Members of the GADD45 Protein Family Show Distinct Propensities to form Toxic Amyloid-Like Aggregates in Physiological Conditions
Abstract
:1. Introduction
2. Results
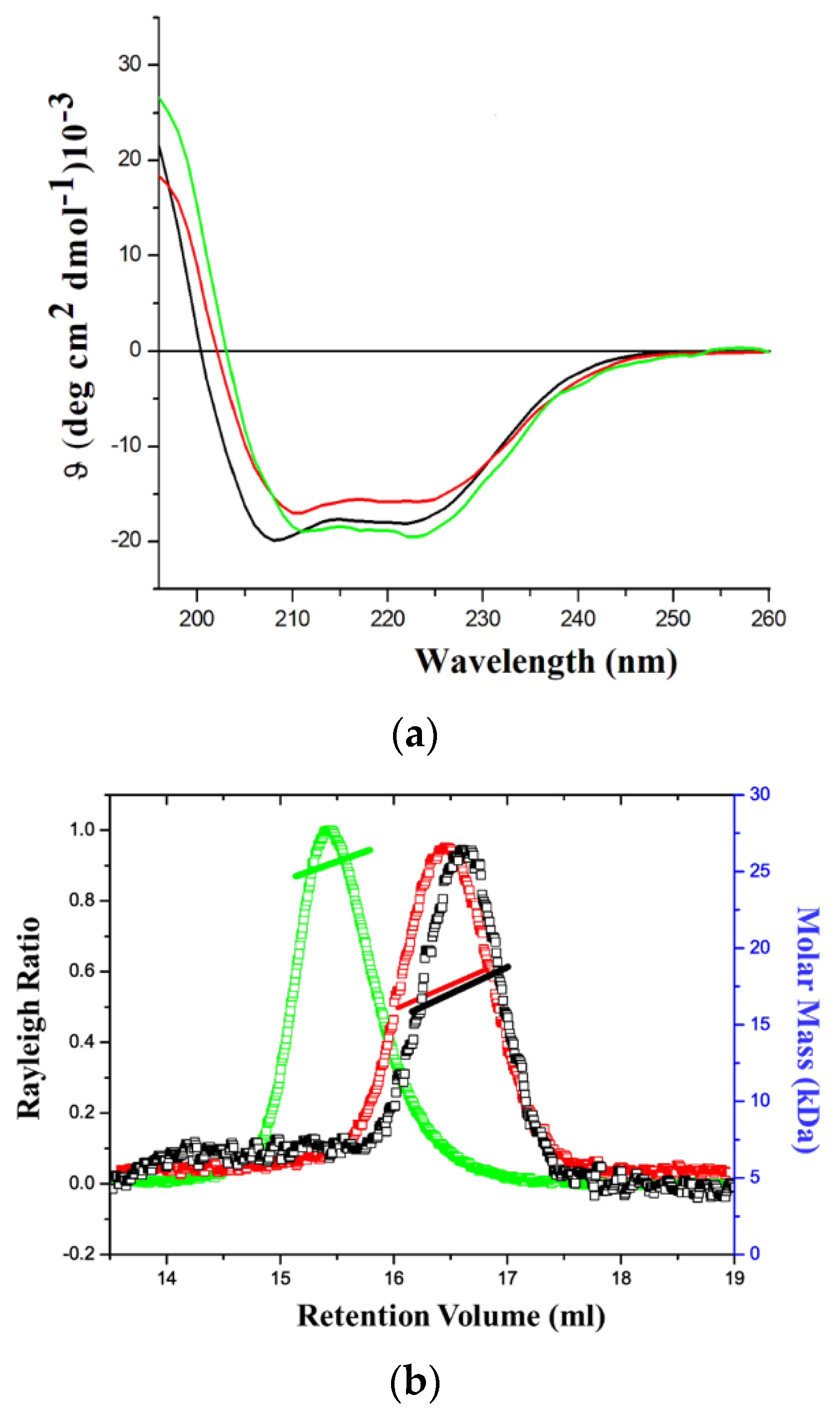
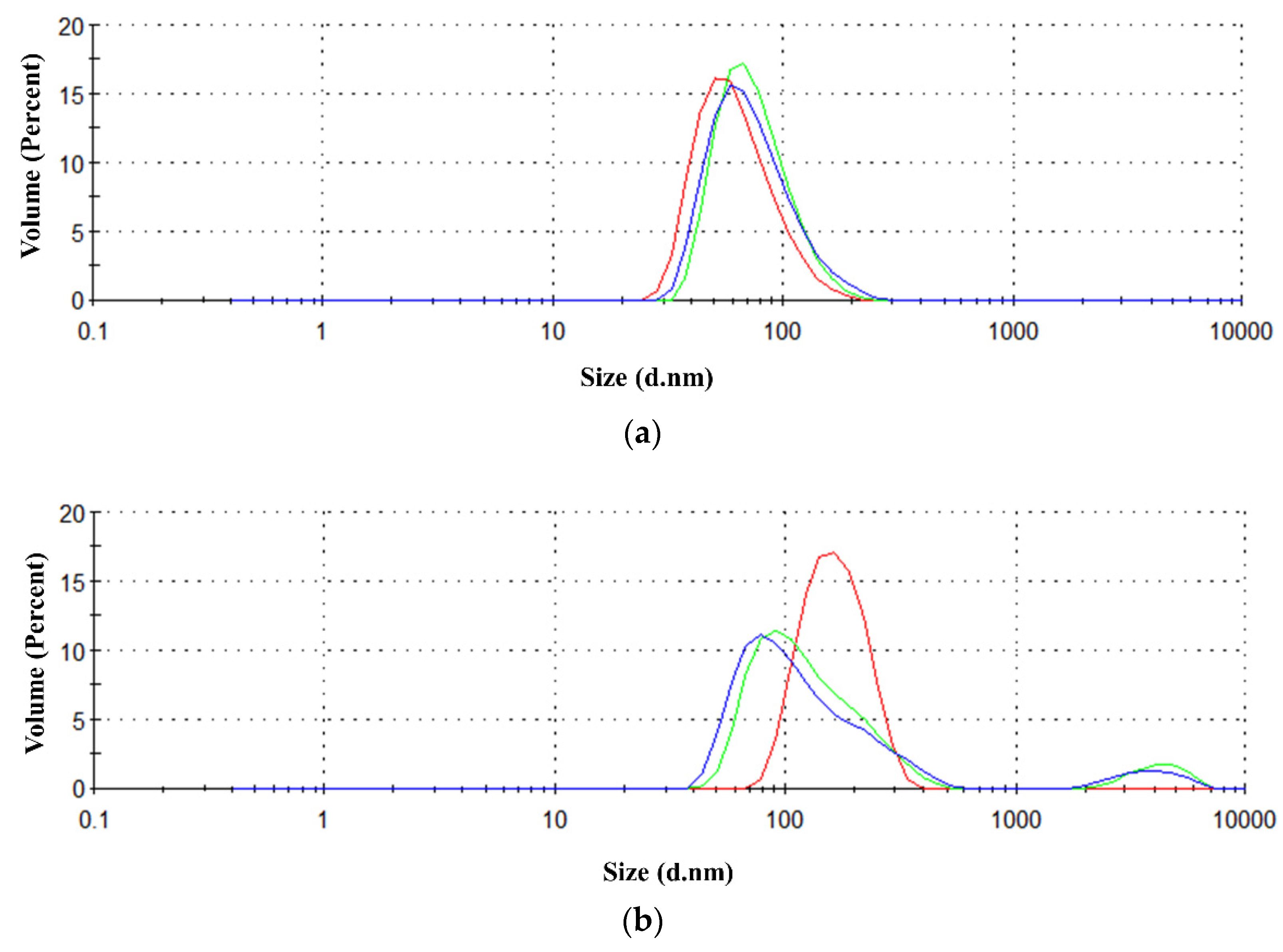
2.1. Production of the Recombinant Proteins and Initial Biophysical Characterizations
2.2. Binding of GADD45 Proteins to MKK7
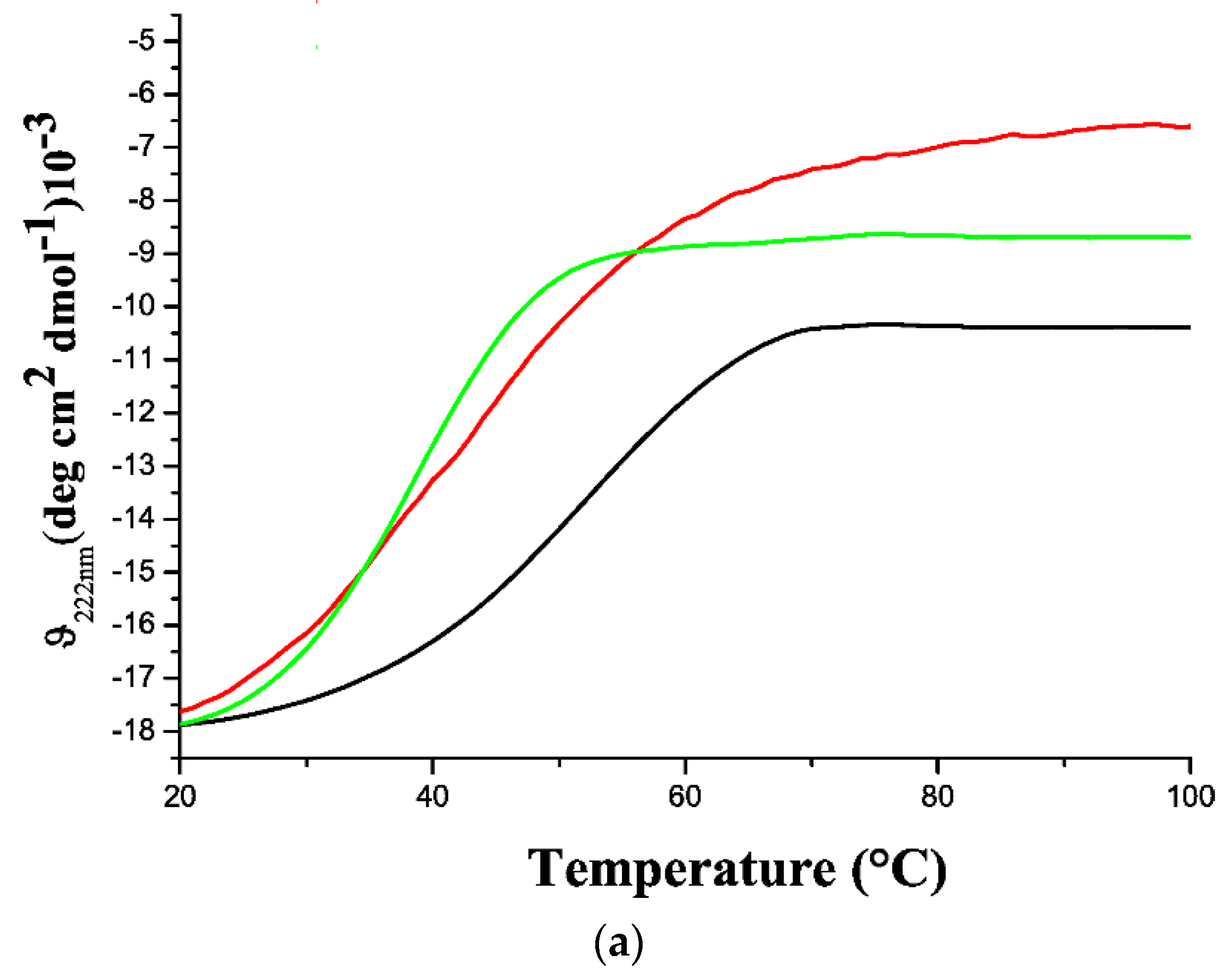
2.3. Thermal Stability of GADD45 Proteins
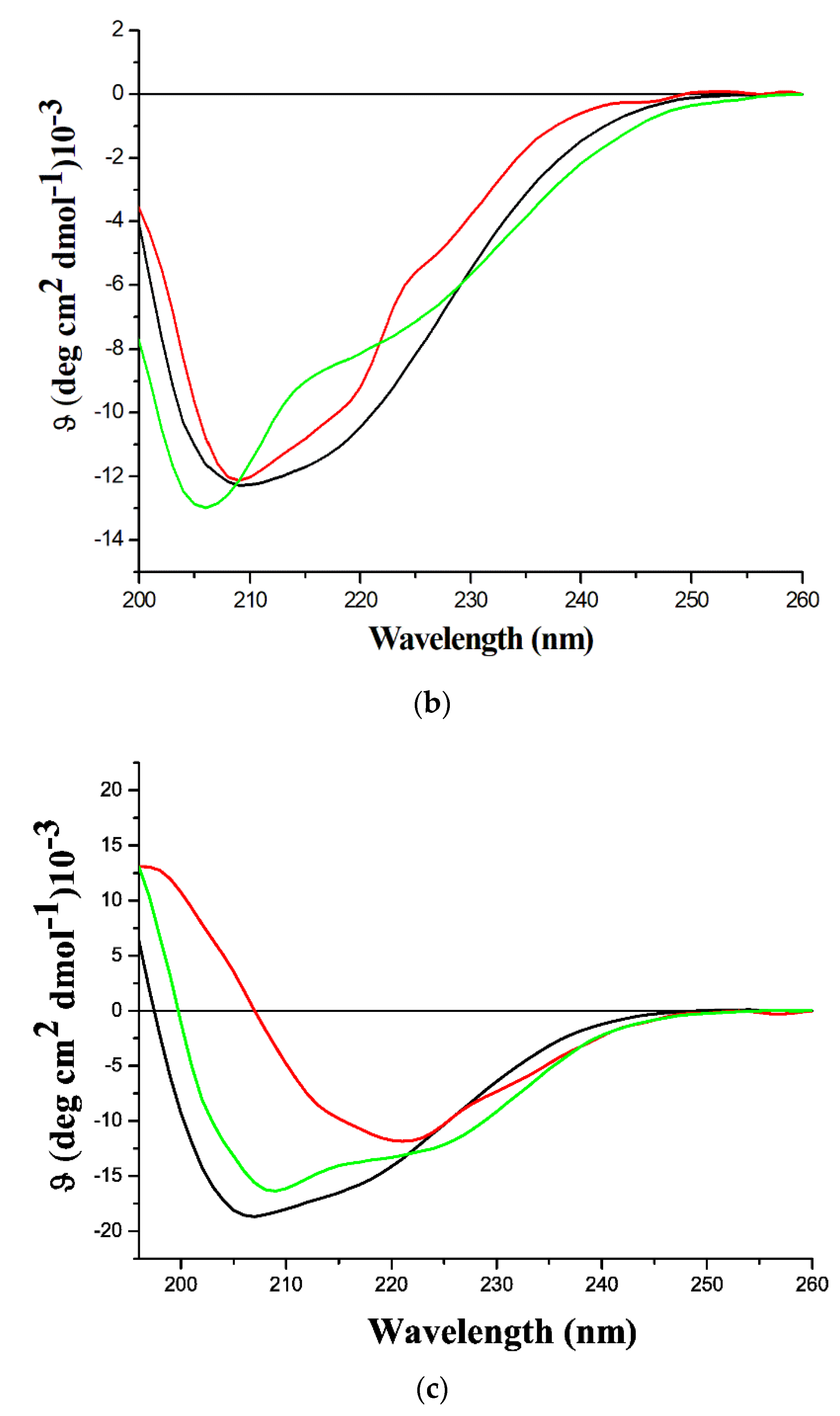
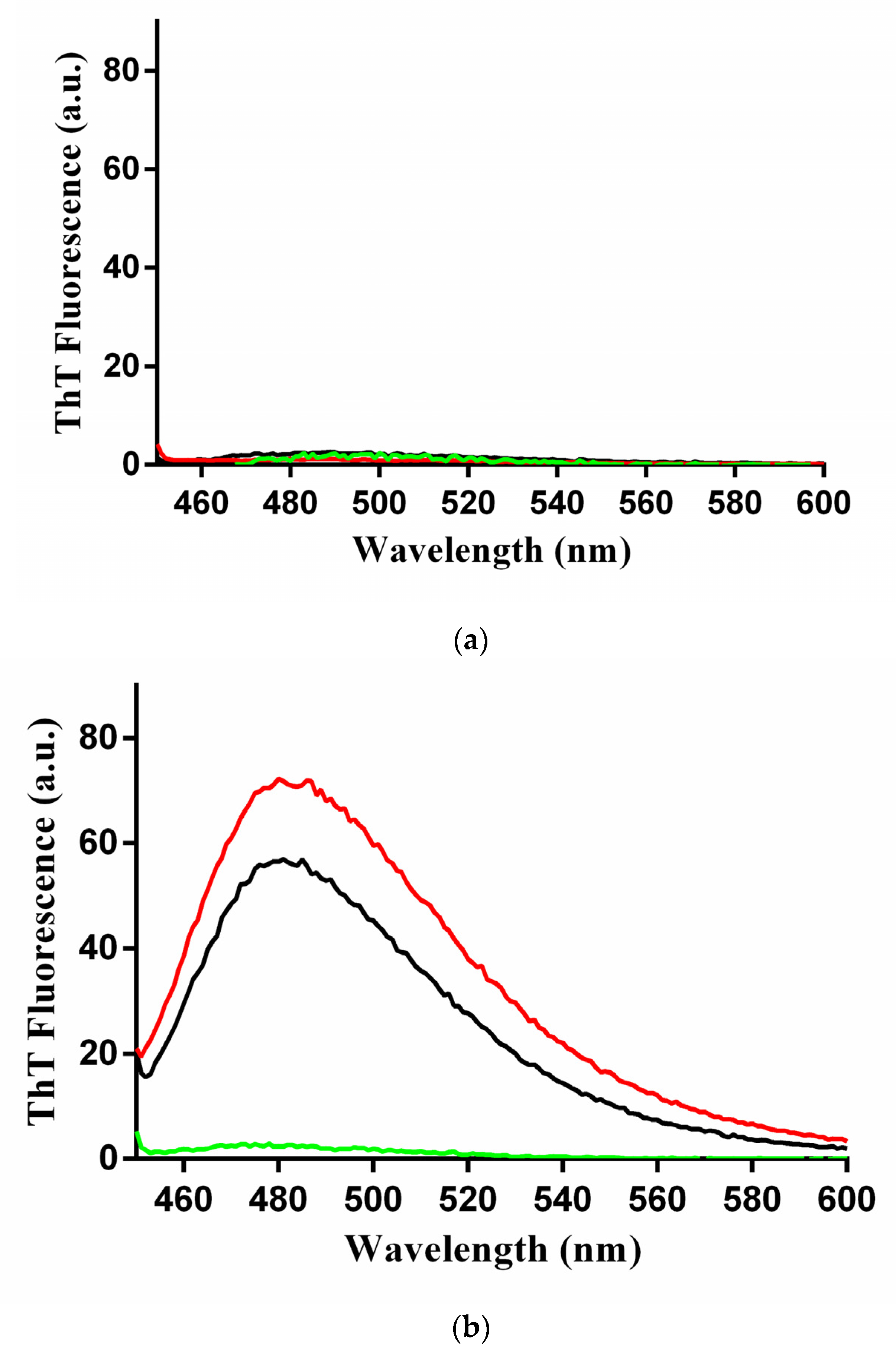
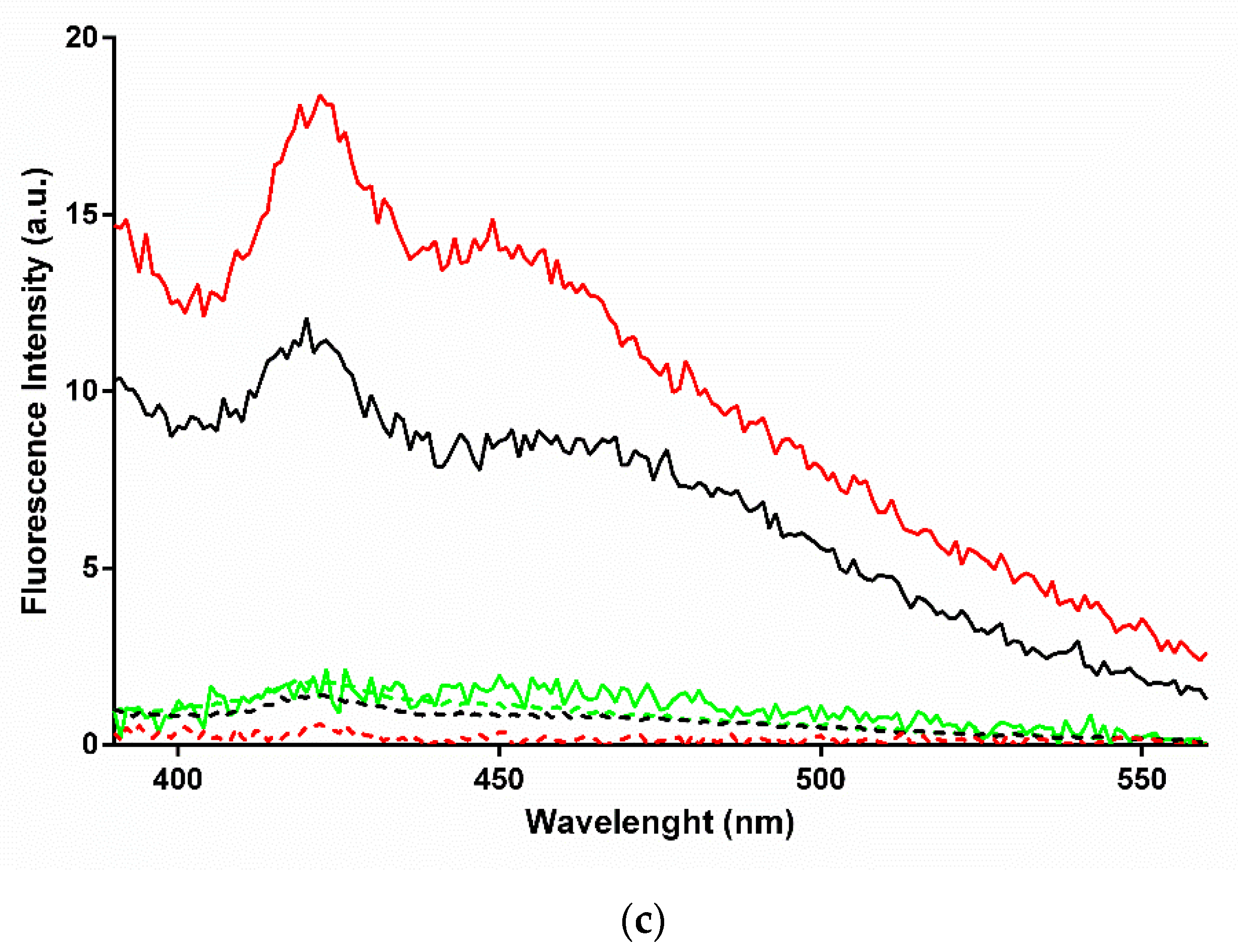
2.4. Characterization of the β-Rich States Formed by GADD45α/GADD45β
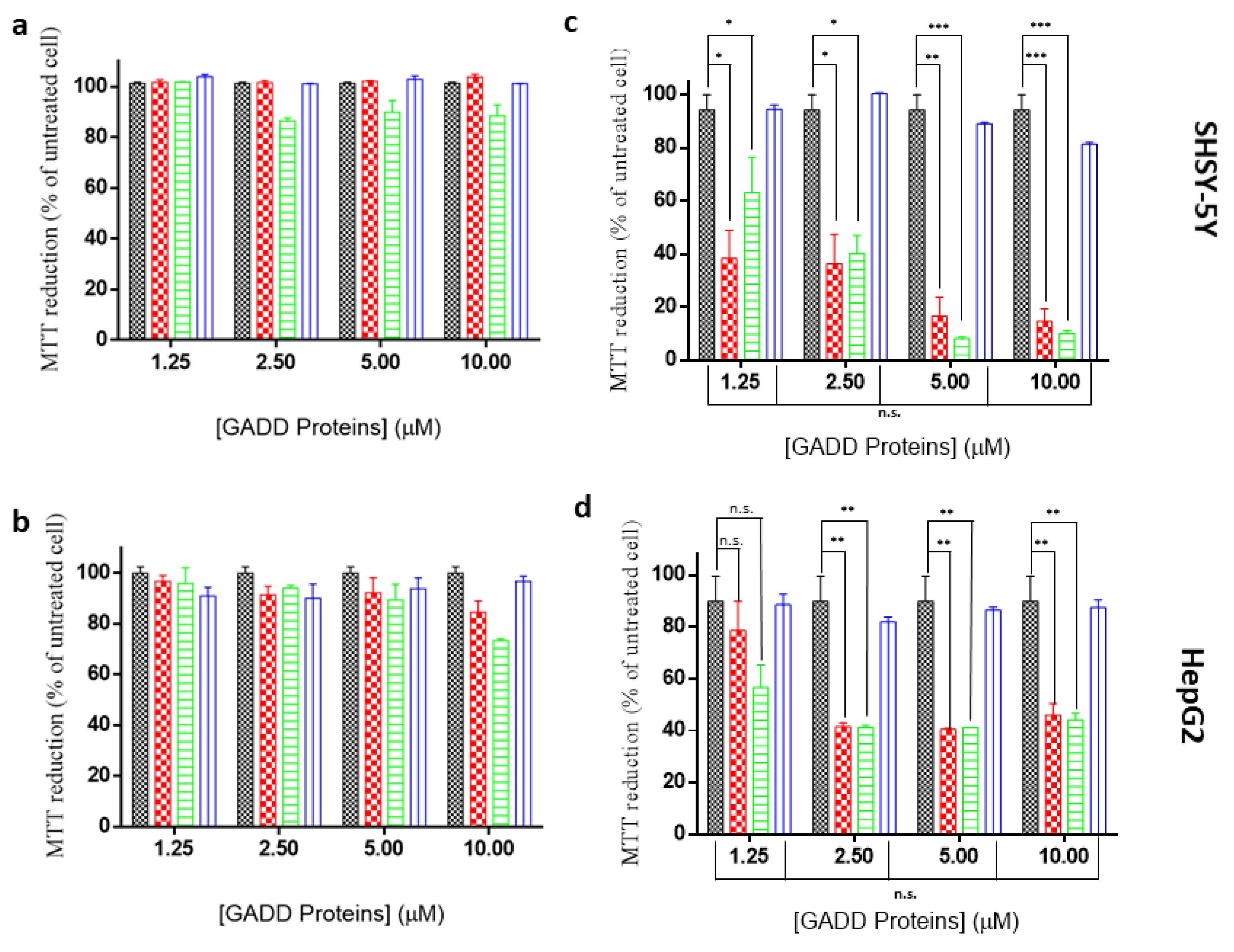
2.5. Toxicity of GADD45α/GADD45β Amyloid-Like Aggregates
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification of GADD45 Proteins
4.2. Scattering (LS) and Dynamic Light Scattering (DLS)
4.3. CD Spectroscopy
4.4. Bio-Layer Interferometry (BLI)
4.5. Thioflavin T (ThT) Assay
4.6. MTT Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Choi, S. Encyclopedia of Signaling Molecules, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-319-67198-7. [Google Scholar]
