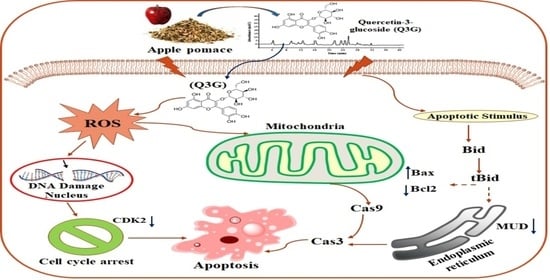
Quercetin-3-Glucoside Extracted from Apple Pomace Induces Cell Cycle Arrest and Apoptosis by Increasing Intracellular ROS Levels
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phenolic Compounds Constituting Apple Pomace
2.2. Antioxidant Potential
2.3. Anti-Inflammatory Effect
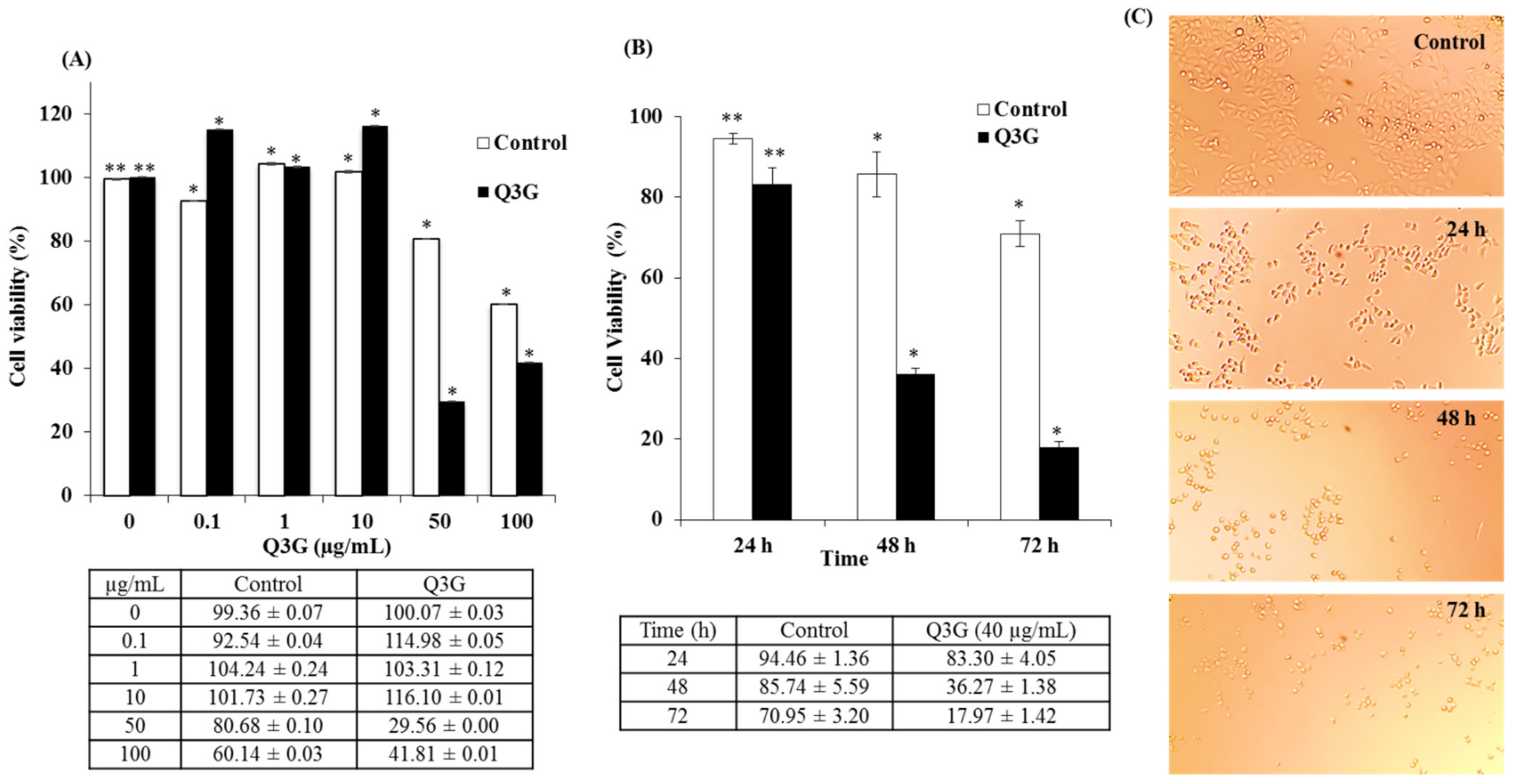
2.4. Dose- and Time-Dependent Cytotoxicity of Q3G
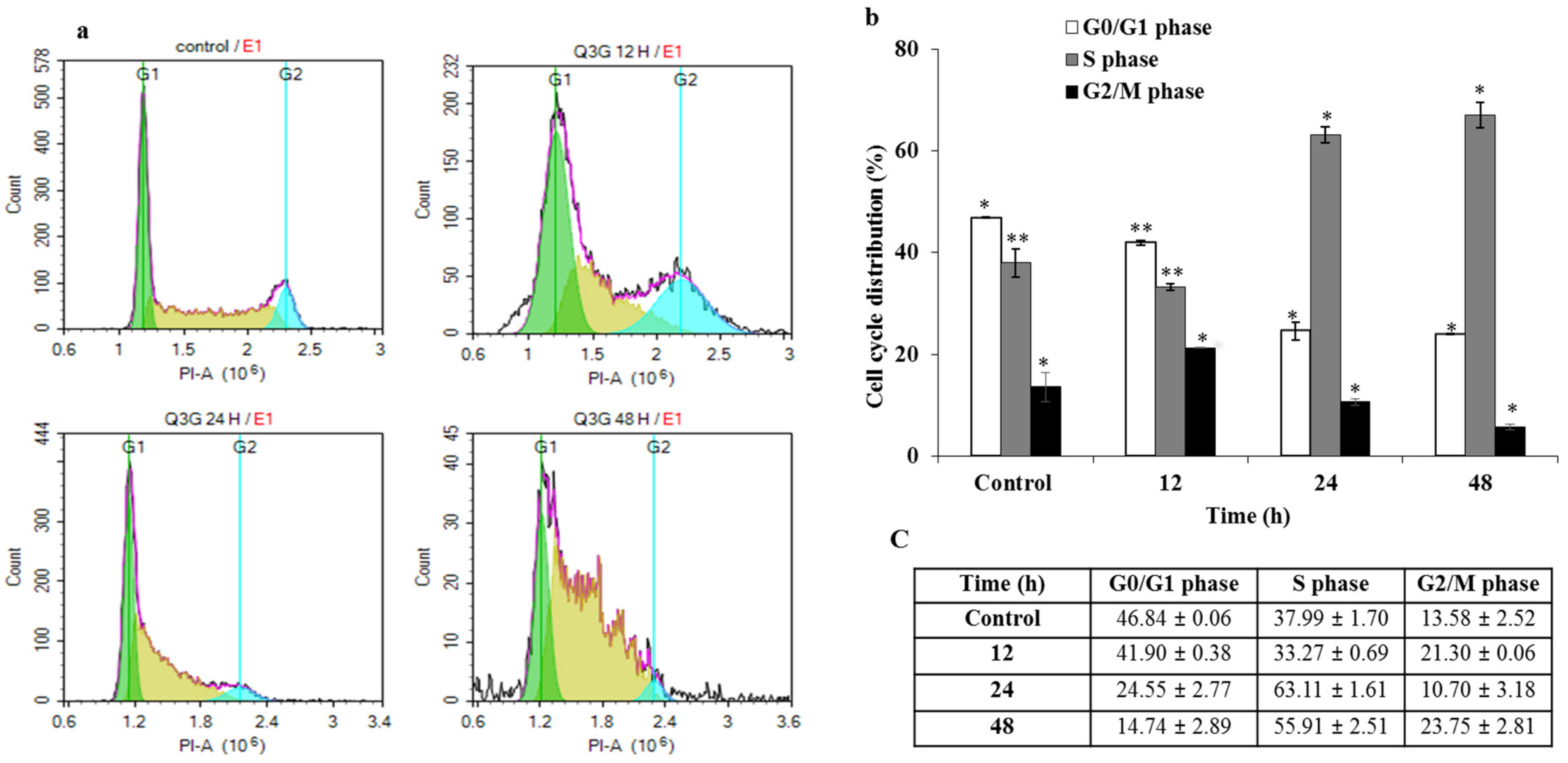
2.5. Q3G Treatment Regulated the Cell Cycle Arrest
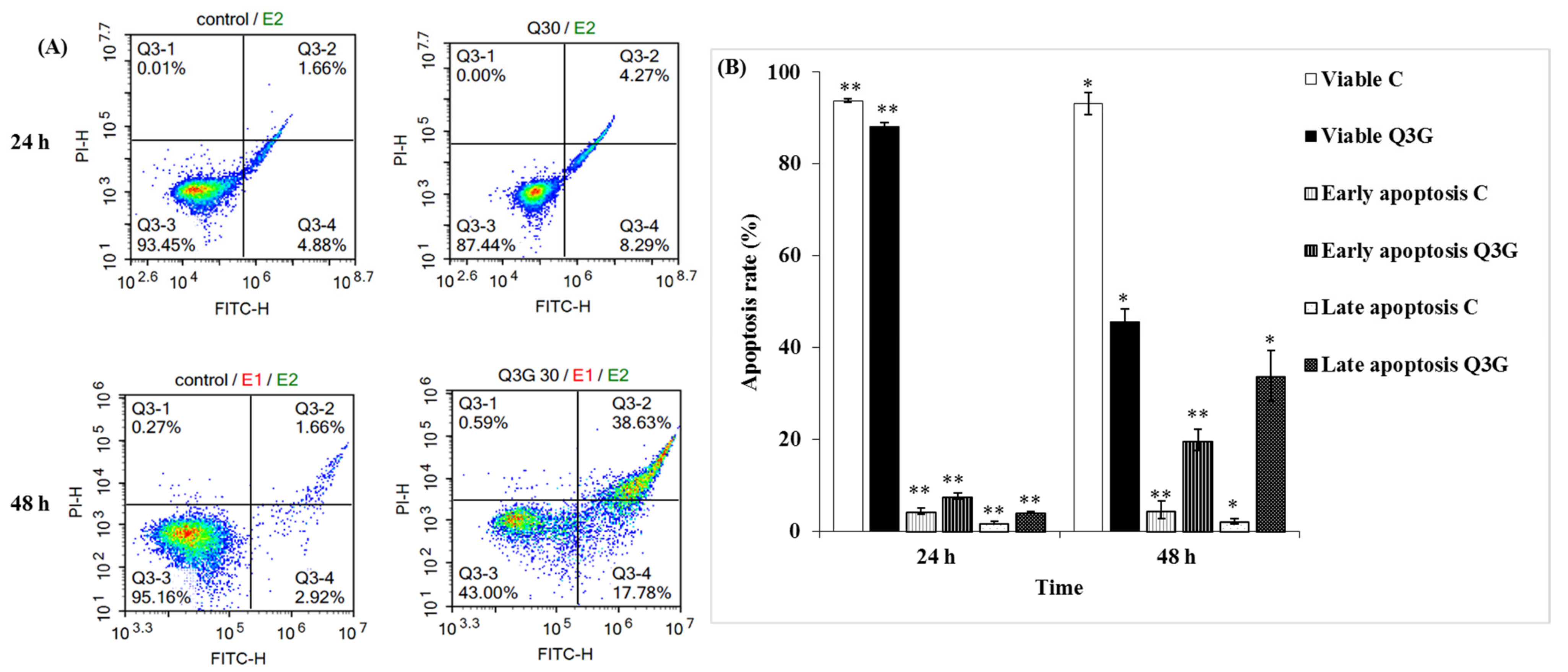
2.6. Q3G Induced Apoptosis in HeLa Cervical Cancer Cells
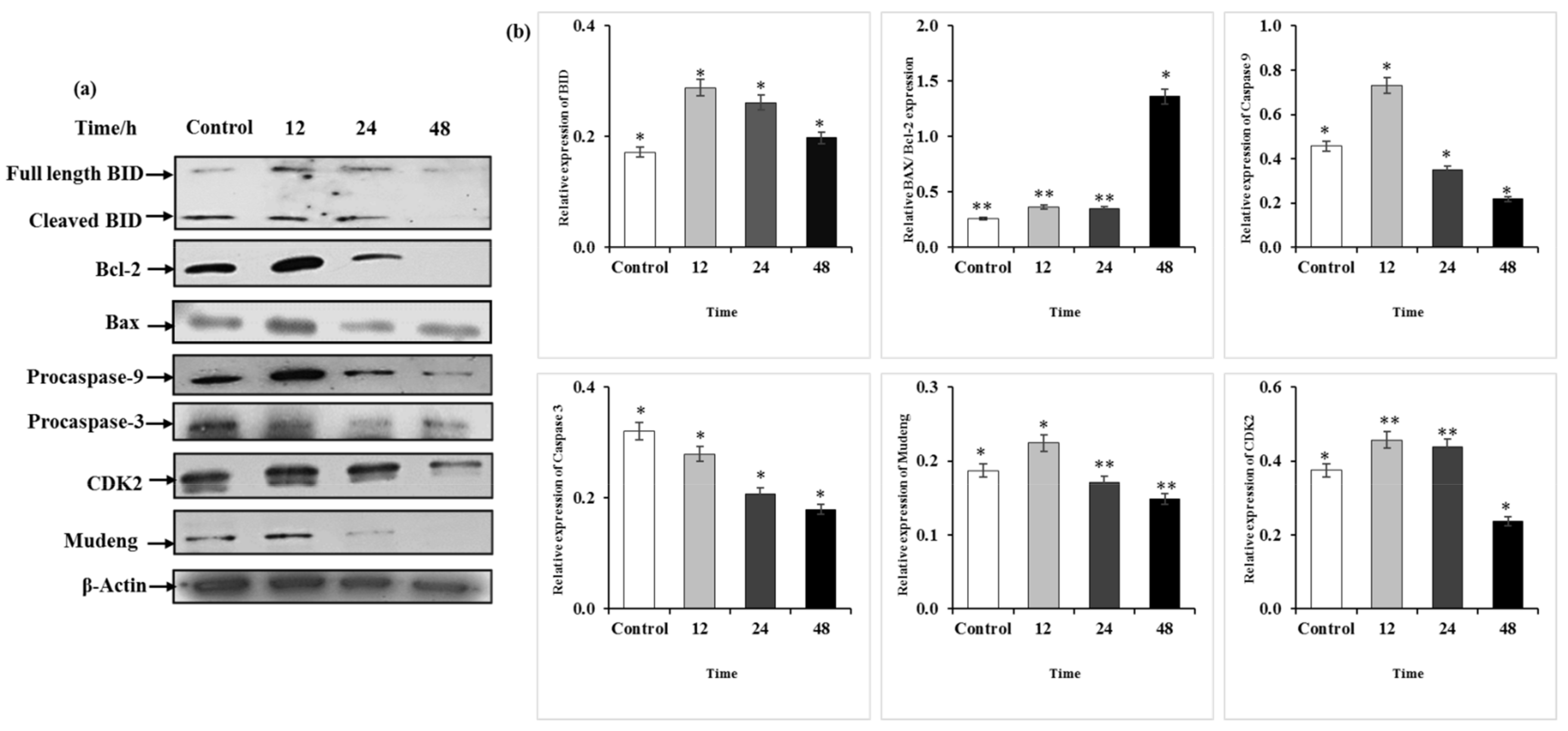
2.7. Q3G Treatment Altered the Apoptosis-Associated Protein Expression in HeLa Cells
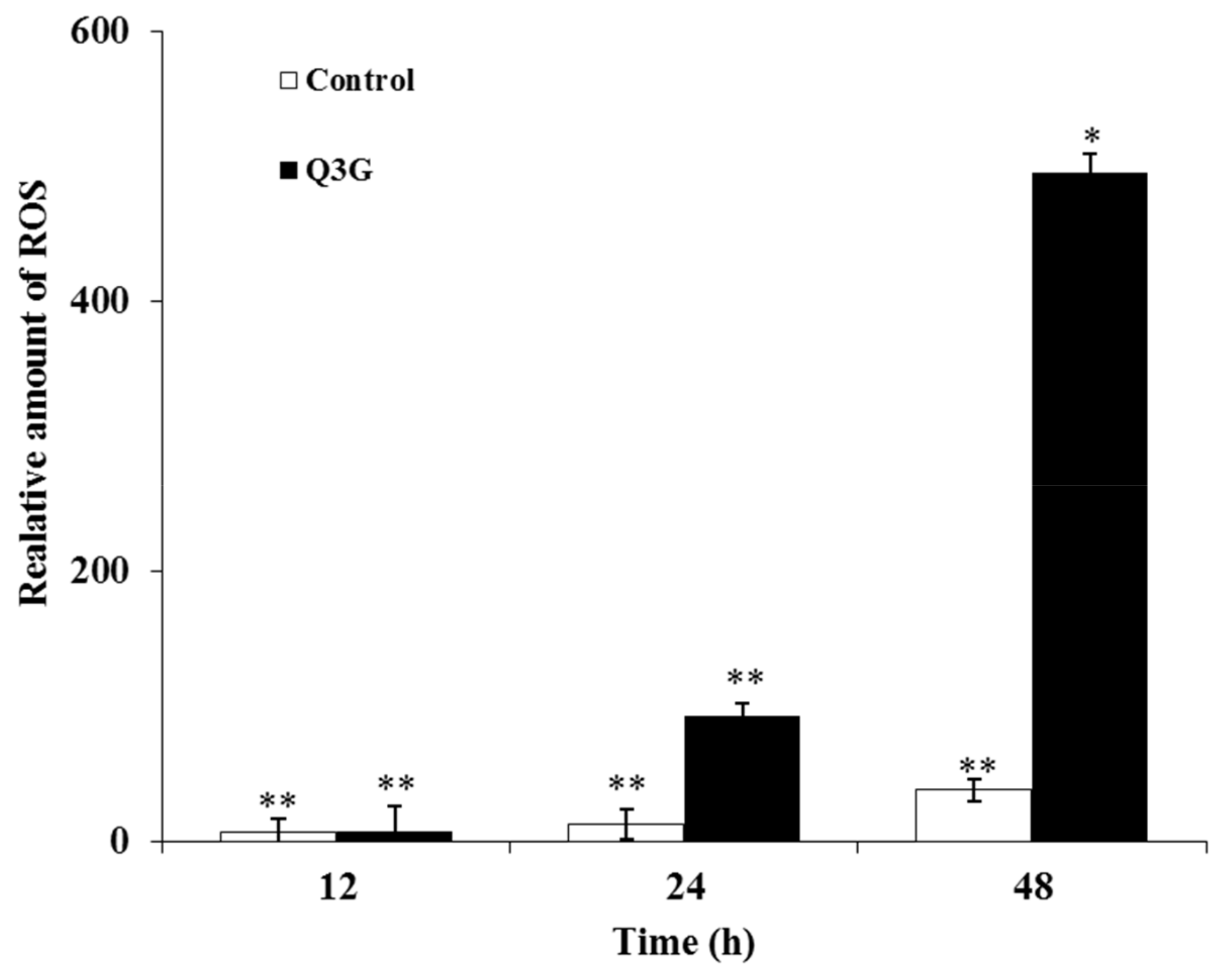
2.8. Q3G Treatment Induced ROS Production in HeLa Cells
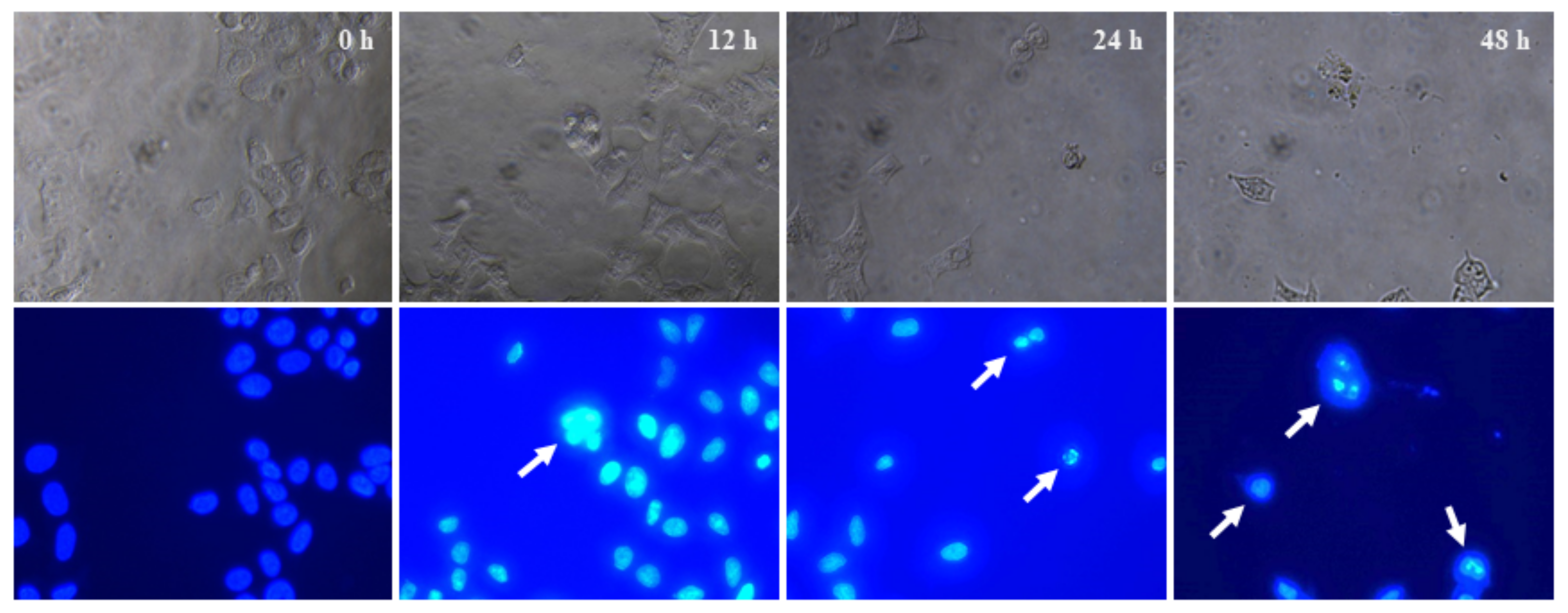
2.9. Cytological Changes in HeLa Cells Eventuated by Q3G
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Extraction of Phenolic Compounds
3.3. HPLC Analysis of Polyphenolic Compounds from Apple Pomace
3.4. In Vitro Antioxidant Activity
3.5. In Vitro Anti-Inflammatory Activity
3.6. Cell Viability Assay
3.7. Assessment of Apoptosis
3.8. Cell Cycle Analysis by Flow Cytometry
3.9. Western Blotting
3.10. Chromatin Condensation Assay
3.11. Intracellular ROS Production
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alberti, A.; Zielinski, A.A.; Zardo, D.M.; Demiate, I.M.; Nogueira, A.; Mafra, L.I. Optimisation of the extraction of phenolic compounds from apples using response surface methodology. Food Chem. 2014, 149, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Perussello, C.A.; Zhang, Z.; Marzocchella, A.; Tiwari, B.K. Valorization of apple pomace by extraction of valuable compounds. Compr. Rev. Food Sci. Food Saf. 2017, 16, 776–796. [Google Scholar] [CrossRef] [Green Version]
- Skrypnik, L.; Novikova, A. Response surface modeling and optimization of polyphenols extraction from apple pomace based on nonionic emulsifiers. Agronomy 2020, 10, 92. [Google Scholar] [CrossRef] [Green Version]
- Uyttebroek, M.; Vandezande, P.; Van Dael, M.; Vloemans, S.; Noten, B.; Bongers, B.; Porto-Carrero, W.; Unamunzaga, M.M.; Bulut, M.; Lemmens, B. Concentration of phenolic compounds from apple pomace extracts by nanofiltration at lab and pilot scale with a techno-economic assessment. J. Food Process Eng. 2018, 41, e12629. [Google Scholar] [CrossRef]
