Abstract
Soil salinization, which is aggravated by climate change and inappropriate anthropogenic activities, has emerged as a serious environmental problem, threatening sustainable agriculture and future food security. Although there has been considerable progress in developing crop varieties by introducing salt tolerance-associated traits, most crop cultivars grown in saline soils still exhibit a decline in yield, necessitating the search for alternatives. Halophytes, with their intrinsic salt tolerance characteristics, are known to have great potential in rehabilitating salt-contaminated soils to support plant growth in saline soils by employing various strategies, including phytoremediation. In addition, the recent identification and characterization of salt tolerance-related genes encoding signaling components from halophytes, which are naturally grown under high salinity, have paved the way for the development of transgenic crops with improved salt tolerance. In this review, we aim to provide a comprehensive update on salinity-induced negative effects on soils and plants, including alterations of physicochemical properties in soils, and changes in physiological and biochemical processes and ion disparities in plants. We also review the physiological and biochemical adaptation strategies that help halophytes grow and survive in salinity-affected areas. Furthermore, we illustrate the halophyte-mediated phytoremediation process in salinity-affected areas, as well as their potential impacts on soil properties. Importantly, based on the recent findings on salt tolerance mechanisms in halophytes, we also comprehensively discuss the potential of improving salt tolerance in crop plants by introducing candidate genes related to antiporters, ion transporters, antioxidants, and defense proteins from halophytes for conserving sustainable agriculture in salinity-prone areas.
1. Introduction
Land degradation resulting from salinity puts global agriculture in jeopardy, as it engulfs 3600 million hectares (Mha) out of 5200 Mha of world agricultural land, leading to the loss of USD 27.5 billion every year [1,2]. Furthermore, reports anticipate that the world population will increase to 9.7 billion by 2050, which would require 70% greater food production by that time, exerting additional pressure on the shrinking areas of agricultural land [3]. To feed the growing human population, many parts of the world in peril of drought are still using salt-contaminated ground water for irrigation [4]. Inappropriate agricultural management practices, such as improper drainage, also exacerbate soil salinity, making the soil untenable for agriculture, as evident in Punjab of Pakistan and Fubei of China [5]. Furthermore, climate-driven impacts, such as rising sea levels, tidal changes, drifting seawater droplets, and disastrous events, such as tsunamis, cause accumulations of large pools of concentrated seawater on agricultural land [6]. Consequently, salinity threatens around 1.5 Mha of land each year, which will increase the risk of losing 50% of cultivable lands by the middle of the 21st century [7,8].
The Harmonized World Soil Database (HWSD) estimated that around 1130 Mha of land is inclined to the incursion of varying salinity levels. Major categories of salinity-affected soils are saline, sodic, and saline-sodic, accounting for 60, 26, and 14%, respectively, of all salinity-affected soils [9] (Figure 1A). Approximately, 736 (65%), 227 (20%), 53 (5%), and 114 (10%) Mha of all salinity-affected soils are further categorized as slightly, moderately, highly, and extremely salinity-affected soils, respectively [9] (Figure 1B). The extent and severity of salinity-affected soils vary by regions across the world. The largest salinity-affected areas are in the Middle East, Australia, North Africa, and the former Soviet Union, which account for 176, 169, 161, and 126 Mha of land, respectively [9] (Figure 1C).
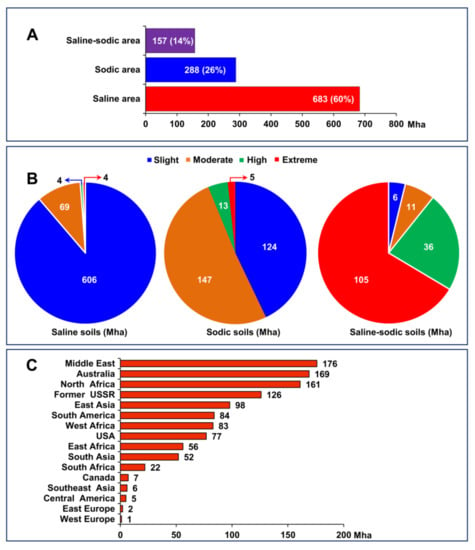
Figure 1.
Overview on soil salinization across the world. (A) Salinity-affected soil categories *. (B) Types and severity levels of salinity-affected soils. (C) Salinity-affected soils in different parts of the world *. * Due to rounding, the sum may not resemble the actual statistics of salinity-affected lands. Mha, million hectares.
Plants grown in salinity-affected areas accumulate higher levels of toxic ions responsible for different physiological abnormalities, including ionic imbalance, impaired gas exchange performance, loss of water homeostasis, alterations in the levels of metabolites, and reactive oxygen species (ROS)-mediated oxidative damage to the cellular compartments [10,11]. To counteract the detrimental effects of salt stress, plants adopt some fundamental mechanisms, including (i) toxic ion (Na+ and Cl−) exclusions or their compartmentation into vacuoles or old tissues; (ii) compatible solute accumulations; (iii) synthesis of numerous stress adaptation-related endogenous metabolites; and (iv) synthesis and activation of antioxidant enzymes [e.g., catalase, (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX), peroxidase (POD), and glutathione S-transferase (GST)], and non-enzymatic antioxidants [e.g., carotenoids, phenolic compounds, flavonoids, ascorbate (AsA), and glutathione (GSH)] [10,12,13,14]. Unfortunately, the traditional crops cannot deploy salt tolerance potential to survive in salinity-affected soils, and a wide range of genetic diversity for salt tolerance in conventional crops, even local landraces, remains elusive. Interestingly, some plants, such as halophytes, comprising only <2% of the world’s flora, can sustain and thrive under extreme salinity through the adoption of intrinsic mechanisms related to selective ion uptake, Na+ compartmentalization, ion homeostasis, production of defense metabolites, salt gland and salt bladder formations, and improved antioxidant properties [14,15]. Given their ability to tolerate high salinity, the cultivation of halophytes on salt-contaminated soils has been introduced as an alternative solution to economical desalination and restoration of highly degraded saline and sodic soils through the process, called phytoremediation [16]. It is worth noting that molecular analyses, such as transcriptome sequencing, have been widely used to investigate the gene expression dynamics of numerous species in response to salt stress [17]. Nevertheless, exploration of salt tolerance mechanisms of halophyte species with the final goal of transferring their salt tolerance-associated traits and/or genes to traditional crops is timely and imperative, which may also contribute to ensuring sustainable agriculture in salinity-affected areas.
Considering the above facts, the present review focuses on the latest update of the adverse effects of salinity both on soils and plants, as well as the intrinsic physiological, biochemical, and molecular mechanisms that halophytes employ to adapt under salinity. In addition, mechanistic aspects of desalinization and phytoremediation potential of halophytes for improving agricultural output from salt-contaminated soils will also be appraised. Finally, a comprehensive update on transcriptomic studies in halophytes and the genetic potential of halophytes for enhancing salt tolerance in traditional crops will be discussed by considering the genes associated with salt tolerance-related traits of halophytes.
2. Adverse Effects of Salinity on Soil Properties
Salinity causes immense alterations in soil properties. For instance, high salt concentrations in soils may result in high bulk density, soil compaction, poor soil structure, clay dispersion, and surface crusting [18]. In addition, salinity-induced poor hydraulic conductivity can severely impede water infiltration, leading to poor growth performance and yield reduction in plants [19]. Accumulation of salt in soils also has detrimental effects on soil microbial communities and their enzymatic activities [20]. Because of their increased sensitivity to environmental perturbations, soil microbiomes and associated enzyme activities are regarded as early indicators for measuring the degree of soil degradation and changes in soil quality [21]. Microbial activities play critical roles in the decomposition of soil organic matter (OM), mineralization of soil, and stabilization of soil aggregates [22,23]. Salinity-induced osmotic imbalance can lead to water leakage from the microbial cells and, consequently, plasmolysis and death of microorganisms [24]. On the other hand, microorganisms may live in low osmotic potential environments by accumulating osmolytes, which is an energy-intensive process [25]. Soil enzymes, including ureases, dehydrogenases, CATs, alkaline phosphatases, and β-glucosidases, derived from soil microbes, plant root exudates, and plant and animal residues, play central roles in the regulation of all the processes involved in OM decomposition, as well as in the cycling of soil nutrients [26]. Despite their immense influence on the alteration of soil quality, only a few studies were conducted to assess the potentiality of these enzymes in changing soil quality in coastal areas. In addition, comprehensive information on how salinity influences soil quality is still ambiguous due to the intricate nature of soils [25]. Therefore, future research should focus on unraveling their intrinsic processes in a spatio-temporal manner.
3. Mechanisms of Salinity-Mediated Adverse Effects on Plants
Elevated soil salinity can hinder plant performance in several ways. Salinity-induced water paucity severely affects seed germination by lowering the osmotic potential of the soil water, which interrupts imbibition in plant seeds [27,28]. Any disruption in seed imbibition can alter enzyme activities and protein metabolism, perturb hormonal balance, retard seed reserve utilization, and very often distort the ultrastructure of cells, tissues, and organs [29,30,31]. Reduction of plant growth and biomass after germination is the foremost negative impact of soil salinity, which depends on salinity levels, duration of salt exposure and type of plant species [27]. Plant cells can lose water after plant exposure to salinity within a few seconds, and as a result, cell shrinkage occurs [32]. Cells regain their original volume over hours, but cell elongation rates are reduced [33]. Over days, cell division rates are also severely reduced, leading to decreased plant growth [34]. Salinity can also cause visible injuries in the oldest leaves, the demise of older and younger leaves, which may result in death of the plants before seed maturation [32,33]. Nevertheless, the reduced growth rate of plants under salinity is predominantly regulated by root responses, as roots are directly exposed to saline soils, and act as a key conduit for mineral translocation to aerial parts of the plant [34]. As mentioned above, salt stress lowers the water potential of soil solution, which also interrupts water conductivity of roots, ultimately leading to a reduction in water and mineral influx to the plant body [35].
High concentrations of Na+ and Cl− in soil solution restrict uptake and accumulation of macronutrients like Ca2+ and Mg2+, which perturbs signal transduction, membrane stability, chlorophyll synthesis, and stomatal opening [34,36]. Furthermore, salinity’s adverse effects on photosynthesis are intricately associated to the destruction of photosynthetic pigments, and the underlying facts are still not adequately elucidated. Exposure of plants to salt stress causes a decrease in stomatal density, stomatal conductance, photosynthetic activity, and carbon assimilation, but an increase in mesophyll resistance, thereby reducing light absorption efficiency of photosystem (PS)-I and PS-II [37]. Salt stress also suppresses the activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), a key enzyme involved in carbon dioxide (CO2) assimilation during the dark phase of photosynthesis [38]. In addition, salinity also induces the production of ROS, including hydroxyl radical (OH•), superoxide (O2•−), hydrogen peroxide (H2O2) and singlet oxygen (1O2), in cellular organelles like chloroplasts, mitochondria, peroxisomes, and endoplasmic reticulum [12,13]. The elevated levels of ROS can negatively affect plant growth and developmental processes by causing protein synthesis reduction, cell membrane destruction, genomic instability, and damage to photosynthetic apparatus [13,39]. Although Cl− is the most abundant anion in saline soils, its effects on plant performance, as well as its uptake and transport mechanisms within plants have still received less attention than those of Na+ [40]. As an essential micronutrient, Cl− plays a putative role in enzyme activation in the cytoplasm, oxygen evolution during photosynthesis, stabilization of membrane potential, and maintenance of turgor pressure and pH of the plant cells [40]. However, once excessively accumulated, Cl− has even more detrimental effects than Na+ in destabilizing the normal functions of the cells [40]. All above-mentioned salinity-induced adverse effects on plants trigger diminution of crop yield, although the intensity and damage vary depending on plant species.
4. Halophytes and Their Classification
Halophytes are plants that have the ability to survive and complete their life cycles in environments with high salinity, without suffering major negative impacts on their growth or development [15]. Based on their levels of tolerance to different types of saline soils, halophytes can broadly be categorized into two groups: obligate halophytes and facultative halophytes. Obligate halophytes can sustain their sufficient growth and development in high saline habitats containing salinity levels similar to seawater [41]. On the other hand, facultative halophytes are generally found in low saline habitats, although they can thrive in salt-devoid environments as well [42]. Additionally, Von Sengbusch [43] used the eco-physiological characteristics to distinguish obligate and facultative halophytes from habitat-independent halophytes. Habitat-independent halophytes are not necessarily native to saline habitats; they usually prefer to grow in salt-free soils but can cope with the saline conditions [43].
