicago/Turabian StyleLi, Congcong, Zhongkui Lu, Min Wang, Siao Chen, Lu Han, and Weiwei Han. 2021. "A Possible Mechanism of Graphene Oxide to Enhance Thermostability of D-Psicose 3-Epimerase Revealed by Molecular Dynamics Simulations" International Journal of Molecular Sciences 22, no. 19: 10813. https://doi.org/10.3390/ijms221910813
APA StyleLi, C., Lu, Z., Wang, M., Chen, S., Han, L., & Han, W. (2021). A Possible Mechanism of Graphene Oxide to Enhance Thermostability of D-Psicose 3-Epimerase Revealed by Molecular Dynamics Simulations. International Journal of Molecular Sciences, 22(19), 10813. https://doi.org/10.3390/ijms221910813