- Shahidi, F. Nutraceuticals, functional foods and dietary supplements in health and disease. J. Food Drug Anal. 2012, 20, 226–230. [Google Scholar]
- Savatović, S.M.; Ćetković, G.S.; Đilas, S.M.; Tumbas, V.; Čanadanović-Brunet, J.M.; Četojević-Simin, D.D.; Mandić, A.I. Antioxidant and antiproliferative activity of granny smith apple pomace. Acta Period. Technol. 2008, 39, 201–212. [Google Scholar] [CrossRef]
- Rana, S.; Gupta, S.; Rana, A.; Bhushan, S. Functional properties, phenolic constituents and antioxidant potential of industrial apple pomace for utilization as active food ingredient. Food Sci. Hum. Wellness 2015, 4, 180–187. [Google Scholar] [CrossRef] [Green Version]
- Lohani, U.C.; Muthukumarappan, K. Application of the pulsed electric field to release bound phenolics in sorghum flour and apple pomace. Innov. Food Sci. Emerg. 2016, 35, 29–35. [Google Scholar] [CrossRef]
- McCann, M.J.; Gill, C.I.R.; O’Brien, G.; Rao, J.R.; McRoberts, W.C.; Hughes, P.; McEntee, R.; Rowland, I.R. Anti-cancer properties of phenolics from apple waste on colon carcinogenesis in vitro. Food Chem. Toxicol. 2007, 45, 1224–1230. [Google Scholar] [CrossRef]
- Muino, I.; Diaz, M.T.; Apeleo, E.; Perez-Santaescolastica, C.; Rivas-Canedo, A.; Perez, C.; Caneque, V.; Lauzurica, S.; de la Fuente, J. Valorisation of an extract from olive oil waste as a natural antioxidant for reducing meat waste resulting from oxidative processes. J. Clean. Prod. 2017, 140, 924–932. [Google Scholar] [CrossRef]
- Yates, M.; Gomez, M.R.; Martin-Luengo, M.A.; Ibanez, V.Z.; Serrano, A.M.M. Multivalorization of apple pomace towards materials and chemicals. Waste to wealth. J. Clean. Prod. 2017, 143, 847–853. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Meng, F.; Reece, E.A.; Zhao, Z. Modulation of nuclear factor-κB signaling and reduction of neural tube defects by quercetin-3-glucoside in embryos of diabetic mice. Am. J. Obstet. Gynecol. 2018, 219, 197.e1–197.e8. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.; Lee, H.; Sung, J. Relative protective activities of quercetin, quercetin-3-glucoside, and rutin in alcohol-induced liver injury. J. Food Biochem. 2019, 43, e13002. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Kim, S.J.; Kim, J.H. Quercetin-3-O-glucoside suppresses pancreatic cancer cell migration induced by tumor-deteriorated growth factors in vitro. Oncol. Rep. 2016, 35, 2473–2479. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Han, S.I.; Yun, J.H.; Kim, J.H. Quercetin 3-O-glucoside suppresses epidermal growth factor-induced migration by inhibiting EGFR signaling in pancreatic cancer cells. Tumour Biol. 2015, 36, 9385–9393. [Google Scholar] [CrossRef] [PubMed]