Nevertheless, the comprehensive classification of halophytes is still ambiguous because scientists defined halophytes by considering different aspects [44], including integration of anatomical observations, intensity of salinity and mechanisms of salt tolerance [45,46]. More recently, Santos et al. [47] developed a database of halophytes, namely eHALOPH (http://www.sussex.ac.uk/affiliates/halophytes/; accessed on 20 July 2021), which accumulates the information of plant species that can survive under 80 mM or higher salt concentrations. The authors classified the halophytes into seven categories: hydro-halophytes, chasmophytes, phreatophytes, psammophiles, xerophytes, weedy halophytes, and xero-halophytes. Hydro-halophytes typically grow in aquatic conditions or on wet soil; chasmophytes are found on cliff-tops, rocky and sandy seashores, and saltmarshes; phreatophytes are deep-rooted plants obtaining water from a deep underground source that may or may not be saline; psammophiles are usually found in sandy soils; xerophytes are adapted to extreme drought-prone areas; weedy halophyte species are predominantly invading and colonize highly disturbed sites or regions; and xero-halophytes are adapted to inland salt desert and saline habitats [47].
In addition, based on the physiological basis of salt tolerance as well as the accumulation and transportation of ions, Breckle et al. [48] classified halophytes into three major categories, including recretohalophytes, euhalophytes, and salt-exclusion halophytes (also referred as salt excluders). Recretohalophytes have unique salt-secreting structures, such as salt glands and salt bladders, whereas euhalophytes have the ability to dilute absorbed salts in their succulent leaves or stems. Salt excluders, on the other hand, can prevent the uptake of salt ions from soils or shed leaves containing toxic levels of salt [49,50]. With these complex classifications, it is important to acquire knowledge on their extent of salt tolerance levels, and what the intrinsic mechanisms they adopt to combat salinity effects.
5. Physiological Mechanisms Associated with Halophyte Adaptation to Soil Salinity
Comprehensive research on halophytic plants is crucial for a better understanding of their salt tolerance mechanisms and their exploration for breeding purposes. To withstand and continue to be grown well in a saline environment, halophytic plants employ different morphophysiological strategies, such as salt exclusion, salt excretion through specialized organs, i.e., salt glands and/or salt bladders, and dilution of salt ions by succulence. We will review these morphophysiological mechanisms in light of their functions in halophytes’ resistance to salt stress (Figure 2 and Figure 3; Table 1).
5.1. Salt Exclusion
Halophytic plants, particularly hydro-halophytes and some phreatophytes, were reported to exclude excessive salts by root system-induced ultrafiltration mechanisms. More specifically, installing apoplastic barriers at the roots improves bypass flow resistance, resulting in effective exclusion of salts from the roots and, as a result, a reduced accumulation of toxic ions in the aboveground shoots through the transpiration stream [51] (Figure 2; Table 1). The root endodermis and exodermis are two distinct cell barrier layers with highly specialized roles ascribed to two particular cell wall features: (a) the Casparian band (CB) and (b) the suberin lamella (SL) (Figure 2). The CB, a paracellular lignin deposition in the endodermis and exodermis cell walls, forces all apoplastic transport into the strictly regulated symplastic system [52,53]. Furthermore, SL, primarily made of long-chain fatty acids, permeates the whole exodermis/endodermis cell wall, providing a hydrophobic barrier that aids in ion and water uptake regulation [54,55]. SL is thought to play an essential role in preventing the direct movement of water and ions from the apoplast into the endodermal protoplasts [56,57]. It was recently discovered that SL acts as an apoplastic barrier at the site of lateral root emergence where CBs have not yet developed [57]. A limited number of studies have been performed on halophytes and shown that apoplastic barriers are crucial for salt adaptation of Suaeda maritima [58], Avicennia officinalis [59], S. salsa [60], and A. marina [61]. However, in comparison with CBs, there is less evidence for the role of SL in plant salt tolerance; and hence, more research is needed to assess the role of SL in ion exclusion from plants under salinity.
Importantly, salt overly sensitive 1 (SOS1) in the cortical cells plays an essential role in exclusion of Na+ from roots (Figure 2). Recently, Liu et al. [62] reported that S. salsa exhibited greater expression of S. salsa SOS1 (SsSOS1) in roots of intertidal S. salsa population than in those of inland S. salsa population, which was correlated with the increased exclusion of Na+ by roots; and thus, it could thrive in a high saline environment (Table 1). Similarly, greater expression of Karelinia caspia SOS1 (KcSOS1) gene in the roots of the recretohalophyte K. caspia was also observed [63] (Table 1). However, the mechanisms underlying root system-induced ultrafiltration under saline conditions remain elusive and require further in-depth investigations. Many reports have recommended that salt exclusion mechanism as a cure to reduce accumulations of toxic levels of Na+ and Cl− in plant bodies [64]. A strong argument could, however, be raised against this mechanism since it may elevate soil salinity levels by restriction of salt uptake by roots [65].
5.2. Salt Excretion
Halophytes, particularly recretohalophytes, directly secrete remarkably higher contents of salt ions onto the leaf surface, for dealing with maintenance of cellular ion homeostasis, through a unique epidermal structure called salt glands, making them superior compared with other classes of halophytes and non-halophytes [50] (Figure 3). Several species of the Limonium genus, including L. bicolor, L. aureum, L. gmelinii, L. otolepis, and L. sinuatum, are good examples of recretohalophytes, which boost salt tolerance by enhancing salt secretion via the salt glands [66,67,68] (Table 1). Species of another recretohalophyte genus, Tamarix, are salt-tolerant by increasing the density of salt glands and the rate of salt secretion per salt gland [69] (Table 1). However, salt glands in different halophytes species possess diverse structural characteristics, and the number of cells that make up salt glands are different [50]. Still, their common features include the followings: (i) salt glands are surrounded by a thickened cuticle; (ii) having many plasmodesmata between the cells, a large number of highly developed mitochondria and many small vesicles in the cytoplasm; and (iii) having no chloroplasts in the cells [50,70].
Multi-cellular salt glands varying from 4 to 40 cells are found in dicotyledonous recretohalophytes, including L. bicolor [66], whereas bi-cellular salt glands are prominent in monocotyledonous recretohalophytes of the Poaceae family [71]. Ions secreted through salt glands include various cations (Na+, K+, Ca2+, Mg2+, Fe2+, Mn2+, and Zn2+) and anions (Cl−, Br−, I−, SO42−, PO43−, and NO3−), depending largely on the environment [50]. The actual pathways of salt secretion from salt glands remain to be determined, although several possible pathways based on recent studies have been suggested by Yuan et al. [70] and Lu et al. [50]. Nevertheless, it was also demonstrated that salt glands might impose detrimental threats to plants by causing leaf dehydration because the glands produce salt crystals on the leaf surface [65]. This problem can be minimized through de novo synthesis of compatible solutes, but it requires high energy, i.e., 30 to 109 molecules of ATP to produce one molecule of any compatible solute, which ultimately leads to a penalty in crop yield [65].
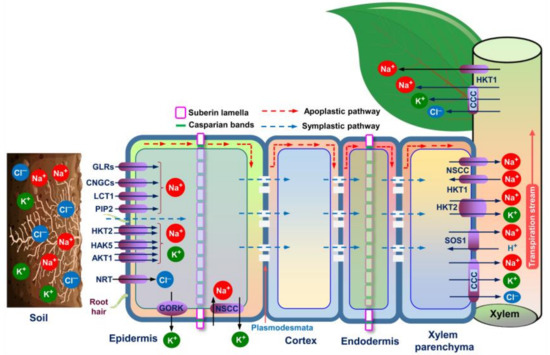
Figure 2.
Putative salt uptake and accumulation mechanisms in an ideal halophyte plant (modified from Zhao et al. [11], Arif et al. [34], and Arbelet-Bonnin et al. [72]. Na+ is transported from soils to/within plant root cells via two major pathways: symplastic and apoplastic. The major transporters/channels embedded in the root epidermis for up-taking Na+ are GLRs, CNGCs, LCT1, PIP2, HKT2, HAK5, and AKT1. In the presence of Na+, K+ is competitively uptaken via HKT2, HAK5, and AKT1. The entrance of Cl− into the root cells is mediated by separate transporters, including NRT. NSCC and GORK channels, on the other hand, are involved in salinity-induced K+ exclusion from the root epidermis. Apoplastic barriers, consisting of Casparian bands and suberin lamellae in the epidermis and endodermis of the roots, can block and/or limit the bypass flow of Na+ and other solutes moving to the aboveground parts. Passive Na+ entry into the xylem is facilitated by NSCCs, while cotransporters like SOS1, CCC, and HKT2 are required for active loading of Na+ into the xylem. Na+ can return to xylem parenchyma using HKT1. The xylem-loaded Na+, K+ and Cl− can be transported up through the transpiration stream, and subsequently enter into the aboveground organs via CCC and HKT1. Abbreviations: AKT1, Arabidopsis K+ transporter 1 (a shaker-type K+ channel); CCC, cation-chloride cotransporter; CNGC, cyclic nucleotide-gated channel; GORK, gated outwardly rectifying K+ channel; GLR, glutamate receptor-like channel; HKT, high-affinity potassium transporter; HAK, high-affinity potassium uptake transporter; LCT, low-affinity cation transporter; NRT, nitrate transporter; NSCC, non-selective cation channel; PIP2, plasma membrane intrinsic protein; SOS1, salt overly sensitive 1.
Epidermal bladder cells (EBCs), also known as salt bladders, can be found in more than half of the halophytes [65,73]. The EBC have a modified non-glandular trichome structure that is divided into an apical and basal cell, and the apical cell gains a globular shape, typically with a cell volume of 500 nL and an average diameter of 1 mm [65,73,74] (Figure 3). The diameter of EBCs often stretches ~10× larger in comparison with normal epidermal cells to sequestrate excessive salts, by as much as 1000× more than the traditional leaf cell vacuoles can do [65].
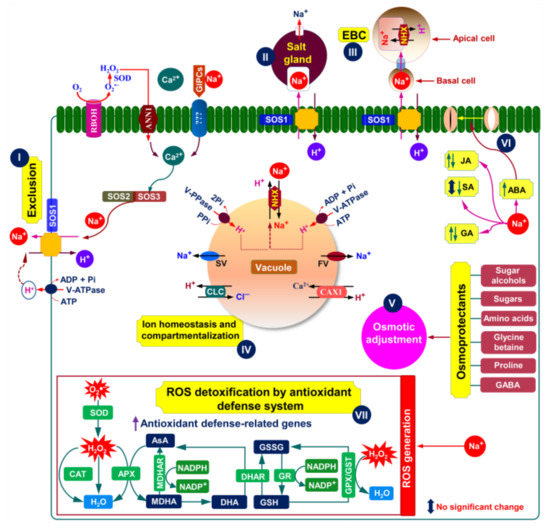
Figure 3.
Putative salt tolerance mechanisms in an ideal halophyte plant (modified from Zhao et al. [11], Arif et al. [34], Arbelet-Bonnin et al. [72], Tran et al. [75], and Himabindu et al. [76]. The absorption of Na+ in plant cells is counterbalanced by active Na+ extrusion through SOS1 Na+/H+ exchanger (I). The RBOH-mediated ROS formation in the plasma membrane can activate ANN1-induced Ca2+-signaling pathway (I). The Na+ bound GIPCs can trigger Ca2+ influx through an unknown Ca2+ channel (I). Ca2+ influx mediated by either ANN1 or GIPCs is essential for activating the SOS-signaling pathway (I). The SOS3 relays salt-triggered Ca2+ signals to SOS2 kinase, forming the SOS3-SOS2 protein complex that phosphorylates SOS1 to induce Na+ efflux (I). To maintain ion homeostasis, halophytes can also sequestrate excess Na+ into the vacuoles and/or excrete Na+ via two unique structures, namely salt glands and salt bladders (II and III). It is also possible to avoid excessive Na+ accumulation in the cytosol by sequestrating it into the vacuoles (IV). Predominantly, Na+-sequestration in the vacuoles is conferred by tonoplast-based Na+/H+ exchangers of the NHX family (e.g., NHX1 or NHX2), fueled by either H+-PPase or H+-ATPase pumps (IV). Another equally important mechanism of vacuolar Na+-sequestration is the efficient control of tonoplast slow vacuolar (SV) and fast vacuolar (FV) activating ion channels, which may allow vacuolar Na+ to leak back into the cytosol (IV). Once, if both channels are downregulated or blocked by choline, Na+ leakage is inhibited, resulting in an increased level of salt tolerance (IV). On the other hand, Cl− influx in the vacuoles is mediated by CLC (IV). To maintain cytosolic Ca2+ homeostasis under saline conditions, the influx of Ca2+ into the vacuole is mediated by CAX1 (IV). In response to salt stress, halophytes often accumulate various osmoprotectants, including proline, total free amino acids, sugars, polyols, glycine betaine, and GABA, which play vital roles in conferring an efficient osmotic adjustment (V). In addition, changes in the levels of different phytohormones may also help in stress adaptation; for instance, increased ABA level aids in controlling stomatal closure to maintain better water status at the tissue level (VI). It is worth mentioning that changes in the levels of phytohormones (e.g., JA, SA, and GA) in halophytes under salt stress varies dependently on the type of halophytes (VI). Furthermore, excessive salt accumulation can trigger oxidative stress via overproduction of ROS, such as O2•– and H2O2 (VII). To fight against ROS-mediated oxidative damage, halophytes induce a vibrant antioxidant defense system by activating both enzymatic (SOD, CAT, APX, MDHAR, DHAR, GR, GPX, and GST) and non-enzymatic (AsA and GSH) antioxidants (VII). Abbreviations: ABA, abscisic acid; ANN1, annexin1; APX, ascorbate peroxidase; AsA, ascorbic acid; CAT, catalase; CAX, Ca2+/H+ exchanger; CLC, chloride channel (H+/Cl– antiporter); DHA, dehydroascorbate; DHAR, dehydroascorbate reductase; EBC, epidermal bladder cell; GA, gibberellic acid; GIPC, glycosyl inositol phosphoryl ceramide; GST, glutathione S-transferase; GPX, glutathione peroxidase; GSH, glutathione; GSSG, oxidized glutathione; GR, glutathione reductase; GABA, gamma amino butyric acid; H2O2, hydrogen peroxide; JA, jasmonic acid; MDHA, monodehydroascorbate; MDHAR, monodehydroascorbate reductase; NHX, Na+/H+ exchanger; O2•–, superoxide; RBOH, respiratory burst oxidase homolog; ROS, reactive oxygen species; SOS, salt overly sensitive system; SA, salicylic acid; SOD, superoxide dismutase; V-PPase, vacuolar pyrophosphatase; V-ATPase, vacuolar H+ ATPase.