- Fornace, A.J.; Alamo, I.; Hollander, M.C. DNA Damage-Inducible Transcripts in Mammalian Cells. Proc. Natl. Acad. Sci. USA 1988, 85, 8800–8804. [Google Scholar] [CrossRef] [Green Version]
- Cretu, A.; Sha, X.; Tront, J.; Hoffman, B.; Liebermann, D.A. Stress Sensor Gadd45 Genes as Therapeutic Targets in Cancer. Cancer Ther. 2009, 7, 268–276. [Google Scholar] [PubMed]
- Schäfer, A. Gadd45 Proteins: Key Players of Repair-Mediated DNA Demethylation. Adv. Exp. Med. Biol. 2013, 793, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Tamura, R.E.; de Vasconcellos, J.F.; Sarkar, D.; Libermann, T.A.; Fisher, P.B.; Zerbini, L.F. GADD45 Proteins: Central Players in Tumorigenesis. Curr. Mol. Med. 2012, 12, 634–651. [Google Scholar] [CrossRef] [PubMed]
- Liebermann, D.A.; Hoffman, B. Gadd45 in Stress Signaling. J. Mol. Signal. 2008, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Tornatore, L.; Sandomenico, A.; Raimondo, D.; Low, C.; Rocci, A.; Tralau-Stewart, C.; Capece, D.; D’Andrea, D.; Bua, M.; Boyle, E.; et al. Cancer-Selective Targeting of the NF-ΚB Survival Pathway with GADD45β/MKK7 Inhibitors. Cancer Cell 2014, 26, 495–508. [Google Scholar] [CrossRef] [Green Version]