- Massias, A.; Boisard, S.; Baccaunaud, M.; Calderon, F.L.; Subra-Paternault, P. Recovery of phenolics from apple peels using CO2 + ethanol extraction: Kinetics and antioxidant activity of extracts. J. Supercrit. Fluid. 2015, 98, 172–182. [Google Scholar] [CrossRef]
- Ramirez-Ambrosi, M.; Abad-Garcia, B.; Viloria-Bernal, M.; Garmon-Lobato, S.; Berrueta, L.A.; Gallo, B. A new ultrahigh performance liquid chromatography with diode array detection coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry analytical strategy for fast analysis and improved characterization of phenolic compounds in apple products. J. Chromatogr. A 2013, 1316, 78–91. [Google Scholar] [CrossRef]
- Barreira, J.C.M.; Arraibi, A.A.; Ferreira, I.C.F.R. Bioactive and functional compounds in apple pomace from juice and cider manufacturing: Potential use in dermal formulations. Trends Food Sci. Tech. 2019, 90, 76–87. [Google Scholar] [CrossRef]
- Tow, W.W.; Premier, R.; Jing, H.; Ajlouni, S. Antioxidant and antiproliferation effects of extractable and nonextractable polyphenols isolated from apple waste using different extraction methods. J. Food Sci. 2011, 76, T163–T172. [Google Scholar] [CrossRef]
- Zhang, T.J.; Wei, X.Y.; Miao, Z.; Hassan, H.; Song, Y.B.; Fan, M.T. Screening for antioxidant and antibacterial activities of phenolics from Golden Delicious apple pomace. Chem. Cent. J. 2016, 10, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, R.; Yang, R.; Xie, S.; Sockovie, E.; Khanizadeh, S. Which polyphenolic compounds contribute to the total antioxidant activities of apple? J. Agric. Food Chem. 2005, 53, 4989–4995. [Google Scholar] [CrossRef]
- Garcia, Y.D.; Valles, B.S.; Lobo, A.P. Phenolic and antioxidant composition of by-products from the cider industry: Apple pomace. Food Chem. 2009, 117, 731–738. [Google Scholar] [CrossRef]
- Gonzalez-Periz, A.; Claria, J. New approaches to the modulation of the cyclooxygenase-2 and 5-lipoxygenase pathways. Curr. Top. Med. Chem. 2007, 7, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Roschek, B.; Fink, R.C.; Li, D.; McMichael, M.; Tower, C.M.; Smith, R.D.; Alberte, R.S. Pro-inflammatory enzymes, cyclooxygenase 1, cyclooxygenase 2, and 5-lipooxygenase, inhibited by stabilized rice bran extracts. J. Med. Food 2009, 12, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Bhujade, A.M.; Talmale, S.; Kumar, N.; Gupta, G.; Reddanna, P.; Das, S.K.; Patil, M.B. Evaluation of Cissus quadrangularis extracts as an inhibitor of COX, 5-LOX, and proinflammatory mediators. J. Ethnopharmacol. 2012, 141, 989–996. [Google Scholar] [CrossRef]