More fundamentally, EBCs have long been considered imperative for maintaining low Na+ levels in leaves, especially in young leaves [77]. Because younger leaves have small and underdeveloped vacuoles in the mesophyll cells, they rely solely on EBCs for internal salt sequestration [77]. Besides serving as a potential hotspot for storage of excessive Na+ and Cl−, EBCs may also store different metabolic compounds, ROS-scavenging metabolites and organic osmoprotectants [78]. A positive correlation between the presence and/or the number of EBCs and salt tolerance was reported in Chenopodium quinoa [74,78]. Some candidate genes related to EBC formation were recently identified using transcriptome analysis in C. quinoa, Mesembryanthemum crystallinum, and Atriplex centralasiatica species [79,80,81] (Table 1). More fundamentally, Imamura et al. [82] first identified a gene, named reduced epidermal bladder cells (REBC), involved in the EBC development in C. quinoa, laying the foundation for elucidating the EBC-associated stress tolerance mechanism in halophytes.
5.3. Succulence
Halophytes also adopt succulence mechanisms for ion homeostasis and accumulation of osmoprotectants to maintain cell turgor pressure under saline conditions [83,84] (Table 1). Succulent plants, popularly referred to as halo-succulents, have been characterized by thick leaves and stems, increased size of mesophyll cells, smaller intercellular space, enhanced tissue water content and high turgor pressure [83]. Succulent leaves possess more and stretched-sized mitochondria, which presumes to provide the excess energy required for salt compartmentalization and sequestration [84]. A large number of halophytes, including Salsola drummondii, Achras sapota, and Sarcocornia fruticosa, were reported to have efficient succulent mechanisms to confer salt tolerance via accumulation of elevated salts in their leaves and stems while maintaining improved photosynthetic efficiency [85,86,87] (Table 1). However, there is still much to learn about the ‘nuts and bolts’ of succulence. Therefore, current research should focus on (i) how the complex structural-functional coordination regulates the physiological processes in succulent plants; (ii) transcriptomics and gene-editing techniques to provide insights into the functional biology of succulent plants; and (iii) genetic engineering to introduce succulence traits into non-succulent plants to enhance their resilience to salinity-caused environmental perturbations.

Table 1.
List of selected halophyte species along with their adaptive mechanisms favorable for salt tolerance.
Table 1.
List of selected halophyte species along with their adaptive mechanisms favorable for salt tolerance.
| Sl. No. | Species Name | Maximum Salt Tolerance Limit (mM) | Salt Tolerance Mechanisms | References |
|---|---|---|---|---|
| 1 | Haloxylon salicornicum | 400 |
| [88] |
| 2 | Scorzonera hieraciifolia | 600 |
| [89] |
| 3 | Salsola crassa | 300 |
| [90] |
| 4 | Aster tripolium | 600 |
| [91] |
| 5 | Lycium humile | 750 |
| [92] |
| 6 | Limonium bicolor, L. gmelinii, L. otolepis, L. aureum, L. sinuatum, and L. gmelinii | 100, 50, 300, 350, 400, and 420, respectively |
| [66,67,68,69] |
| Tamarix gallica, T. ramosissima, and T. laxa | 300 | |||
| 7 | Chenopodium quinoa and Atriplex centralasiatica | 300–400 |
| [73,78,80,81] |
| 8 | Nitraria sibirica | 200 |
| [93] |
| 9 | Mesembryanthemum crystallinum | 400 |
| [75] |
| 10 | Lobularia maritima | 400 |
| [94] |
| 11 | C. quinoa | 400 |
| [95] |
| 12 | Urochondra setulosa | 500 |
| [96] |
| 13 | Cakile maritima | 400 |
| [97] |
| 14 | Karelinia caspia | 200 |
| [63] |
| 15 | Suaeda salsa | 200 |
| [98] |
| 16 | Hordeum brevisubulatum | 750 |
| [99] |
| 17 | Zoysia macrostachya | 300 |
| [100] |
| 18 | Sarcocornia fruticosa | 60 |
| [85,86,87] |
| Achras sapota | 120 | |||
| S. drummondii | 500 | |||
| 19 | H. salicornicum | 400 |
| [101] |
| 20 | A. tripolium | 300 |
| [102] |
| 21 | Leptochloa fusca | 450 |
| [103] |
| 22 | C. maritima | 400 |
| [104] |
| 23 | Eutrema parvula | 300 |
| [105] |
| 24 | Salvadora persica | 750 |
| [106] |
| 25 | Pongamia pinnata | 500 |
| [107] |
| 26 | A. halimus | 171 |
| [108] |
| 27 | Sesuvium portulacastrum | 120 |
| [109] |
| 28 | Zygophyllum xanthoxylum | 50 |
| [110] |
| 29 | Puccinellia tenuiflora | 150 |
| [111] |
| 30 | Kosteletzkya virginica | 390 |
| [112,113] |
Abbreviations: ABA, abscisic acid; AKT, Arabidopsis K+ transporter; AsA, ascorbate; APX, ascorbate peroxidase; BR, brassinosteroids; CAT, catalase; CCC, cation-chloride cotransporter; CLC, chloride channel; EC, electrical conductivity; GA3/GA4, gibberellic acid; GPX, glutathione peroxidase; GORK, gated outwardly rectifying K+ channel; GR, glutathione reductase; GSH, glutathione; HKT, high affinity potassium transporter; HAK, high affinity potassium uptake transporter; IAA, indole acetic acid; JA, jasmonic acid; KT, potassium transporter; Kmt, Shaker type-potassium channel; lmt, myo-inositol O-methyl transferase; NADP, nicotinamide adenine dinucleotide phosphate; NCED, 9-cis-epoxycarotenoid dioxygenase; NHX, Na+/H+ exchanger; NRT, nitrate transporter; OAT, ornithine-δ-aminotransferase; PMA, plasma membrane H+-ATPase; POD, peroxidase; PYL, pyrabactin resistance 1-like; P5CS, delta 1-pyrroline-5-carboxylate synthase; ROS, reactive oxygen species; SKOR, stelar K+ outward rectifier; SOS, salt overly sensitive; SOD, superoxide dismutase; TCA, tricarboxylic acid; TPK, two pore K+ channel; VAP, vesicle-associated protein; VAMP, vesicle-associated membrane protein; VHA, vacuolar H+-ATPase; Vmac, V-ATPase subunit c; VP, vacuolar H(+)-pyrophosphatase; ZR, zeatin riboside.
6. Biochemical Mechanisms Associated with Halophyte Adaptation to Salinity
Plants, including halophytes, adopt several biochemical strategies to adapt against salinity-caused adverse effects. For instance, ion homeostasis, osmotic adjustment, and ROS-detoxification are among the important biochemical mechanisms used by plants to protect their cellular constituents from the detrimental effects of excessive salt [76] (Figure 2 and Figure 3; Table 1). Here, we summarize these three biochemical mechanisms associated with halophyte responses and tolerance to salt stress.
6.1. Ion Homeostasis
Various channels and transporters are actively involved in accommodating Na+, Cl−, and K+ in the tissues of halophytic plants. It is well known that the uptake of Na+ from soils to roots occurs mainly via the apoplastic and symplastic pathways [34]. To transport salt ions via the symplastic pathway, transporters and channels are needed. Generally, non-selective cation channels (NSCCs), including cyclic nucleotide-gated channels (CNGCs) and glutamate receptor-like channels (GLRs), high-affinity K+ transporters (e.g., HKT2 and HAK5) and aquaporins, such as plasma membrane intrinsic protein (PIP) isoforms, are mainly responsible for root-Na+ uptake (Figure 2). The influx of Na+ can also be aided by low-affinity cation transporter (LCT1) and Arabidopsis K+ transporter 1 (AKT1) (Figure 2) [34]. The inward movement of Cl− in root cells is mediated by H+/Cl− symporters, Cl−/H+ co-transporters, and nitrate transporters (NRTs). During high salinity, several anion channels, including slow anion channels (SLAC channels), are involved in the passive transport of Cl− [114]. Cation chloride cotransporters (CCCs) help in Cl− influx in the xylem and shoot tissues [115]. The chloride channels (CLCs) found on tonoplast are responsible for Cl− sequestration [75] (Figure 3). In the presence of Na+, K+ is competitively uptaken via HKT2, HAK5, and AKT1 [11].
To prevent the accumulation of excess salt in their cytosol, halophytes mainly adopt two strategies: (i) prevention of salt entrance to the roots and (ii) vacuolar sequestration of salt ions [76] (Figure 3). However, these mechanisms are not mutually exclusive, and a particular plant may employ multiple strategies depending on the circumstances. For example, ion homeostasis in the cytosol is a unique strategy that plants follow to limit salt ion accumulation by compartmentalizing them effectively, which is essential for plant survival under salinity-triggered hostile conditions. The SOS system is crucial for perceiving salt-mediated Ca2+ signals, as well as for the exclusion of excess Na+ by roots, loading of Na+ into the xylem and sequestration of excess Na+ into the vacuoles using Na+/H+ exchangers (NHXs) [11,116] (Figure 3). The SOS system is predominantly comprised of SOS3/SOS3-like calcium-binding proteins (SCaBPs), SOS2 (a serine/threonine-protein kinase) and SOS1 (a plasma membrane Na+/H+ antiporter) [11,76] (Figure 3). Importantly, SOS4 (pyridoxal kinase) and SOS5 have recently been discovered, with SOS4 being involved in the biosynthesis of pyridoxal-5-phosphate (vitamin B6) that plays a key role in Na+/K+ homeostasis in plants, and SOS5 being a putative cell surface adhesion protein that is required for normal cell expansion [116]. Generally, SOS3 relays salt-triggered Ca2+ signals to SOS2 kinase, forming an SOS3–SOS2 protein complex that phosphorylates SOS1 to induce Na+ efflux [11,84] (Figure 3). The elevated expression of the SOS1 gene and its correlation with the improvement of salt tolerance have been reported in the obligate halophyte Cakile maritima [104], ornamental halophyte Lobularia maritima [94], and recretohalophyte Karelinia caspia [63] (Table 1). Likewise, Marriboina et al. [107] and Arbelet-Bonnin et al. [104] reported that the simultaneous expression of SOS1, SOS2, and SOS3 contributed to high salinity tolerance in the phreatophyte Pongamia pinnata and obligate halophyte C. maritima species, respectively (Table 1).
NHXs are also helpful in controlling vacuolar Na+ influx and maintaining optimum intracellular Na+/K+ homeostasis [76] (Figure 3; Table 1). For instance, under saline conditions, the perennial woody halophyte Nitraria sibirica could maintain an optimal K+/Na+ balance at the tissue- and cell-specific levels via vacuolar Na+ sequestration by boosting the activities of both vacuolar H+-ATPase and H+-PPase, and upregulating the expression of NsVHA, NsVP1, and NsNHX1 [93]. Meanwhile, N. sibirica increased the expression of two-pore K+ (NsTPK) at the transcriptional level, promoting K+ efflux from the vacuoles to the cytoplasm for cytosolic K+ homeostasis [93] (Table 1). In addition, Tran et al. [75] found that salt-exposed M. crystallinum enhanced the expression of genes encoding tonoplast antiporters H+/Cl− (i.e., McCLC1), Na+/H+ exchangers (i.e., McNHX1), and V-ATPase subunit c (i.e., McVmac1) for sequestration of Cl− and Na+ into the vacuoles for conferring salt tolerance in M. crystallinum (Table 1).