- Tornatore, L.; Capece, D.; Sandomenico, A.; Verzella, D.; Vecchiotti, D.; Zazzeroni, F.; Ruvo, M.; Franzoso, G. The Screening of Combinatorial Peptide Libraries for Targeting Key Molecules or Protein-Protein Interactions in the NF-ΚB Pathway. Methods Mol. Biol. Clifton NJ 2021, 2366, 343–356. [Google Scholar] [CrossRef]
- Rega, C.; Russo, R.; Focà, A.; Sandomenico, A.; Iaccarino, E.; Raimondo, D.; Milanetti, E.; Tornatore, L.; Franzoso, G.; Pedone, P.V.; et al. Probing the Interaction Interface of the GADD45β/MKK7 and MKK7/DTP3 Complexes by Chemical Cross-Linking Mass Spectrometry. Int. J. Biol. Macromol. 2018, 114, 114–123. [Google Scholar] [CrossRef]
- Zhao, Z.; Gao, Y.; Guan, X.; Liu, Z.; Jiang, Z.; Liu, X.; Lin, H.; Yang, M.; Li, C.; Yang, R.; et al. GADD45B as a Prognostic and Predictive Biomarker in Stage II Colorectal Cancer. Genes 2018, 9, 361. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Zhao, Y.; Wang, N.; You, N.; Zhu, W.; Zhang, P.; Ren, Q.; Yin, J.; Cheng, T.; Ma, X. GADD45g Acts as a Novel Tumor Suppressor and Its Activation Confers New Combination Regimens for the Treatment of AML. Blood 2021, 138, 464–479. [Google Scholar] [CrossRef]
