A 4-Gene Signature of CDKN1, FDXR, SESN1 and PCNA Radiation Biomarkers for Prediction of Patient Radiosensitivity
Abstract
1. Introduction
2. Results
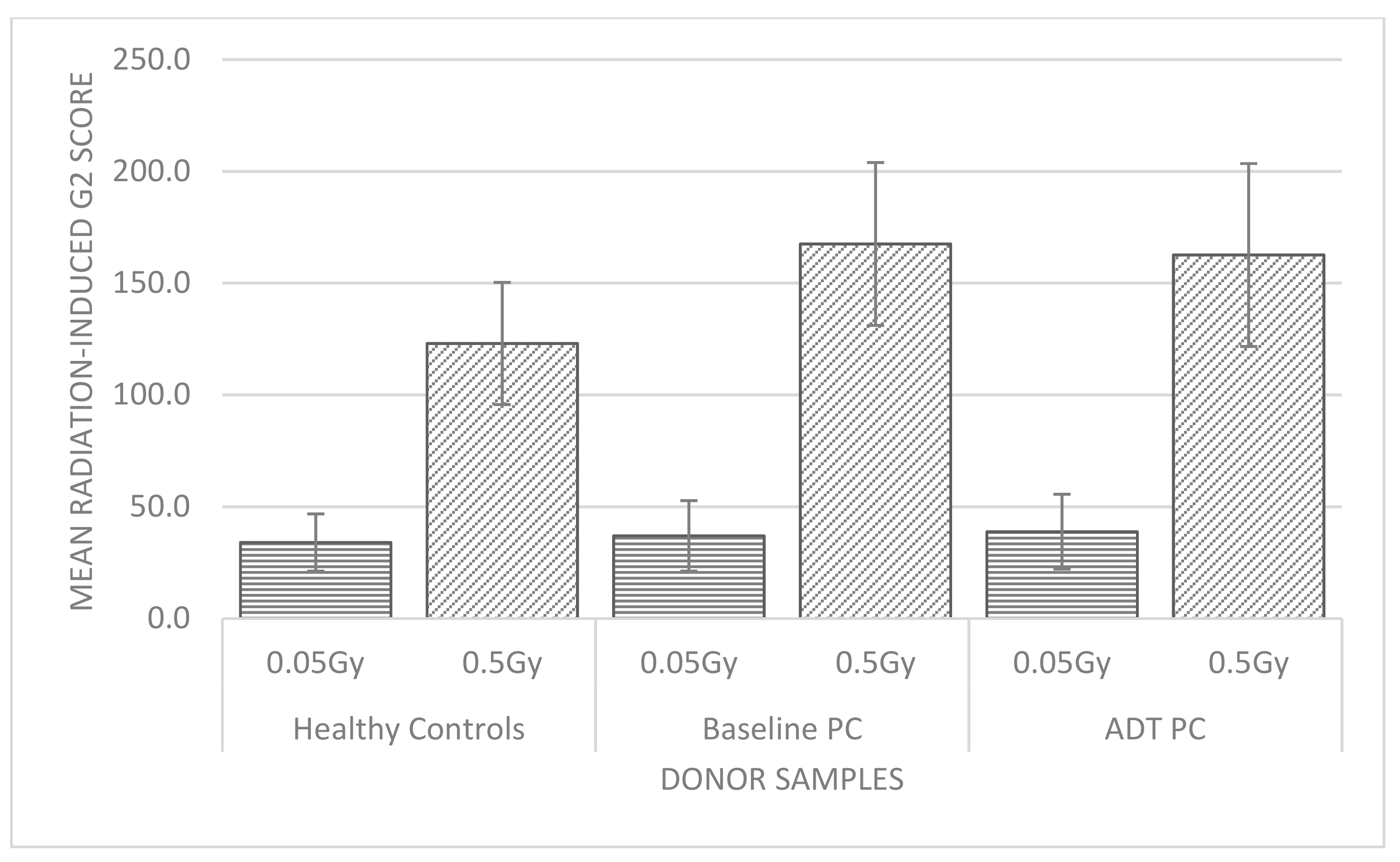