6.2. Osmotic Adjustment
Plants can attain osmotic adjustment by employing two strategies: (i) increased uptake of inorganic ions, and (ii) de novo synthesis of organic osmolytes to maintain turgor pressure and organellar volume within the growing plant cells [11]. Fundamentally, Na+ and Cl− in vacuoles, as well as K+ and compatible organic solutes like proline and sugars in the cytoplasm, are crucial to assure osmotic adjustment in the cells in a salt-caused osmotic environment (Figure 3) [117]. Generally, salinity-induced K+ efflux from the cell takes place via two major types of channels, including guard cell outward rectifying K+ (GORK) channels and/or ROS-activated NSCCs [11] (Figure 2). Although GORK channels play a central role in stress-induced K+ loss from the cytosol, comprehensive research into the molecular background of GORK channel-mediated osmotic balance in halophytes is still elusive. Nonetheless, by studying two contrasting C. quina accessions, Kiani-Pouya et al. [95] have recently revealed that the transcript levels of CqGORK in roots remained unchanged or decreased in salt-tolerant accession but increased in salt-sensitive accession, implying GORK’s role in K+ retention for better osmotic adjustment in the cells of salt-sensitive C. quina. Furthermore, the ion selectivity approach of HAKs/HKTs has been reported to confer greater uptake of K+ in Eutrema parvula [105] and Hordeum brevisubulatum [99] and, thus, contributed to improved K+/Na+ homeostasis (Table 1). Nonetheless, future studies should be taken to decipher how the movement of K+ between intra-cellular and inter-cellular membranes by K+ transporters contribute to the adjustment of the osmotic environment in salt-exposed halophytic plants.
Halophytes often produce a multitude of osmoprotectant molecules, including amino acids, soluble sugars, sugar alcohols and glycine betaine to maintain osmotic balance under salinity [118] (Figure 3; Table 1). The accumulation of proline and the elevated expression of delta 1-pyrroline-5-carboxylate synthase (P5CS) gene involved in proline biosynthesis, were associated with improved salt tolerance in M. crystallinum [75] and Urochondra setulosa [96] (Table 1). In addition, salt tolerance by glycine betaine has also been recorded in Atriplex halimus [108] and Sesuvium portulacastrum [109] (Table 1). It is worth mentioning that metabolite profiling in halophytes has recently gained momentum as a means of gaining a comprehensive understanding of the levels of accumulated osmolytes in relation to salt tolerance. For instance, Panda et al. [88] and Kumari and Parida [106] uncovered the salt-responsive metabolites and metabolic pathways in xero-halophyte Haloxylon salicornicum and facultative halophyte Salvadora persica plant, respectively (Table 1). They found that salt-exposed H. salicornicum and S. persica had greater levels of sugars, sugar alcohols, and amino acids, indicating that these osmolytes might contribute to the osmotic adjustment, resulting in salt tolerance in these plants (Table 1). Furthermore, metabolite profiling of C. maritima exposed to 400 mM sodium chloride (NaCl) showed that proline, glycine betaine and gamma amino butyric acid (GABA) significantly accumulated, while sugar levels surprisingly decreased, indicating that amino acids and amino acids-derived molecules played the primary roles in osmoprotection to cope with salt stress in this halophyte [97] (Table 1). Nonetheless, further exploration of metabolite changes in other halophytes should be undertaken to deepen our existing knowledge on putative salt tolerance mechanisms in halophytes.
6.3. ROS-Detoxification
Plants rely on efficient antioxidant defense mechanisms to protect them from oxidative stress, induced by the overaccumulation of ROS under elevated salinity [119]. To alleviate ROS-induced damage, plants possess a robust antioxidant defense system, which comprises of non-enzymatic antioxidants like flavonoids, AsA, GSH, and tocopherols, and enzymatic antioxidants, such as SOD, CAT, APX, glutathione reductase (GR), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), GPX, and GST [39] (Figure 3). The improved salt tolerance of various halophytes is associated with their ability to maintain ROS homeostasis by a proper activation of their antioxidant system under salt stress [89,90,94,100,101,120]. For example, in H. salicornicum plant, the higher activity of SOD, higher level of GSH and higher ratios of AsA/dehydroascorbate (DHA) and GSH/oxidized glutathione (GSSG) play a pivotal role in scavenging of ROS to withstand oxidative stress and improve salt tolerance [101] (Table 1).
In response to salt stress, halophytic plants modulate the levels of endogenous phytohormones in order to regulate various defense mechanisms, including antioxidant defense [91,121]. Detailed roles of phytohormones in halophyte responses to abiotic stresses have been reviewed in-depth elsewhere [121]. In halophytes, phytohormone accumulation varies depending on the types of halophytes exposed to salt stress. For instance, jasmonic acid (JA) content increased in S. persica [106] but dropped in Aster tripolium [91] and H. salicornicum [88] under salt stress (Figure 3, Table 1). The levels of salicylic acid did not alter significantly in S. persica [106] and H. salicornicum [88], while being remarkably dropped in A. tripolium [91] (Figure 3). The content of gibberellic acid (GA) increased noticeably in S. salsa [98], while it decreased in H. salicornicum [88] (Figure 3, Table 1). Because JA can be produced via linolenic acid oxidation [122], the JA level in plants is linked to the amount of ROS and the rate of ROS-induced lipid peroxidation [123]. Indeed, a lower level of JA was positively correlated with the reduced level of ROS under salt stress [88]. On the other hand, the level of abscisic acid (ABA), which acts as a critical hormone in stomatal closure to reduce transpiration, significantly increased in Lycium humile, S. persica, H. salicornicum and A. tripolium in response to salt stress [88,91,92,106] (Table 1). Panda et al. [88] reported that enhanced ABA accumulation helped in reduction of oxidative stress in salt-exposed H. salicornicum. The generation of ROS and subsequent redox processing are an integral part of hormone regulation and have a role in controlling plant development and stress tolerance [124]. Therefore, future studies should focus on spatial-temporal regulation of ROS generation and identification of proteins that could detect changes in ROS. Such information may help us further understand the crosstalk among multiple hormones signaling pathways in halophytes, and their hormone-modulated stress-tolerant phenotype.
7. Halophyte-Mediated Phytoremediation of Salinity-Affected Soils
Extensively used methods to reclaim salt-affected areas are leaching and adding chemicals and organic amendments [125]. Leaching facilitates the movement of salt particles from the surface horizon to the deeper horizon. However, lack of water during leaching and its effects on reduced soil stability, contents of total nitrogen and total organic carbon, microbial functions and overall soil fertility limit its practice in salt-contaminated areas [126]. Chemical approach involves an ion exchange process through existing and/or adding calcium carbonate (CaCO3) or other chemical substitutes to the soils; however, poor solubility and quality and high prices restrain further expansion of chemical uses in reclaiming of saline soils [126,127]. Interestingly, optimal rates of the different organic amendment (not more than 50 T ha−1) can efficiently increase dissolution of CaCO3 [128], as well as improve soil-physical (e.g., structure, permeability, water holding capacity, etc.) and chemical properties (e.g., pH, cation exchange capacity, etc.) that favors plant growth and microbial activities [129]. However, benefits are restrained due to the risk of releasing various chemicals, including heavy metals, phenolic compounds, ethylene and ammonia, excess salts, and organic acids from uncategorized organic wastage [129,130,131,132]. All of these constraints have forced the farmers to look for alternative options that are environment-friendly, cost-effective, and aesthetically pleasing.
Phytoremediation, popularly known as vegetative bioremediation or biological reclamation, is a promising option over other remediation techniques, which involves cultivating certain salt-tolerant species to remove salt ions from the soil to restore salinity-degraded soils [20,126,133]. Its acceptance is increasing progressively due to numerous benefits. For example, phytoremediation (i) is economically viable, as it does not require the purchase of chemicals; (ii) enhances nutrient availability for plants in soil, which helps boost crop yield; (iii) removes excess salts more uniformly, even from higher depth when compared with gypsum; and (iv) is environment-friendly as it sequestrates carbon in both above- and below-ground biomasses [47]. Unfortunately, information regarding mechanistic insights into phytoremediation of saline soils using halophytes is still elusive. Because of complicated interactions, it is challenging to visualize plants’ complete and holistic mode-of-action in the saline soil remediation process, and their beneficial effects on soil properties. According to our current knowledge, we have illustrated a well-established and recognized underlying phytoremediation mechanism of halophytes in Figure 4.
First, halophytic plants can help remove salts by taking salt ions through root system and store them in aboveground plant parts (Figure 4). S. salsa has recently been successfully used to reclaim saline soil in the northwest of China due to its salt extraction potential, which ranges from 3.75 to 3.91 T ha−1 year−1 [133]. It is important to note that S. salsa can theoretically remove 3.0–3.8 T Na+ ha−1 with a population density of 15 plants m−2 [134]. Liang and Shi [134] also reported that the aboveground biomass of S. salsa plants accumulated approximately 27 mg of salt g−1 dry weight. Shaygan et al. [135] demonstrated that the shoots of Tecticornia pergranulata could accumulate 98 g of salt Kg−1 dry weight. Sodium chloride is often used to lower the freezing point of water on North American highways to prevent the risk of vehicle crashes and injuries [136]. The same study [136] showed that A. patula would take six years to remove all the salt from an area contaminated with 1540 g Cl− per m2, if salt application was halted. On the other hand, since salt is applied every winter, A. patula would be a potential candidate for the management of salt amount accumulated over time in the roadside soil and groundwater.
Second, some studies suggest that planting halophytes could reduce salt ion accumulation in soils by improving soil physical properties. Briefly, the deep-rooted halophytic plants have the potential to act as tillage tools. This is popularly referred to as biological drilling and has been reported to stabilize soil structure by reducing soil bulk density, thereby increasing soil porosity and hydraulic properties that augment Na+ leaching and replace it with other cations, besides inducing higher nutrient use efficiency [137,138] (Figure 4). In line with this finding, planting S. salsa with cotton (Gossypium hirsutum) as an intercropping system significantly decreased soil salinity and bulk density, while increasing soil porosity, soil organic carbon, root growth, total aboveground biomass, and cotton yield, when compared with the traditional monocropping system [134]. In addition, A. nummularia showed improvement of hydraulic conductivity and reduction in bulk density, which led to enhanced macropores and water infiltration rate under 40.86 dS m−1 salinity level [138]. Ashraf et al. [139] also reported that planting halophytes reduced bulk density, while it increased soil porosity and hydraulic conductivity, which eventually improved salt ion leaching from saline-alkali soil.
Third, root respiration and decomposition of organic matter play a pivotal role in reducing Na+ content in the saline-sodic soils because they can increase CO2 concentration in the soil atmosphere [140]. In brief, irrigation water and soil moisture-induced availability of soil water react with CO2 to form carbonic acid (H2CO3) (Figure 4). The H2CO3 dissociation and N2-fixation in halophyte roots lead to the release of the proton (H+), which causes dissolution of calcite (CaCO3) to produce Ca2+, H2O and CO2 [140] (Figure 4). The released Ca2+ in the soil could facilitate the removal of Na+ from the cation exchange sites of soil colloid [140].The exchanged Na+ content in soils is then eliminated via two routes: (i) uptake through halophytic plant roots, and (ii) leaching as a result of flooding irrigation prior to sowing, which ultimately contributes to the reduction in soil sodicity and salinity (Figure 4).
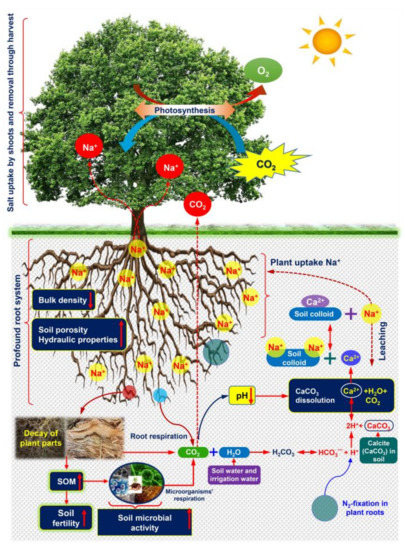
Figure 4.
Putative phytoremediation process by halophyte plants in salinity-affected areas, and their beneficial effects on soil properties (modified from Qadir et al. [140] and Jesus et al. [126]. Deep-rooted plants help reduce soil bulk density and increase soil porosity, enabling storage of excessive salt ions in deeper soil layers through leaching. On the other hand, the reclamation of saline soils needs a source of Ca2+ that replaces excessive Na+ from the cation exchange sites of soil colloid. Decay of plant parts enhances soil organic matter (SOM) content, leading to the improvement of soil fertility. The microbes-mediated SOM decomposition and root respiration increase the concentration of carbon dioxide (CO2) in the soil atmosphere. Water (H2O) from irrigation and soil moisture sources reacts with CO2 to generate carbonic acid (H2CO3). In halophyte roots, H2CO3 dissociation and N2-fixation release proton (H+) that enhances dissolution of calcium carbonate (CaCO3) to release Ca2+, H2O and CO2. The released Ca2+ could help remove Na+ from cation exchange sites of soil colloid. Finally, the exchanged Na+ can be uptaken by roots, and compartmentalized in aboveground shoots or leached into the deeper layer of soils via irrigation. In non-calcareous soils, an increase in CO2 induces the release of H+, which results in a decrease in the pH of the soil. The acidic nature of soil enhances the dissolution of CaCO3, which helps reduce the amount of salt ions in soils via Ca2+ exchange with Na+.