- Greenfield, N.J. Methods to Estimate the Conformation of Proteins and Polypeptides from Circular Dichroism Data. Anal. Biochem. 1996, 235, 1–10. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Spectra to Estimate Protein Secondary Structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Holzwarth, G.; Doty, P. THE ULTRAVIOLET CIRCULAR DICHROISM OF POLYPEPTIDES. J. Am. Chem. Soc. 1965, 87, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, R.; Pantoja-Uceda, D.; Prieto, J.; Diercks, T.; Marcaida, M.J.; Montoya, G.; Campos-Olivas, R.; Blanco, F.J. Solution Structure of Human Growth Arrest and DNA Damage 45α (Gadd45α) and Its Interactions with Proliferating Cell Nuclear Antigen (PCNA) and Aurora A Kinase. J. Biol. Chem. 2010, 285, 22196–22201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Fu, S.; Liu, X.; Zhao, X.; Zhang, W.; Peng, W.; Wu, C.; Li, Y.; Li, X.; Bartlam, M.; et al. Crystal Structure of Human Gadd45γ [Corrected] Reveals an Active Dimer. Protein Cell 2011, 2, 814–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrag, J.D.; Jiralerspong, S.; Banville, M.; Jaramillo, M.L.; O’Connor-McCourt, M.D. The Crystal Structure and Dimerization Interface of GADD45γ. Proc. Natl. Acad. Sci. USA 2008, 105, 6566–6571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornatore, L.; Marasco, D.; Dathan, N.; Vitale, R.M.; Benedetti, E.; Papa, S.; Franzoso, G.; Ruvo, M.; Monti, S.M. Gadd45β Forms a Homodimeric Complex That Binds Tightly to MKK7. J. Mol. Biol. 2008, 378, 97–111. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular Mechanism of Thioflavin-T Binding to Amyloid Fibrils. Biochim. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [Green Version]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Balasco, N.; Diaferia, C.; Morelli, G.; Vitagliano, L.; Accardo, A. Amyloid-Like Aggregation in Diseases and Biomaterials: Osmosis of Structural Information. Front. Bioeng. Biotechnol. 2021, 9, 641372. [Google Scholar] [CrossRef]
- Chan, F.T.S.; Schierle, G.S.K.; Kumita, J.R.; Bertoncini, C.W.; Dobson, C.M.; Kaminski, C.F. Protein Amyloids Develop an Intrinsic Fluorescence Signature during Aggregation. Analyst 2013, 138, 2156–2162. [Google Scholar] [CrossRef] [Green Version]
- Monti, A.; Bruckmann, C.; Blasi, F.; Ruvo, M.; Vitagliano, L.; Doti, N. Amyloid-like Prep1 Peptides Exhibit Reversible Blue-Green-Red Fluorescence in Vitro and in Living Cells. Chem. Commun. 2021, 57, 3720–3723. [Google Scholar] [CrossRef]
- Diaferia, C.; Sibillano, T.; Altamura, D.; Roviello, V.; Vitagliano, L.; Giannini, C.; Morelli, G.; Accardo, A. Structural Characterization of PEGylated Hexaphenylalanine Nanostructures Exhibiting Green Photoluminescence Emission. Chem. Weinh. Bergstr. Ger. 2017, 23, 14039–14048. [Google Scholar] [CrossRef]