- Jaramillo, S.; Lopez, S.; Varela, L.M.; Rodriguez-Arcos, R.; Jimenez, A.; Abia, R.; Guillen, R.; Muriana, F.J. The flavonol isorhamnetin exhibits cytotoxic effects on human colon cancer cells. J. Agric. Food Chem. 2010, 58, 10869–10875. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, S.K.; Kim, B.S.; Lee, S.H.; Park, Y.S.; Park, B.K.; Kim, S.J.; Kim, J.; Choi, C.; Kim, J.S.; et al. Apoptotic effect of quercetin on HT-29 colon cancer cells via the AMPK signaling pathway. J. Agric. Food Chem. 2010, 58, 8643–8650. [Google Scholar] [CrossRef]
- Yeh, S.L.; Lin, Y.C.; Lin, Y.L.; Li, C.C.; Chuang, C.H. Comparing the metabolism of quercetin in rats, mice and gerbils. Eur. J. Nutr. 2016, 55, 413–422. [Google Scholar] [CrossRef]
- Ahn, H.J.; You, H.J.; Park, M.S.; Li, Z.P.; Choe, D.; Johnston, T.V.; Ku, S.; Ji, G.E. Microbial biocatalysis of quercetin-3-glucoside and isorhamnetin-3-glucoside in Salicornia herbacea and their contribution to improved anti-inflammatory activity. RSC Adv. 2020, 10, 5339–5350. [Google Scholar] [CrossRef] [Green Version]
- Ezzati, M.; Yousefi, B.; Velaei, K.; Safa, A. A review on anti-cancer properties of Quercetin in breast cancer. Life Sci. 2020, 248, 117463. [Google Scholar] [CrossRef]
- Yang, J.; Liu, R.H. Synergistic effect of apple extracts and quercetin 3-β-d-glucoside combination on antiproliferative activity in MCF-7 human breast cancer cells in vitro. J. Agric. Food Chem. 2009, 57, 8581–8586. [Google Scholar] [CrossRef]
- Choi, J.A.; Kim, J.Y.; Lee, J.Y.; Kang, C.M.; Kwon, H.J.; Yoo, Y.D.; Kim, T.W.; Lee, Y.S.; Lee, S.J. Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int. J. Oncol. 2001, 19, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Cheong, E.; Ivory, K.; Doleman, J.; Parker, M.L.; Rhodes, M.; Johnson, I.T. Synthetic and naturally occurring COX-2 inhibitors suppress proliferation in a human oesophageal adenocarcinoma cell line (OE33) by inducing apoptosis and cell cycle arrest. Carcinogenesis 2004, 25, 1945–1952. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Ebermann, R.; Marian, B. Quercetin-induced apoptosis in colorectal tumor cells: Possible role of EGF receptor signaling. Nutr. Cancer 1999, 34, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Jia, P.; Yan, Z.; Liu, X.; Li, X.; Liu, H. Quercetin induces cell cycle G1 arrest through elevating Cdk inhibitors p21 and p27 in human hepatoma cell line (HepG2). Methods Find. Exp. Clin. Pharmacol. 2007, 29, 179–183. [Google Scholar] [CrossRef]