Nonetheless, in non-calcareous soils, augmentation of CO2 results in the lowering of soil pH that enhances dissolution of CaCO3 to make Ca2+ available to interchange with Na+ [140]. Planting two facultative halophytes, S. verrucosum and Bacopa monnieri, for 240 days reduced the electrical conductivity (EC) value from 11.13 to 7.97 dS m−1, pH from 7.84 to 7.42, while improving soil porosity from 54.71 to 57.23%. Notably, the combined plantation of these two halophytes had a phytodesalination capacity of 1.21 T Na+ ha−1, which helped prepare the soil for Zea mays to thrive and boosted yields from 7.2 to 8.5 T ha−1 [141]. S. monoica, a salt marsh halophyte, has been shown to lower the soil EC value from 4.75 to 2.10 dS m−1 and pH from 8.40 to 6.73 [142].
However, the use of halophytes as a desalinization tool cannot be guaranteed in all situations, since this approach is affected by environmental inconsistencies, such as rainfall and temperature, or cultivation modes, such as irrigation and non-irrigation, as well as the risk of recycling back of the salt particles through plant parts [143]. Therefore, robust assessments of the potential of various halophytes by analyzing their mode-of-action are needed prior to their application in the desalinization of salt-affected areas. In addition, further research will be required to testify the suitability of the plant-uptake-mediated phytoremediation process, and if found significant, it will open a new avenue for restoring salinity-affected areas, notably non-calcareous soils and soils under non-leaching circumstances.
8. Potential of Halophytes as Genetic Resources for the Engineering of Crops with Improved Salt Tolerance
Halophytes can survive better under ambient salinity than glycophytes, which might be attributable to proper modulation of genes that play crucial roles in triggering their salt tolerance mechanisms [76,144]. High-throughput transcriptome sequencing is increasingly being used to understand the responses of plants to abiotic stresses at gene expression level [145]. Recently, Guo et al. [146] conducted a comparative transcriptome analysis of the model halophyte Puccinellia tenuiflora and several glycophytic Gramineae plants to uncover their underlying molecular responses to excessive salt stress. They found that the greater expression of the gene families involved in K+ uptake, sucrose biosynthesis, pentose phosphate pathway and flavonoid biosynthesis might be associated with better growth of P. tenuiflora under high salinity conditions. In Salicornia persica, Aliakbari et al. [147] identified 1595 differentially expressed genes (DEGs) between the control and salt-treated plant shoots, including those encoding transcription factors, transporters, and protein kinases. Functional annotation analyses indicated that improved energy homeostasis and the synthesis of primary metabolites might play pivotal roles in salt tolerance of S. persica. Gene network analysis revealed that Ca+ signaling, ABA signaling, and Na+ compartmentalization are essential components in conferring salt tolerance in S. persica. Han et al. [148] identified 399 upregulated and 327 downregulated genes in S. rigida plants in responses to different regimes of soil salinity (100, 300, and 500 mM NaCl). Gene ontology (GO) enrichment analysis suggested that the downregulated genes were related to organic substance transport, nitrogen compound transport and intracellular protein transport. In contrast, the upregulated genes were involved in cell wall biogenesis, such as cell wall assembly, extracellular matrix organization, and cell wall formation [148].
Moreover, RNA-seq identified 9144 DEGs in A. centralasiatica leaves under salt stress, with 3819 upregulated and 5325 downregulated genes [81]. GO enrichment analysis showed that salt stress dramatically increased the expression of genes associated with ion transport, ABA-dependent signaling pathway, ROS-scavenging and transcriptional regulation (HB-7 and MYB78) pathway [81]. A de novo transcriptome profiling in the halophyte U. setulosa revealed the upregulation of genes encoding photosynthetic enzymes, transcription factors, mitogen-activated protein kinases (MAPKs), transporter proteins, cell membrane proteins, antioxidant enzymes, and enzymes for synthesis of compatible solutes by salinity [96]. Wang et al. [100] revealed many DEGs, specifically 4903 upregulated and 3800 downregulated genes in Z. macrostachya, in response to 300 mM NaCl. Such expression change in a large number of genes may contribute to improved plant growth performance by modulating plant hormone signal transduction, ion homeostasis via salt secretion, and osmoregulatory substance accumulation and prevention of oxidative damage through the activation of an efficient ROS-scavenging system.
Because they are genetically equipped with salt tolerance potential, halophytes could be a vital genetic resource for developing transgenic plants with better adaptability to salt stress. In fact, many transgenic plants have been developed via the introduction of genes from halophytic plants to enhance salt tolerance. Most of these genes encode antiporters, ion transporters, ROS scavengers, antioxidants, and proteins that play crucial roles in plants′ signal transduction and functional stabilization [149,150]. For example, transgenic poplar plant ectopically expressing a N. sibirica vacuolar Na+/H+ antiporter gene (NsNHX1) displayed a more remarkable improvement in salt tolerance through improved compartmentalization of Na+, more efficient photosynthesis, greater activity of antioxidant enzymes, and enhanced osmotic adjustment [151] (Table 2). Ectopic overexpression of a high-affinity K+ transporter-encoding gene, SbHKT1, from S. bigelovii in cotton improved salt tolerance of transgenic plants by enhancing K+ uptake capacity, K+/Na+ homeostasis, and the scavenging of ROS through increased activities of antioxidant enzymes, including SOD, POD, and CAT [152]. On the other hand, transgenic Arabidopsis ectopically expressing Zygophyllum xanthoxylum nitrate transporter 1-encoding gene ZxNRT1.5 displayed improvement in NO3− uptake and root-to-shoot NO3− translocation, which eventually led to increased biosynthesis of amino acids in the transgenic plants that helped in an osmotic adjustment under salt stress [153,154]. Further studies are required to uncover the comprehensive molecular mechanisms underlying ZxNRT1.5 involvement in regulating Na+, K+, and Cl− transport. In addition, ectopic overexpression of the N. tangutorum calcineurin B-like protein-interacting protein kinase 11 (NtCIPK11) gene from N. tangutorum in Arabidopsis improved seed germination and root length, as well as altered the transcription of genes encoding key enzymes involved in proline metabolism and K+ transportation in transgenic plants subjected to salt stress [155,156] (Table 2). Transgenic tobacco plants ectopically expressing several novel genes from S. brachiata, such as S. brachiata stress-related protein (SbSRP), S. brachiata salt inducible-1 (SbSI-1), S. brachiata myeloblastoma15 (SbMYB15), S. brachiata dehydration-responsive element-binding 2A (SbDREB2A), and S. brachiata RNA poly III complex 5-like subunit (SbRPC5L), and Aeluropus lagopoides, such as AlRab7 (AlRabring7), displayed higher accumulations of phytohormones (e.g., indole acetic acid) and compatible solutes (e.g., proline), improved gas exchange features, reduced membrane damage, improved antioxidant capacity, lower accumulations of toxic ions, ultimately resulting in better biomass and growth performance of transgenic plants under salt stress [157,158,159,160,161,162] (Table 2). In addition, ectopic expression of L. maritima vacuolar H+-ATPase subunit E1 (LmVHA-E1) from L. maritima, Tamarix hispida dehydration-responsive element-binding (ThDREB) from T. hispida, PutNHX1 or PutAPX from P. tenuiflora, or SeNHX1 from S. europaea in Arabidopsis plants improved ROS-scavenging, as well as compartmentalization of toxic Na+ into vacuoles to enhance salt tolerance of transgenic plants [150,163,164] (Table 2). Furthermore, transgenic tobacco harboring Iris lactea tonoplast Na+/H+ antiporter gene IlNHX showed elevation in the compartmentalization of Na+ into vacuoles via enhanced vacuolar V-ATPase activity, which contributed to the maintenance of K+/Na+ homeostasis [165] (Table 2).

Table 2.
Improvement of salt tolerance in plants by overexpressing various genes isolated from different halophytes.
Halophytes with intrinsic and efficient salt tolerance abilities can be used as models for improving crop and/or plant responses to salt stress (Table 2). Furthermore, because crops are predominantly glycophytes, it is critical to highlight the molecular differences between these two plant groups. Such findings could be the key to guiding and optimizing glycophyte transformation using halophytic candidate genes, thereby providing a new avenue for study aimed at preparing crops to cope with elevated soil salinity. However, despite their impressive implication, information regarding identification, metabolic functions, and isolation of crucial genes from halophytes that are associated with salt tolerance is still limited. Therefore, identification and characterization of the novel salt tolerance-associated genes from halophytes, and their subsequent utilization in genetic engineering are important areas of plant research, which deserves more attention to sustainably increase crop production for achieving global food security under the era of climate change.
9. Conclusions and Future Perspectives
Salt-induced land degradation restrains crop production and, thus, endangers sustainable food production and food security in the world. Climate-driven changes and anthropogenic activities have aggravated salinity problems, pressing the need to cultivate plant species that can withstand ambient salinity. Halophytes are a diverse group of plants with excellent adaptabilities to high salinity at both cell and whole-plant levels, and thus, have significant potential to meet this such cultivation challenge and be successfully integrated into future farming systems. Several halophytes are able to grow naturally in ambient salinity through the adoption of intrinsic salt tolerance mechanisms. These halophytes could be used as potent commercial alternatives to conventional crops, although commercialization of halophytic products has just started. At the same time, the cultivation of halophytic species in salinity-affected areas can reduce salt contamination of soils. Thus, this phytoremediation technique could be the most potent and viable strategy to remediate salinity-affected lands and facilitate crop production in salt-prone areas.
Despite its tremendous potential in soil salinity management, phytoremediation has limitations since it is a time-consuming process and requires screening out of suitable species. Therefore, future research should focus on sorting out halophytic plants with multiple salt tolerance mechanisms, which can produce higher biomass for removing the maximum quantity of salts from contaminated soils. Furthermore, despite tremendous research advances on halophytes (for more than 100 years), some basic physiological, biochemical and molecular mechanisms are still unclear. Hence, in-depth research is eminently needed to unravel halophytes’ underlying salt tolerance mechanisms for better soil conservation. Furthermore, recent genetic and omic studies showed that salt tolerance is a complex trait that depends on whole plant responses rather than the action of a single gene or protein. Therefore, exploration of the salt tolerance potential of various halophytes and the development of transgenic plants with superior remediation features are crucial for sustainable agriculture in salinity-affected areas to reduce the curse of global climate change.