- Verzella, D.; Bennett, J.; Fischietti, M.; Thotakura, A.K.; Recordati, C.; Pasqualini, F.; Capece, D.; Vecchiotti, D.; D’Andrea, D.; Di Francesco, B.; et al. GADD45β Loss Ablates Innate Immunosuppression in Cancer. Cancer Res. 2018, 78, 1275–1292. [Google Scholar] [CrossRef] [Green Version]
- Leuenberger, P.; Ganscha, S.; Kahraman, A.; Cappelletti, V.; Boersema, P.J.; von Mering, C.; Claassen, M.; Picotti, P. Cell-Wide Analysis of Protein Thermal Unfolding Reveals Determinants of Thermostability. Science 2017, 355, eaai7825. [Google Scholar] [CrossRef]
- Doti, N.; Monti, A.; Bruckmann, C.; Calvanese, L.; Smaldone, G.; Caporale, A.; Falcigno, L.; D’Auria, G.; Blasi, F.; Ruvo, M.; et al. Identification and Characterization of Cytotoxic Amyloid-like Regions in Human Pbx-Regulating Protein-1. Int. J. Biol. Macromol. 2020, 163, 618–629. [Google Scholar] [CrossRef]
- Lim, J.; Yue, Z. Neuronal Aggregates: Formation, Clearance, and Spreading. Dev. Cell 2015, 32, 491–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Natural Polyphenols Inhibit Different Steps of the Process of