- Chou, C.C.; Yang, J.S.; Lu, H.F.; Ip, S.W.; Lo, C.; Wu, C.C.; Lin, J.P.; Tang, N.Y.; Chung, J.G.; Chou, M.J.; et al. Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells. Arch. Pharm. Res. 2010, 33, 1181–1191. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Wlodkowic, D.; Telford, W.; Skommer, J.; Darzynkiewicz, Z. Apoptosis and beyond: Cytometry in studies of programmed cell death. Methods Cell Biol. 2011, 103, 55–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudan, S.; Rupasinghe, H.P. Quercetin-3-O-glucoside induces human DNA topoisomerase II inhibition, cell cycle arrest and apoptosis in hepatocellular carcinoma cells. Anticancer Res. 2014, 34, 1691–1699. [Google Scholar]
- Teijido, O.; Dejean, L. Upregulation of Bcl2 inhibits apoptosis-driven BAX insertion but favors BAX relocalization in mitochondria. FEBS Lett. 2010, 584, 3305–3310. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, E.J. Selective inhibitors of Bcl-2 and Bcl-xL: Balancing antitumor activity with on-target toxicity. Bioorg. Med. Chem. Lett. 2016, 26, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Eskes, R.; Desagher, S.; Antonsson, B.; Martinou, J.C. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 2000, 20, 929–935. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Che, X.; Zheng, Q.; Wu, A.; Pan, K.; Shao, A.; Wu, Q.; Zhang, J.; Hong, Y. Caspases: A molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 2014, 10, 1072–1083. [Google Scholar] [CrossRef]
- Tang, D.; Wang, C.; Gao, Y.; Pu, J.; Long, J.; Xu, W. Deep hypothermia-enhanced autophagy protects PC12 cells against oxygen glucose deprivation via a mitochondrial pathway. Neurosci. Lett. 2016, 632, 79–85. [Google Scholar] [CrossRef]
- Choi, J.H.; Lim, J.B.; Wickramanayake, D.D.; Wagley, Y.; Kim, J.; Lee, H.C.; Seo, H.G.; Kim, T.H.; Oh, J.W. Characterization of MUDENG, a novel anti-apoptotic protein. Oncogenesis 2016, 5, e221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.R.; Shin, J.N.; Moon, A.R.; Park, S.Y.; Hong, G.; Lee, M.J.; Yun, C.W.; Seol, D.W.; Piya, S.; Bae, J.; et al. A novel protein, MUDENG, induces cell death in cytotoxic T cells. Biochem. Biophys. Res. Commun. 2008, 370, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Muthu, M.; Chun, S.; Gopal, J.; Park, G.S.; Nile, A.; Shin, J.; Shin, J.; Kim, T.H.; Oh, J.W. The MUDENG augmentation: A genesis in anti-cancer therapy? Int. J. Mol. Sci. 2020, 21, 5583. [Google Scholar] [CrossRef]