Author Contributions
Conceptualization, M.M.R., M.G.M., and L.S.-P.T.; data curation, M.N.S., M.M.U.A. and M.A.R.; writing—original draft preparation, M.M.R. and S.S.K.; writing—review and editing, M.M.R., M.G.M. and L.S.-P.T. Figures and tables, M.M.R., A.K.D. and S.S.K.; supervision, M.G.M. and L.S.-P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors gratefully acknowledge Sharmin Sultana, the Institute of Biotechnology and Genetic Engineering, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur 1706, Bangladesh, for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, J.; Shi, H. Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 2013, 115, 1–22. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Calone, R.; Bregaglio, S.; Sanoubar, R.; Noli, E.; Lambertini, C.; Barbanti, L. Physiological adaptation to water salinity in six wild halophytes suitable for Mediterranean agriculture. Plants 2021, 10, 309. [Google Scholar] [CrossRef]
- Minhas, P.; Qadir, M.; Yadav, R. Groundwater irrigation induced soil sodification and response options. Agric. Water Manag. 2019, 215, 74–85. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, D.; Li, Y.; Li, X. Soil salinity evolution and its relationship with dynamics of groundwater in the oasis of inland river basins: Case study from the Fubei region of Xinjiang Province, China. Environ. Monit. Assess. 2007, 140, 291–302. [Google Scholar] [CrossRef]
- Omuto, C.T.V.; Vargas, R.R.; El Mobarak, A.M.; Mohamed, N.; Viatkin, K.; Yigini, Y. Mapping of Salt-Affected Soils: Technical Manual; FAO: Rome, Italy, 2020. [Google Scholar]
- Rahman, M.; Haque, A.; Nihad, S.A.I.; Howlader, R.A.; Akand, M.H. Morpho-physiological response of Acacia auriculiformis as influenced by seawater induced salinity stress. For. Syst. 2016, 25, e071. [Google Scholar] [CrossRef] [Green Version]
- Karakas, S.; Bolat, I.; Dikilitas, M. The use of halophytic companion plant (Portulaca oleracea L.) on some growth, fruit, and biochemical parameters of strawberry plants under salt stress. Horticulturae 2021, 7, 63. [Google Scholar] [CrossRef]
- Wicke, B.; Smeets, E.; Dornburg, V.; Vashev, B.; Gaiser, T.; Turkenburg, W.; Faaij, A. The global technical and economic potential of bioenergy from salt-affected soils. Energy Environ. Sci. 2011, 4, 2669–2681. [Google Scholar]
- Rahman, M.; Mostofa, M.G.; Rahman, A.; Islam, R.; Keya, S.S.; Das, A.K.; Miah, G.; Kawser, A.Q.M.R.; Ahsan, S.M.; Hashem, A.; et al. Acetic acid: A cost-effective agent for mitigation of seawater-induced salt toxicity in mung bean. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1. [Google Scholar] [CrossRef]
- Zaid, A.; Wani, S.H. Reactive oxygen species generation, scavenging and signaling in plant defense responses. In Bioactive Molecules in Plant Defense; Springer Science and Business Media LLC: Berlin, Germany, 2019; pp. 111–132. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Rahman, M.; Ansary, M.U.; Keya, S.S.; Abdelrahman, M.; Miah, G.; Tran, L.-S.P. Silicon in mitigation of abiotic stress-induced oxidative damage in plants. Crit. Rev. Biotechnol. 2021, 41, 918–934. [Google Scholar] [CrossRef]
- Patel, M.K.; Kumar, M.; Li, W.; Luo, Y.; Burritt, D.J.; Alkan, N.; Tran, L.-S.P. Enhancing salt tolerance of plants: From metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cells 2020, 9, 2492. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Litalien, A.; Zeeb, B. Curing the earth: A review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.-W.; Wong, F.-L.; Yung, W.-S.; Ku, Y.-S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.-Y.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2018, 42, 98–114. [Google Scholar] [CrossRef] [Green Version]
- Rengasamy, P. Soil chemistry factors confounding crop salinity tolerance—A review. Agronomy 2016, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Feng, S.; Wang, J.; Huo, Z.; Ji, Q. Effects of irrigation water salinity on soil salt content distribution, soil physical properties and water use efficiency of maize for seed production in arid Northwest China. Int. J. Agric. Biol. Eng. 2018, 11, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Tian, L.; Chang, C.; Wang, S.; Zhang, J.; Zhou, X.; Li, X.; Tran, L.-S.P.; Tian, C. Grass and maize vegetation systems restore saline-sodic soils in the Songnen Plain of northeast China. Land Degrad. Dev. 2018, 29, 1107–1119. [Google Scholar] [CrossRef]
- Guangming, L.; Xuechen, Z.; Xiuping, W.; Hongbo, S.; Jingsong, Y.; Xiangping, W. Soil enzymes as indicators of saline soil fertility under various soil amendments. Agric. Ecosyst. Environ. 2017, 237, 274–279. [Google Scholar] [CrossRef]
- Zhang, W.-w.; Chong, W.; Rui, X.; Wang, L.-j. Effects of salinity on the soil microbial community and soil fertility. J. Integr. Agric. 2019, 18, 1360–1368. [Google Scholar]
- Tian, L.; Lin, X.; Tian, J.; Ji, L.; Chen, Y.; Tran, L.-S.P.; Tian, C. Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 2020, 21, 1792. [Google Scholar] [CrossRef] [Green Version]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef] [Green Version]
- Rath, K.M.; Rousk, J. Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: A review. Soil Biol. Biochem. 2015, 81, 108–123. [Google Scholar] [CrossRef]
- Xie, X.; Pu, L.; Wang, Q.; Zhu, M.; Xu, Y.; Zhang, M. Response of soil physicochemical properties and enzyme activities to long-term reclamation of coastal saline soil, Eastern China. Sci. Total Environ. 2017, 607–608, 1419–1427. [Google Scholar] [CrossRef]
- Uçarlı, C. Effects of salinity on seed germination and early seedling stage. In Abiotic Stress in Plants; IntechOpen: London, UK, 2020. [Google Scholar]
- El-Keblawy, A.; Aljasmi, M.; Gairola, S.; Mosa, K.A.; Hameed, A. Provenance determines salinity tolerance and germination requirements of the multipurpose tree Prosopis juliflora seeds. Arid Land Res. Manag. 2021, 1–16. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Alencar, N.L.M.; Gadelha, C.G.; Gallão, M.I.; Dolder, M.A.H.; Prisco, J.T.; Gomes-Filho, E. Ultrastructural and biochemical changes induced by salt stress in Jatropha curcas seeds during germination and seedling development. Funct. Plant Biol. 2015, 42, 865–874. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B.; Zhang, X.-Q.; Li, C. Genome- wide association study of salinity tolerance during germination in barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Basu, S.; Kumar, A.; Benazir, I.; Kumar, G. Reassessing the role of ion homeostasis for improving salinity tolerance in crop plants. Physiol. Plant. 2020, 171, 502–519. [Google Scholar] [CrossRef]
- Geilfus, C.-M. Chloride in soil: From nutrient to soil pollutant. Environ. Exp. Bot. 2018, 157, 299–309. [Google Scholar] [CrossRef]
- Gururani, M.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [Green Version]
- Miteva, T.; Zhelev, N.Z.; Popova, L. Effect of salinity on the synthesis of ribulose-1,5-bisphosphate carboxylase/oxygenase in barley leaves. J. Plant Physiol. 1992, 140, 46–51. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Geilfus, C.-M. Chloride: From nutrient to toxicant. Plant Cell Physiol. 2018, 59, 877–886. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef] [Green Version]
- Parida, A.K.; Jha, B. Salt tolerance mechanisms in mangroves: A review. Trees 2010, 24, 199–217. [Google Scholar] [CrossRef]
- Von Sengbusch, P. Halophytes Botanik Online; University of Hamburg: Hamburg, Germany, 2003. [Google Scholar]
- Grigore, M.N.; Toma, C.; Boscaiu, M. Dealing with halophytes: An old problem, the same continuous exciting challenge. Analele Stiintifice ale Universitatii” Alexandru Ioan Cuza” din Iasi. Biol. Veg. 2010, 56, 21–32. [Google Scholar]
- Grigore, M.-N.; Toma, C. A proposal for a new halophytes classification, based on integrative anatomy observations. Muz. Olten. Craiova Stud. Com. Şt. Nat. 2010, 26, 45–50. [Google Scholar]
- Breckle, S.-W. Salinity, halophytes and salt affected natural ecosystems. In Salinity: Environment-Plants-Molecules; Springer: Dordrecht, The Netherlands, 2002; pp. 53–77. [Google Scholar]
- Santos, J.; Al-Azzawi, M.; Aronson, J.; Flowers, T.J. eHALOPH a database of salt-tolerant plants: Helping put halophytes to work. Plant Cell Physiol. 2015, 57, e10. [Google Scholar] [CrossRef] [Green Version]
- Breckle, S. How do halophytes overcome salinity. Biol. Salt Toler. Plants 1995, 23, 199–203. [Google Scholar]
- Chen, M.; Yang, Z.; Liu, J.; Zhu, T.; Wei, X.; Fan, H.; Wang, B. Adaptation mechanism of salt excluders under saline conditions and its applications. Int. J. Mol. Sci. 2018, 19, 3668. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Yuan, F.; Guo, J.; Han, G.; Wang, C.; Chen, M.; Wang, B. Current understanding of role of vesicular transport in salt secretion by salt glands in recretohalophytes. Int. J. Mol. Sci. 2021, 22, 2203. [Google Scholar] [CrossRef]
- Hussain, S.; Khalid, M.F.; Sohail, M.; Anjum, M.A.; Ejaz, S.; Nafees, M.; Zakir, I.; Ahmad, M.; Ali, S.; Ahmad, S. Role of transporters in accumulating salt ions by halophytes. Approaches Remediat. Inorg. Pollut. 2021, 11–40. [Google Scholar] [CrossRef]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Naseer, S.; Lee, Y.; Lapierre, C.; Franke, R.; Nawrath, C.; Geldner, N. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. USA 2012, 109, 10101–10106. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, P.; Ranathunge, K.; Nayak, S.; Schreiber, L.; Mathew, M.K. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 4215–4228. [Google Scholar] [CrossRef]
- Zhang, S.; Quartararo, A.; Betz, O.K.; Madahhosseini, S.; Heringer, A.S.; Le, T.; Shao, Y.; Caruso, T.; Ferguson, L.; Jernstedt, J. Root vacuolar sequestration and suberization are prominent responses of Pistacia spp. rootstocks during salinity stress. Plant Direct 2021, 5, e00315. [Google Scholar]
- Cui, B.; Liu, R.; Flowers, T.J.; Song, J. Casparian bands and Suberin lamellae: Key targets for breeding salt tolerant crops? Environ. Exp. Bot. 2021, 191, 104600. [Google Scholar]
- Wang, P.; Wang, C.-M.; Gao, L.; Cui, Y.-N.; Yang, H.-L.; De Silva, N.D.G.; Ma, Q.; Bao, A.-K.; Flowers, T.J.; Rowland, O.; et al. Aliphatic suberin confers salt tolerance to Arabidopsis by limiting Na+ influx, K+ efflux and water backflow. Plant Soil 2020, 448, 603–620. [Google Scholar] [CrossRef]
- Hajibagheri, M.A.; Yeo, A.R.; Flowers, T.J. Salt tolerance in Suaeda Maritima (L.) Dum. fine structure and ion concentrations in the apical region of roots. New Phytol. 1985, 99, 331–343. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Jyothi-Prakash, P.A.; Qin, L.; He, J.; Lin, Q.; Loh, C.-S.; Kumar, P.P. Role of root hydrophobic barriers in salt exclusion of a mangrove plant Avicennia officinalis. Plant Cell Environ. 2014, 37, 1656–1671. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, Y.; Zhang, X.; Liu, R.; Song, J. Chloride allocation in the euhalophyte Suaeda salsa from different habitats in field and controlled saline conditions. Aquat. Bot. 2020, 167, 103292. [Google Scholar] [CrossRef]
- Cheng, H.; Inyang, A.; Li, C.-D.; Fei, J.; Zhou, Y.-W.; Wang, Y.-S. Salt tolerance and exclusion in the mangrove plant Avicennia marina in relation to root apoplastic barriers. Ecotoxicology 2020, 29, 676–683. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, R.; Ma, Y.; Song, J. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 2018, 146, 1–7. [Google Scholar] [CrossRef]
- Guo, Q.; Meng, L.; Han, J.; Mao, P.; Tian, X.; Zheng, M.; Mur, L.A. SOS1 is a key systemic regulator of salt secretion and K+/Na+ homeostasis in the recretohalophyte Karelinia caspia. Environ. Exp. Bot. 2020, 177, 104098. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Shabala, S.; Bose, J.; Hedrich, R. Salt bladders: Do they matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar]
- Leng, B.; Yuan, F.; Dong, X.; Wang, J.; Wang, B. Distribution pattern and salt excretion rate of salt glands in two recretohalophyte species of Limonium (Plumbaginaceae). S. Afr. J. Bot. 2018, 115, 74–80. [Google Scholar] [CrossRef]