Transthyretin (TTR) Amyloid Fibril Formation. FEBS Lett. 2011, 585, 2424–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, N.; Pereira-Henriques, A.; Almeida, M.R. Transthyretin Chemical Chaperoning by Flavonoids: Structure-Activity Insights towards the Design of Potent Amyloidosis Inhibitors. Biochem. Biophys. Rep. 2015, 3, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Uncovering the Neuroprotective Mechanisms of Curcumin on Transthyretin Amyloidosis. Int. J. Mol. Sci. 2019, 20, 1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahn, T.R.; Parker, M.J.; Homans, S.W.; Radford, S.E. Amyloid Formation under Physiological Conditions Proceeds via a Native-like Folding Intermediate. Nat. Struct. Mol. Biol. 2006, 13, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Sandomenico, A.; Di Rienzo, L.; Calvanese, L.; Iaccarino, E.; D’Auria, G.; Falcigno, L.; Chambery, A.; Russo, R.; Franzoso, G.; Tornatore, L.; et al. Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches. Biomedicines 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smaldone, G.; Caruso, D.; Sandomenico, A.; Iaccarino, E.; Focà, A.; Ruggiero, A.; Ruvo, M.; Vitagliano, L. Members of the GADD45 Protein Family Show Distinct Propensities to form Toxic Amyloid-Like Aggregates in Physiological Conditions. Int. J. Mol. Sci. 2021, 22, 10700. https://doi.org/10.3390/ijms221910700