- Taverne, Y.J.; Bogers, A.J.; Duncker, D.J.; Merkus, D. Reactive oxygen species and the cardiovascular system. Oxid. Med. Cell. Longev. 2013, 2013, 862423. [Google Scholar] [CrossRef]
- Schumacker, P.T. Reactive oxygen species in cancer cells: Live by the sword, die by the sword. Cancer Cell 2006, 10, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Needs, P.W.; Lu, Y.; Kroon, P.A.; Ren, D.; Yang, X. Different antitumor effects of quercetin, quercetin-3′-sulfate and quercetin-3-glucuronide in human breast cancer MCF-7 cells. Food Funct. 2018, 9, 1736–1746. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Chidambara Murthy, K.N.; Kim, J.; Vikram, A.; Patil, B.S. Differential inhibition of human colon cancer cells by structurally similar flavonoids of citrus. Food Chem. 2012, 132, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Marchissio, M.J.; Frances, D.E.; Carnovale, C.E.; Marinelli, R.A. Evidence for necrosis, but not apoptosis, in human hepatoma cells with knockdown of mitochondrial aquaporin-8. Apoptosis 2014, 19, 851–859. [Google Scholar] [CrossRef]
- Huang, A.C.; Lin, T.P.; Weng, Y.S.; Ho, Y.T.; Lin, H.J.; Huang, L.J.; Kuo, S.C.; Chung, J.G. Ethyl 2-[N-m-chlorobenzyl-(2′-methyl)] anilino-4-oxo-4,5-dihydrofuran-3-carboxylate (JOT01006) induces apoptosis in human cervical cancer HeLa cells. Anticancer Res. 2007, 27, 2505–2514. [Google Scholar]
- Malhotra, P.; Adhikari, M.; Singh, S.K.; Kumar, R. N-acetyl tryptophan glucopyranoside (NATG) provides radioprotection to murine macrophage J774A.1 cells. Free Radic. Res. 2015, 49, 1488–1498. [Google Scholar] [CrossRef]
- Wagley, Y.; Choi, J.H.; Wickramanayake, D.D.; Choi, G.Y.; Kim, C.K.; Kim, T.H.; Oh, J.W. A monoclonal antibody against human MUDENG protein. Monoclon. Antib. Immunodiagn. Immunother. 2013, 32, 277–282. [Google Scholar] [CrossRef] [Green Version]
- Nile, S.H.; Park, S.W. Chromatographic analysis, antioxidant, anti-inflammatory, and xanthine oxidase inhibitory activities of ginger extracts and its reference compounds. Indus. Crop. Prod. 2015, 70, 238–244. [Google Scholar] [CrossRef]