- Li, J.; Yuan, F.; Liu, Y.; Zhang, M.; Liu, Y.; Zhao, Y.; Wang, B.; Chen, M. Exogenous melatonin enhances salt secretion from salt glands by upregulating the expression of ion transporter and vesicle transport genes in Limonium bicolor. BMC Plant Biol. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Mi, P.; Yuan, F.; Guo, J.; Han, G.; Wang, B. Salt glands play a pivotal role in the salt resistance of four recretohalophyte Limonium Mill. species. Plant Biol. 2021. [Google Scholar] [CrossRef]
- Wei, X.; Yan, X.; Yang, Z.; Han, G.; Wang, L.; Yuan, F.; Wang, B. Salt glands of recretohalophyte Tamarix under salinity: Their evolution and adaptation. Ecol. Evol. 2020, 10, 9384–9395. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.; Wang, B. Progress in studying salt secretion from the salt glands in recretohalophytes: How do plants secrete salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar]
- Céccoli, G.; Ramos, J.; Pilatti, V.; Dellaferrera, I.; Tivano, J.C.; Taleisnik, E.; Vegetti, A.C. Salt glands in the Poaceae family and their relationship to salinity tolerance. Bot. Rev. 2015, 81, 162–178. [Google Scholar] [CrossRef] [Green Version]
- Arbelet-Bonnin, D.; Ben-Hamed-Louati, I.; Laurenti, P.; Abdelly, C.; Ben-Hamed, K.; Bouteau, F. Cakile maritima, a promising model for halophyte studies and a putative cash crop for saline agriculture. Adv. Agron. 2019, 155, 45–78. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.S.; Lutz, A.; Rupasinghe, T.; Bazihizina, N.; Bohm, J.; Alharbi, S.; Hedrich, R.; Shabala, S. Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 2017, 40, 1900–1915. [Google Scholar] [CrossRef] [Green Version]
- Barkla, B.J.; Rhodes, T.; Tran, K.-N.T.; Wijesinghege, C.; Larkin, J.C.; Dassanayake, M. Making epidermal bladder cells bigger: Developmental- and salinity-induced endopolyploidy in a model halophyte. Plant Physiol. 2018, 177, 615–632. [Google Scholar] [CrossRef]
- Tran, D.Q.; Konishi, A.; Cushman, J.C.; Morokuma, M.; Toyota, M.; Agarie, S. Ion accumulation and expression of ion homeostasis-related genes associated with halophilism, NaCl-promoted growth in a halophyte Mesembryanthemum crystallinum L. Plant Prod. Sci. 2019, 23, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Himabindu, Y.; Chakradhar, T.; Reddy, M.C.; Kanygin, A.; Redding, K.E.; Chandrasekhar, T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016, 124, 39–63. [Google Scholar] [CrossRef] [Green Version]
- Bonales-Alatorre, E.; Shabala, S.; Chen, Z.-H.; Pottosin, I. Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, Quinoa. Plant Physiol. 2013, 162, 940–952. [Google Scholar] [CrossRef] [Green Version]
- Otterbach, S.L.; Khoury, H.; Rupasinghe, T.; Mendis, H.; Kwan, K.H.; Lui, V.; Natera, S.H.A.; Klaiber, I.; Allen, N.M.; Jarvis, D.E.; et al. Characterization of epidermal bladder cells in Chenopodium quinoa. Plant Cell Environ. 2021. [Google Scholar] [CrossRef]
- Roeurn, S.; Hoshino, N.; Soejima, K.-T.; Inoue, Y.; Cushman, J.C.; Agarie, S. MYB and HD-ZIP IV homologs related to trichome formation are involved in epidermal bladder cell development in the halophyte Mesembryanthemum crystallinum L. Plant Prod. Sci. 2017, 20, 72–82. [Google Scholar] [CrossRef]
- Böhm, J.; Messerer, M.; Müller, H.M.; Scholz-Starke, J.; Gradogna, A.; Scherzer, S.; Maierhofer, T.; Bazihizina, N.; Zhang, H.; Stigloher, C.; et al. Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr. Biol. 2018, 28, 3075–3085.e7. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Zhang, X.; Wang, N.; Cui, Y.; Zhang, L.; Fan, S. Transcriptome analysis of salt stress response in halophyte Atriplex centralasiatica leaves. Acta Physiol. Plant. 2019, 42, 3. [Google Scholar] [CrossRef]
- Imamura, T.; Yasui, Y.; Koga, H.; Takagi, H.; Abe, A.; Nishizawa, K.; Mizuno, N.; Ohki, S.; Mizukoshi, H.; Mori, M. A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun. Biol. 2020, 3, 1–14. [Google Scholar] [CrossRef]
- Lokhande, V.H.; Suprasanna, P. Prospects of halophytes in understanding and managing abiotic stress tolerance. In Environmental Adaptations and Stress Tolerance of Plants in the Era of Climate Change; Springer: Berlin, Germany, 2012; pp. 29–56. [Google Scholar]
- Nikalje, G.C.; Srivastava, A.K.; Pandey, G.K.; Suprasanna, P. Halophytes in biosaline agriculture: Mechanism, utilization, and value addition. Land Degrad. Dev. 2017, 29, 1081–1095. [Google Scholar] [CrossRef]
- García-Caparrós, P.; Llanderal, A.; Pestana, M.; Correia, P.J.; Lao, M.T. Nutritional and physiological responses of the dicotyledonous halophyte Sarcocornia fruticosa to salinity. Aust. J. Bot. 2017, 65, 573–581. [Google Scholar] [CrossRef]
- Elnaggar, A.; Mosa, K.A.; El-Keblawy, A.; Tammam, A.; El-Naggar, M. Physiological and biochemical insights for salt stress tolerance in the habitat-indifferent halophyte Salsola drummondii during the vegetative stage. Botany 2020, 98, 1–17. [Google Scholar] [CrossRef]
- Rahman, M.; Mostofa, M.G.; Rahman, A.; Miah, G.; Saha, S.R.; Karim, M.A.; Keya, S.S.; Akter, M.; Islam, M.; Tran, L.-S.P. Insight into salt tolerance mechanisms of the halophyte Achras sapota: An important fruit tree for agriculture in coastal areas. Protoplasma 2018, 256, 181–191. [Google Scholar] [CrossRef]
- Panda, A.; Rangani, J.; Parida, A.K. Unraveling salt responsive metabolites and metabolic pathways using non-targeted metabolomics approach and elucidation of salt tolerance mechanisms in the xero-halophyte Haloxylon salicornicum. Plant Physiol. Biochem. 2020, 158, 284–296. [Google Scholar] [CrossRef]
- Altuntaş, C.; Terzi, R. Concomitant accumulations of ions, osmoprotectants and antioxidant system-related substances provide salt tolerance capability to succulent extreme-halophyte Scorzonera hieraciifolia. Authorea Prepr. 2021. [Google Scholar] [CrossRef]
- Yıldız, M.; Terzi, H. Comparative analysis of salt-induced changes in the root physiology and proteome of the xero-halophyte Salsola crassa. Braz. J. Bot. 2021, 44, 33–42. [Google Scholar] [CrossRef]
- Wiszniewska, A.; Koźmińska, A.; Hanus-Fajerska, E.; Dziurka, K.; Dziurka, M. Insight into phytohormonal modulation of defense mechanisms to salt excess in a halophyte and a glycophyte from Asteraceae family. Plant Soil 2021, 463, 55–76. [Google Scholar] [CrossRef]
- Palchetti, M.V.; Reginato, M.; Llanes, A.; Hornbacher, J.; Papenbrock, J.; Barboza, G.E.; Luna, V.; Cantero, J.J. New insights into the salt tolerance of the extreme halophytic species Lycium humile (Lycieae, Solanaceae). Plant Physiol. Biochem. 2021, 163, 166–177. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, H.; Shabala, S.; Li, H.; Yang, X.; Zhang, H. Tissue tolerance mechanisms conferring salinity tolerance in a halophytic perennial species Nitraria sibirica Pall. Tree Physiol. 2020. [Google Scholar] [CrossRef]
- Ben Hsouna, A.; Ghneim-Herrera, T.; Ben Romdhane, W.; Dabbous, A.; Ben Saad, R.; Brini, F.; Abdelly, C.; Ben Hamed, K. Early effects of salt stress on the physiological and oxidative status of the halophyte Lobularia maritima. Funct. Plant Biol. 2020, 47, 912–924. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Rasouli, F.; Shabala, L.; Tahir, A.T.; Zhou, M.; Shabala, S. Understanding the role of root-related traits in salinity tolerance of quinoa accessions with contrasting epidermal bladder cell patterning. Planta 2020, 251, 1–11. [Google Scholar] [CrossRef]
- Mann, A.; Kumar, N.; Kumar, A.; Lata, C.; Kumar, A.; Meena, B.L.; Mishra, D.; Grover, M.; Gaba, S.; Parameswaran, C.; et al. De novo transcriptomic profiling of differentially expressed genes in grass halophyte Urochondra setulosa under high salinity. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Arbelet-Bonnin, D.; Blasselle, C.; Palm, E.R.; Redwan, M.; Ponnaiah, M.; Laurenti, P.; Meimoun, P.; Gilard, F.; Gakière, B.; Mancuso, S.; et al. Metabolism regulation during salt exposure in the halophyte Cakile maritima. Environ. Exp. Bot. 2020, 177, 104075. [Google Scholar] [CrossRef]
- Guo, J.; Lu, C.; Zhao, F.; Gao, S.; Wang, B. Improved reproductive growth of euhalophyte Suaeda salsa under salinity is correlated with altered phytohormone biosynthesis and signal transduction. Funct. Plant Biol. 2020, 47, 170–183. [Google Scholar] [CrossRef]
- Zhang, H.; Xiao, W.; Yu, W.; Jiang, Y.; Li, R. Halophytic Hordeum brevisubulatum HbHAK1 facilitates potassium retention and contributes to salt tolerance. Int. J. Mol. Sci. 2020, 21, 5292. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Liu, K.; Zhang, X.-J.; Zhang, L.-Y.; Fan, S.-J. Transcriptome analysis of halophyte Zoysia macrostachya in response to salinity stress. Plants 2020, 9, 458. [Google Scholar] [CrossRef] [Green Version]
- Panda, A.; Rangani, J.; Parida, A.K. Cross talk between ROS homeostasis and antioxidative machinery contributes to salt tolerance of the xero-halophyte Haloxylon salicornicum. Environ. Exp. Bot. 2019, 166, 103799. [Google Scholar] [CrossRef]
- Wiszniewska, A.; Koźmińska, A.; Hanus-Fajerska, E.; Dziurka, M.; Dziurka, K. Insight into mechanisms of multiple stresses tolerance in a halophyte Aster tripolium subjected to salinity and heavy metal stress. Ecotoxicol. Environ. Saf. 2019, 180, 12–22. [Google Scholar] [CrossRef]
- Mohammadi, F.; Kavousi, H.R.; Mansouri, M. Effects of salt stress on physio-biochemical characters and gene expressions in halophyte grass Leptochloa fusca (L.) Kunth. Acta Physiol. Plant. 2019, 41, 1–10. [Google Scholar] [CrossRef]
- Arbelet-Bonnin, D.; Ben Hamed-Laouti, I.; Laurenti, P.; Abdelly, C.; Ben Hamed, K.; Bouteau, F. Cellular mechanisms to survive salt in the halophyte Cakile maritima. Plant Sci. 2018, 272, 173–178. [Google Scholar] [CrossRef]
- Ali, A.; Khan, I.U.; Jan, M.; Khan, H.A.; Hussain, S.; Nisar, M.; Chung, W.S.; Yun, D.-J. The High-affinity potassium transporter EpHKT1;2 from the extremophile Eutrema parvula mediates salt tolerance. Front. Plant Sci. 2018, 9, 1108. [Google Scholar] [CrossRef] [Green Version]
- Kumari, A.; Parida, A. Metabolomics and network analysis reveal the potential metabolites and biological pathways involved in salinity tolerance of the halophyte Salvadora persica. Environ. Exp. Bot. 2018, 148, 85–99. [Google Scholar] [CrossRef]
- Marriboina, S.; Sengupta, D.; Kumar, S.; Reddy, A.R. Physiological and molecular insights into the high salinity tolerance of Pongamia pinnata (L.) pierre, a potential biofuel tree species. Plant Sci. 2017, 258, 102–111. [Google Scholar] [CrossRef]
- Hamdani, F.; Derridj, A.; Rogers, H.J. Multiple mechanisms mediate growth and survival in young seedlings of two populations of the halophyte Atriplex halimus (L.) subjected to long single-step salinity treatments. Funct. Plant Biol. 2017, 44, 761–773. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Physiological responses of the halophyte Sesuvium portulacastrum to salt stress and their relevance for saline soil bio-reclamation. Flora 2016, 224, 96–105. [Google Scholar] [CrossRef]
- Ma, Q.; Hu, J.; Zhou, X.-R.; Yuan, H.-J.; Kumar, T.; Luan, S.; Wang, S.-M. ZxAKT1 is essential for K+ uptake and K+/Na+ homeostasis in the succulent xerophyte Zygophyllum xanthoxylum. Plant J. 2017, 90, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.-D.; Wang, P.; Bao, Z.; Ma, Q.; Duan, L.-J.; Bao, A.-K.; Zhang, J.-L.; Wang, S.-M. SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Xia, Y.; Ma, B.L.; Feng, C.; Qin, P. Culture of seashore mallow under different salinity levels using plastic nutrient-rich matrices and transplantation. Agron. J. 2010, 102, 395–402. [Google Scholar] [CrossRef]
- Wang, H.; Tang, X.; Wang, H.; Shao, H.-B. Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 2015, 6, 792. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Li, Z. The importance of Cl− exclusion and vacuolar Cl− sequestration: Revisiting the role of Cl− transport in plant salt tolerance. Front. Plant Sci. 2019, 10, 1418. [Google Scholar] [CrossRef]
- Li, B.; Tester, M.; Gilliham, M. Chloride on the move. Trends Plant Sci. 2017, 22, 236–248. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Jamdade, R.; Gairola, S. Recent advances on cellular signaling paradigm and salt stress responsive genes in halophytes. Handb. Halophytes Mol. Ecosyst. Towards Biosaline Agric. 2020, 1–26. [Google Scholar] [CrossRef]