Smaldone G, Caruso D, Sandomenico A, Iaccarino E, Focà A, Ruggiero A, Ruvo M, Vitagliano L. Members of the GADD45 Protein Family Show Distinct Propensities to form Toxic Amyloid-Like Aggregates in Physiological Conditions. International Journal of Molecular Sciences. 2021; 22(19):10700. https://doi.org/10.3390/ijms221910700
Chicago/Turabian StyleSmaldone, Giovanni, Daniela Caruso, Annamaria Sandomenico, Emanuela Iaccarino, Annalia Focà, Alessia Ruggiero, Menotti Ruvo, and Luigi Vitagliano. 2021. "Members of the GADD45 Protein Family Show Distinct Propensities to form Toxic Amyloid-Like Aggregates in Physiological Conditions" International Journal of Molecular Sciences 22, no. 19: 10700. https://doi.org/10.3390/ijms221910700
APA StyleSmaldone, G., Caruso, D., Sandomenico, A., Iaccarino, E., Focà, A., Ruggiero, A., Ruvo, M., & Vitagliano, L. (2021). Members of the GADD45 Protein Family Show Distinct Propensities to form Toxic Amyloid-Like Aggregates in Physiological Conditions. International Journal of Molecular Sciences, 22(19), 10700. https://doi.org/10.3390/ijms221910700