- Sun, C.T.; Nile, S.H.; Zhang, Y.T.; Qin, L.P.; El-Seedi, H.R.; Daglia, M.; Kai, G.Y. Novel insight into utilization of flavonoid glycosides and biological properties of saffron (Crocus sativus L.) flower byproducts. J. Agric. Food Chem. 2020, 68, 10685–10696. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Babu, N.K.; Gupta, A.D.; Reddanna, P. Evaluation of anti-inflammatory and immunomodulatory effects of Premna integrifolia extracts and assay-guided isolation of a COX-2/5-LOX dual inhibitor. Fitoterapia 2018, 131, 189–199. [Google Scholar] [CrossRef]
- Reddanna, P.; Whelan, J.; Maddipati, K.R.; Reddy, C.C. Purification of arachidonate 5-lipoxygenase from potato-tubers. Method Enzymol. 1990, 187, 268–277. [Google Scholar]
- Nile, A.; Gansukh, E.; Park, G.S.; Kim, D.H.; Hariram Nile, S. Novel insights on the multi-functional properties of flavonol glucosides from red onion (Allium cepa L) solid waste—In vitro and in silico approach. Food Chem. 2021, 335, 127650. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Jung, S.; Park, G.S.; Shin, J.; Oh, J.W. Piperine synergistically enhances the effect of temozolomide against temozolomide-resistant human glioma cell lines. Bioengineered 2020, 11, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.S.; Chang, C.H.; Chou, Y.R.; Yeh, M.Y.; Au, M.K.; Lu, H.F.; Chu, Y.L.; Chou, H.M.; Chou, H.C.; Shih, Y.L.; et al. Curcumin causes DNA damage and affects associated protein expression in HeLa human cervical cancer cells. Oncol. Rep. 2016, 36, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.N.; Shin, D.S.; Kim, H.N.; Jeon, Y.J.; Yun, J.; Lee, Y.J.; Kang, J.S.; Han, D.C.; Kwon, B.M. Sugiol inhibits STAT3 activity via regulation of transketolase and ROS-mediated ERK activation in DU145 prostate carcinoma cells. Biochem. Pharmacol. 2015, 97, 38–50. [Google Scholar] [CrossRef]

| Phenolic Compounds | Peak | Rt (min) | λ Max (nm) | Amount (μg/mg DW) |
|---|---|---|---|---|
| Chlorogenic acid | 1 | 9 | 330 | 5.2 ± 0.082 |
| Epicatechin | 2 | 11 | 285 | 2.6 ± 0.056 |
| Caffeic acid | 3 | 12.5 | 325 | 3.8 ± 0.036 |
| Quercetin-3-glucoside | 4 | 16 | 360 | 8.6 ± 0.091 |
| Coumaric acid | 5 | 21.2 | 312 | 11.5 ± 0.055 |
| Phloridzin | 6 | 23.8 | 290 | 10.2 ± 0.091 |
| Quercetin | 7 | 24.2 | 375 | 8.2 ± 0.81 |
| Phloretin | 8 | 25.1 | 290 | 9.8 ± 0.52 |
| Samples | Radical Scavenging Activity (%) | Enzyme Inhibitory Effect IC50 (μg/mL) | |||||
|---|---|---|---|---|---|---|---|
| DPPH | FRAP | ABTS | ORAC | COX-1 | COX-2 | LOX-5 | |