- Munns, R.; Passioura, J.B.; Colmer, T.D.; Byrt, C.S. Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 2019, 225, 1091–1096. [Google Scholar] [CrossRef] [Green Version]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef] [Green Version]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2013, 65, 1241–1257. [Google Scholar] [CrossRef]
- Llanes, A.; Reginato, M.; Devinar, G.; Luna, V. What is known about phytohormones in halophytes? A review. Biologia 2018, 73, 727–742. [Google Scholar] [CrossRef]
- Gupta, A.; Bhardwaj, M.; Tran, L.-S.P. Jasmonic acid at the crossroads of plant immunity and Pseudomonas syringae virulence. Int. J. Mol. Sci. 2020, 21, 7482. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samól, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, N. Reclamation of saline and sodic soil through phytoremediation. In Environmental Concerns and Sustainable Development: Volume 2: Biodiversity, Soil and Waste Management; Shukla, V., Kumar, N., Eds.; Springer: Singapore, 2020; pp. 279–306. [Google Scholar]
- Jesus, J.; Danko, A.S.; Fiuza, A.; Borges, M.T. Phytoremediation of salt-affected soils: A review of processes, applicability, and the impact of climate change. Environ. Sci. Pollut. Res. 2015, 22, 6511–6525. [Google Scholar] [CrossRef]
- Qadir, M.; Tubeileh, A.; Akhtar, J.; Larbi, A.; Minhas, P.S.; Khan, M.A. Productivity enhancement of salt-affected environments through crop diversification. Land Degrad. Dev. 2008, 19, 429–453. [Google Scholar] [CrossRef]
- Yazdanpanah, N.; Mahmoodabadi, M. Effect of some amendments on leachate properties of a calcareous saline- sodic soil: A laboratory experiment. Int. Agrophys. 2010, 25, 14738. [Google Scholar]
- Leogrande, R.; Vitti, C. Use of organic amendments to reclaim saline and sodic soils: A review. Arid Land Res. Manag. 2018, 33, 1–21. [Google Scholar] [CrossRef]
- Makni, H.; Ayed, L.; Ben Khedher, M.; Bakhrouf, A. Evaluation of the maturity of organic waste composts. Waste Manag. Res. 2009, 28, 489–495. [Google Scholar] [CrossRef]
- Lakhdar, A.; Scelza, R.; Scotti, R.; A Rao, M.; Jedidi, N.; Gianfreda, L.; Abdelly, C. The effect of compost and sewage sludge on soil biologic activities in salt affected soil. Rev. Cienc. Suelo Nutr. Veg. 2010, 10, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Ouni, Y.; Lakhdar, A.; Scelza, R.; Scotti, R.; Abdelly, C.; Barhoumi, Z.; Rao, M.A. Effects of two composts and two grasses on microbial biomass and biological activity in a salt-affected soil. Ecol. Eng. 2013, 60, 363–369. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Jiang, L.; Zhang, K.; Tanveer, M.; Tian, C.; Zhao, Z. Reclamation of saline soil by planting annual euhalophyte Suaeda salsa with drip irrigation: A three-year field experiment in arid northwestern China. Ecol. Eng. 2020, 159, 106090. [Google Scholar] [CrossRef]
- Liang, J.; Shi, W. Cotton/halophytes intercropping decreases salt accumulation and improves soil physicochemical properties and crop productivity in saline-alkali soils under mulched drip irrigation: A three-year field experiment. Field Crop. Res. 2020, 262, 108027. [Google Scholar] [CrossRef]
- Shaygan, M.; Mulligan, D.; Baumgartl, T. The potential of three halophytes (Tecticornia pergranulata, Sclerolaena longicuspis, and Frankenia serpyllifolia) for the rehabilitation of brine-affected soils. Land Degrad. Dev. 2018, 29, 2002–2014. [Google Scholar] [CrossRef]
- Mann, E.; Rutter, A.; Zeeb, B. Evaluating the efficacy of Atriplex spp. in the phytoextraction of road salt (NaCl) from contaminated soil. Environ. Pollut. 2020, 265, 114963. [Google Scholar] [CrossRef]
- Arevalo-Gardini, E.; Canto, M.; Alegre, J.; Loli, O.; Julca, A.; Baligar, V. Changes in soil physical and chemical properties in long term improved natural and traditional agroforestry management systems of cacao genotypes in Peruvian Amazon. PLoS ONE 2015, 10, e0132147. [Google Scholar] [CrossRef]
- Silva, Y.J.A.B.; Freire, M.G.; Lopes, E.A.P.; Santos, M.A. Atriplex nummularia Lindl. as alternative for improving salt-affected soils conditions in semiarid environments: A field experiment. Chil. J. Agric. Res. 2016, 76, 343–348. [Google Scholar]
- Ashraf, M.Y.; Mahmood, K.; Akhter, J.; Hussain, F.; Arshad, M. Phytoremediation of saline soils for sustainable agricultural productivity. In Plant Adaptation and Phytoremediation; Ashraf, M., Ozturk, M., Ahmad, M.S.A., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 335–355. [Google Scholar]
- Qadir, M.; Noble, A.; Oster, J.D.; Schubert, S.; Ghafoor, A. Driving forces for sodium removal during phytoremediation of calcareous sodic and saline-sodic soils: A review. Soil Use Manag. 2005, 21, 173–180. [Google Scholar] [CrossRef]
- Lastiri-Hernández, M.A.; Álvarez-Bernal, D.; Ochoa-Estrada, S.; Contreras-Ramos, S.M. Potential of Bacopa monnieri (L.) Wettst and Sesuvium verrucosum Raf. as an agronomic management alternative to recover the productivity of saline soils. Int. J. Phytoremediation 2019, 22, 343–352. [Google Scholar] [CrossRef]
- Malik, Z.; Ravindran, K. Bioaccumulation of salts and heavy metals by Suaeda monoica: A salt marsh halophyte from paper mill effluent contaminated soil. Int. J. Sci. Technol. Res. 2020, 9, 7248–7254. [Google Scholar]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Barros, N.L.F.; Marques, D.N.; Tadaiesky, L.B.A.; de Souza, C.R.B. Halophytes and other molecular strategies for the generation of salt-tolerant crops. Plant Physiol. Biochem. 2021, 162, 581–591. [Google Scholar] [CrossRef]
- Ye, W.; Wang, T.; Wei, W.; Lou, S.; Lan, F.; Zhu, S.; Li, Q.; Ji, G.; Lin, C.; Wu, X.; et al. The full-length transcriptome of Spartina alterniflora reveals the complexity of high salt tolerance in monocotyledonous halophyte. Plant Cell Physiol. 2020, 61, 882–896. [Google Scholar]
- Guo, R.; Zhao, L.; Zhang, K.; Lu, H.; Bhanbhro, N.; Yang, C. Comparative genomics and transcriptomics of the extreme halophyte Puccinellia tenuiflora provides insights into salinity tolerance differentiation between halophytes and glycophytes. Front. Plant Sci. 2021, 12, 767. [Google Scholar] [CrossRef]
- Aliakbari, M.; Razi, H.; Alemzadeh, A.; Tavakol, E. RNA-seq transcriptome profiling of the halophyte Salicornia persica in response to salinity. J. Plant Growth Regul. 2020, 40, 707–721. [Google Scholar] [CrossRef]
- Han, Z.-J.; Sun, Y.; Zhang, M.; Zhai, J.-T. Transcriptomic profile analysis of the halophyte Suaeda rigida response and tolerance under NaCl stress. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Xiang, D.; Man, L. EhEm1, a novel Em-like protein from Eutrema halophilum, confers tolerance to salt and drought stresses in rice. Mol. Breed. 2018, 38, 17. [Google Scholar] [CrossRef]
- Liu, X.; Cai, S.; Wang, G.; Wang, F.; Dong, F.; Mak, M.; Holford, P.; Ji, J.; Salih, A.; Zhou, M.; et al. Halophytic NHXs confer salt tolerance by altering cytosolic and vacuolar K+ and Na+ in Arabidopsis root cell. Plant Growth Regul. 2017, 82, 333–351. [Google Scholar] [CrossRef]
- Geng, X.; Chen, S.; Yilan, E.; Zhang, W.; Mao, H.; Qiqige, A.; Wang, Y.; Qi, Z.; Lin, X. Overexpression of a tonoplast Na+/H+ antiporter from the halophytic shrub Nitraria sibirica improved salt tolerance and root development in transgenic poplar. Tree Genet. Genomes 2020, 16, 1–14. [Google Scholar] [CrossRef]
- Guo, Q.; Meng, S.; Tao, S.; Feng, J.; Fan, X.; Xu, P.; Xu, Z.; Shen, X. Overexpression of a samphire high-affinity potassium transporter gene SbHKT1 enhances salt tolerance in transgenic cotton. Acta Physiol. Plant. 2020, 42, 36. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Yuan, J.-Z.; Cui, Y.-N.; Li, X.-T.; Wang, F.-Z.; He, Z.-H.; Li, X.-Y.; Bao, A.-K.; Wang, S.; Ma, Q. ZxNPF7.3/NRT1.5 from the xerophyte Zygophyllum xanthoxylum modulates salt and drought tolerance by regulating NO3-, Na+ and K+ transport. Environ. Exp. Bot. 2020, 177, 104123. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.; Zhu, L.; Li, M.; Zhang, J.; Yang, X.; Wang, P.; Lu, Y.; Cheng, T.; Shi, J.; et al. NtCIPK9: A calcineurin B-like protein-interacting protein kinase from the halophyte Nitraria tangutorum, enhances Arabidopsis salt tolerance. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.; Wang, P.; Lu, Y.; Zhang, J.; Yang, X.; Cheng, T.; Shi, J.; Chen, J. CIPK11: A calcineurin B-like protein-interacting protein kinase from Nitraria tangutorum, confers tolerance to salt and drought in Arabidopsis. BMC Plant Biol. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Mishra, A.; Jha, B. Overexpression of a plasma membrane-localized SbSRP-like protein enhances salinity and osmotic stress tolerance in transgenic tobacco. Front. Plant Sci. 2017, 8, 582. [Google Scholar] [CrossRef] [Green Version]
- Kumari, J.; Udawat, P.; Dubey, A.K.; Haque, I.; Rathore, M.S.; Jha, B. Overexpression of SbSI-1, A nuclear protein from Salicornia brachiata confers drought and salt stress tolerance and maintains photosynthetic efficiency in transgenic tobacco. Front. Plant Sci. 2017, 8, 1215. [Google Scholar] [CrossRef] [Green Version]
- Shukla, P.S.; Gupta, K.; Agarwal, P.; Jha, B.; Agarwal, P.K. Overexpression of a novel SbMYB15 from Salicornia brachiata confers salinity and dehydration tolerance by reduced oxidative damage and improved photosynthesis in transgenic tobacco. Planta 2015, 242, 1291–1308. [Google Scholar] [CrossRef]
- Gupta, K.; Jha, B.; Agarwal, P.K. A dehydration-responsive element binding (DREB) transcription factor from the succulent halophyte Salicornia brachiata enhances abiotic stress tolerance in transgenic tobacco. Mar. Biotechnol. 2014, 16, 657–673. [Google Scholar] [CrossRef]
- Kumari, A.; Jha, B. Engineering of a novel gene from a halophyte: Potential for agriculture in degraded coastal saline soil. Land Degrad. Dev. 2018, 30, 595–607. [Google Scholar] [CrossRef]
- Agarwal, P.; Baraiya, B.M.; Joshi, P.S.; Patel, M.; Parida, A.K.; Agarwal, P.K. AlRab7 from Aeluropus lagopoides ameliorates ion toxicity in transgenic tobacco by regulating hormone signaling and reactive oxygen species homeostasis. Physiol. Plant. 2021. [Google Scholar] [CrossRef]
- Dabbous, A.; Ben Saad, R.; Brini, F.; Farhat-Khemekhem, A.; Zorrig, W.; Abdely, C.; Ben Hamed, K. Over-expression of a subunit E1 of a vacuolar H+-ATPase gene (Lm VHA-E1) cloned from the halophyte Lobularia maritima improves the tolerance of Arabidopsis thaliana to salt and osmotic stresses. Environ. Exp. Bot. 2017, 137, 128–141. [Google Scholar] [CrossRef]
- Yang, G.; Yu, L.; Zhang, K.; Zhao, Y.; Guo, Y.; Gao, C. A ThDREB gene from Tamarix hispida improved the salt and drought tolerance of transgenic tobacco and T. hispida. Plant Physiol. Biochem. 2017, 113, 187–197. [Google Scholar] [CrossRef]
- Guo, Q.; Tian, X.; Mao, P.; Meng, L. Overexpression of Iris lactea tonoplast Na+/H+ antiporter gene IlNHX confers improved salt tolerance in tobacco. Biol. Plant. 2020, 64, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, R.; Mukherjee, A.; Bandyopadhyay, S.; Mukherjee, S.; Sengupta, S.; Ray, S.; Majumder, A.L. Selective manipulation of the inositol metabolic pathway for induction of salt-tolerance in indica rice variety. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Chen, S.; Xie, L.; Lu, Z.; Liu, M.; Han, X.; Qiao, G.; Jiang, J.; Zhuo, R.; Qiu, W.; et al. Overexpression of cysteine protease gene from Salix matsudana enhances salt tolerance in transgenic Arabidopsis. Environ. Exp. Bot. 2018, 147, 53–62. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.; Ma, B.; Du, C.; Zheng, L.; Wang, Y. Expression of a Na+/H+ antiporter RtNHX1 from a recretohalophyte Reaumuria trigyna improved salt tolerance of transgenic Arabidopsis thaliana. J. Plant Physiol. 2017, 218, 109–120. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Chang, W.; Liu, X.; Zhu, J.; Fan, W.; Zhang, Z. An aquaporin gene from halophyte Sesuvium portulacastrum, SpAQP1, increases salt tolerance in transgenic tobacco. Plant Cell Rep. 2015, 35, 385–395. [Google Scholar] [CrossRef]
- Chen, Y.; Li, L.; Zong, J.; Chen, J.; Guo, H.; Guo, A.; Liu, J. Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 91, 49–55. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).