| Chlorogenic acid | 81.56 ± 2.24 d | 75.82 ± 3.35 d | 60.62 ± 1.88 d | 58.62 ± 2.33 d | 14.08 ± 1.11 e | 26.69 ± 1.32 e | 9.98 ± 0.94 e |
| Epicatechin | 55.18 ± 1.57 g | 52.36 ± 1.11 g | 48.98 ± 0.82 g | 41.10 ± 0.99 g | 18.29 ± 1.13 f | 24.13 ± 1.31 f | 10.09 ± 1.03 f |
| Caffeic acid | 59.11 ± 1.49 f | 55.92 ±1.23 f | 50.55 ± 1.62 f | 45.11 ± 1.70 f | 20.32 ± 1.44 g | 30.87 ± 1.56 g | 12.26 ± 0.91 g |
| Quercetin-3-glucoside | 90.75 ± 2.81 b | 88.22 ± 3.21 ab | 82.76 ± 1.94 a | 75.29 ± 1.22 ab | 3.62 ± 0.84 b | 5.66 ± 0.95 b | 2.31 ± 0.32 b |
| Coumaric acid | 60.45 ± 1.39 e | 58.63 ± 1.64 e | 51.99 ± 1.61 e | 47.69 ± 1.22 e | 9.09 ± 0.88 d | 15.3 ± 0.91 d | 7.96 ± 0.76 d |
| Phloridzin | 40.13 ± 1.12 i | 37.91 ± 0.95 i | 32.11 ± 0.88 i | 30.11 ± 0.86 i | 28.63 ± 1.06 i | 36.21 ± 1.33 i | 18.98 ± 0.85 i |
| Quercetin | 88.28 ± 3.12 c | 85.37 ± 1.10 c | 80.22 ± 2.17 bc | 70.95 ± 1.88 c | 6.11 ± 0.91 c | 11.3 ± 1.11 c | 5.31 ± 0.86 c |
| Phloretin | 45.62 ± 1.45 h | 40.91 ± 1.02 h | 36.32 ± 1.31 h | 38.12 ± 0.98 h | 25.14 ± 1.02 h | 33.18 ± 1.81 h | 16.86 ± 1.22 h |
| Ascorbic acid * | 92.12 ± 3.15 a | 87.69 ± 1.36 ab | 80.16 ± 3.24 bc | 75.32 ± 1.52 ab | NA | NA | NA |
| Indomethacin * | NA | NA | NA | NA | 1.25 ± 0.15 a | 1.38 ± 0.12 a | NA |
| Zileuton * | NA | NA | NA | NA | NA | NA | 1.14 ± 0.21 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nile, A.; Nile, S.H.; Shin, J.; Park, G.; Oh, J.-W. Quercetin-3-Glucoside Extracted from Apple Pomace Induces Cell Cycle Arrest and Apoptosis by Increasing Intracellular ROS Levels. Int. J. Mol. Sci. 2021, 22, 10749. https://doi.org/10.3390/ijms221910749
Nile A, Nile SH, Shin J, Park G, Oh J-W. Quercetin-3-Glucoside Extracted from Apple Pomace Induces Cell Cycle Arrest and Apoptosis by Increasing Intracellular ROS Levels. International Journal of Molecular Sciences. 2021; 22(19):10749. https://doi.org/10.3390/ijms221910749
Chicago/Turabian StyleNile, Arti, Shivraj Hariram Nile, Juhyun Shin, Gyunseok Park, and Jae-Wook Oh. 2021. "Quercetin-3-Glucoside Extracted from Apple Pomace Induces Cell Cycle Arrest and Apoptosis by Increasing Intracellular ROS Levels" International Journal of Molecular Sciences 22, no. 19: 10749. https://doi.org/10.3390/ijms221910749
APA StyleNile, A., Nile, S. H., Shin, J., Park, G., & Oh, J.-W. (2021). Quercetin-3-Glucoside Extracted from Apple Pomace Induces Cell Cycle Arrest and Apoptosis by Increasing Intracellular ROS Levels. International Journal of Molecular Sciences, 22(19), 10749. https://doi.org/10.3390/ijms221910749