2.1. G2 Chromosomal Radiosensitivity
2.1.1. Healthy Control Donors
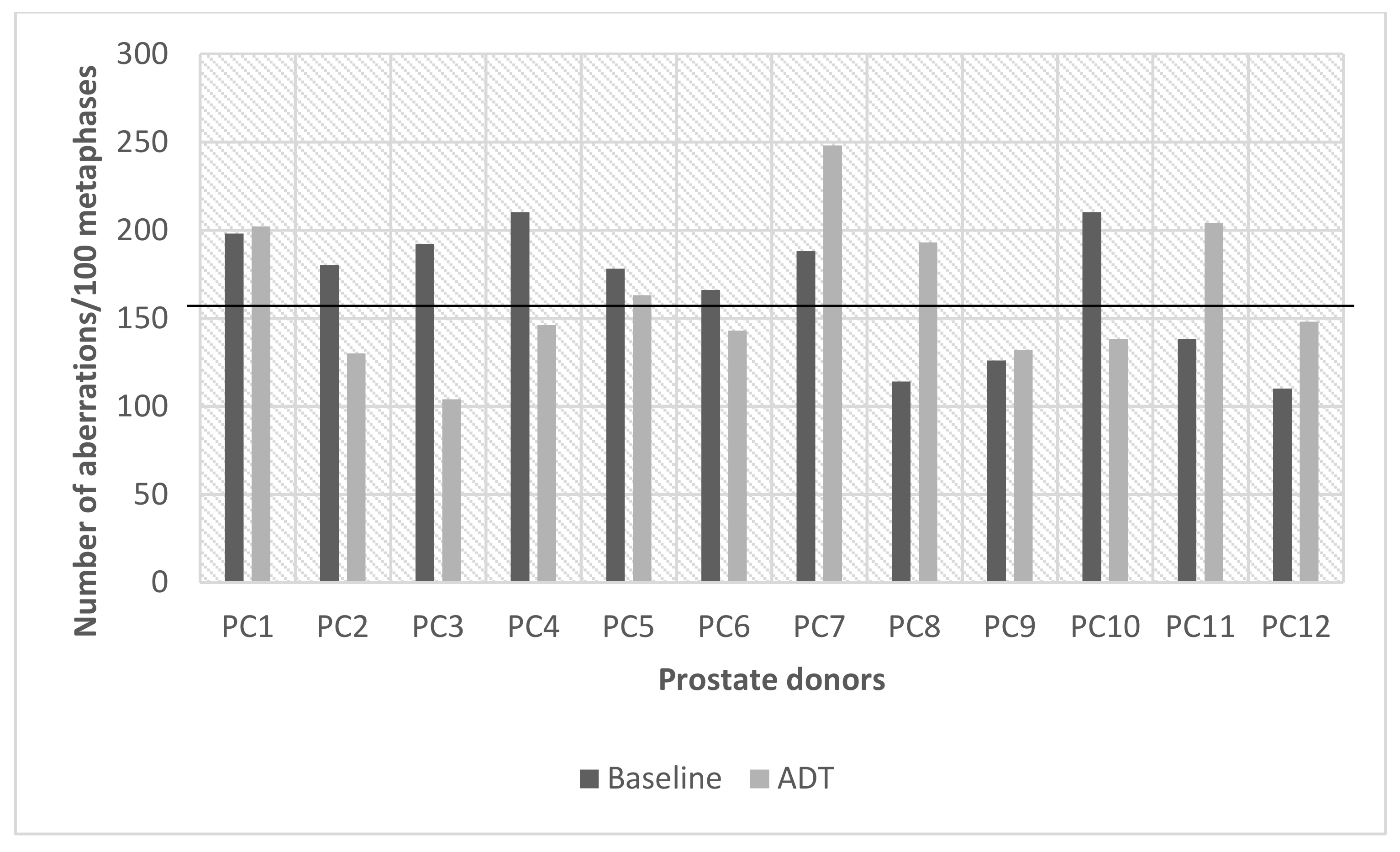
2.1.2. Prostate Cancer Donors
2.1.3. G2 Chromosomal Radiosensitivity Shifts in Prostate Cancer Donors
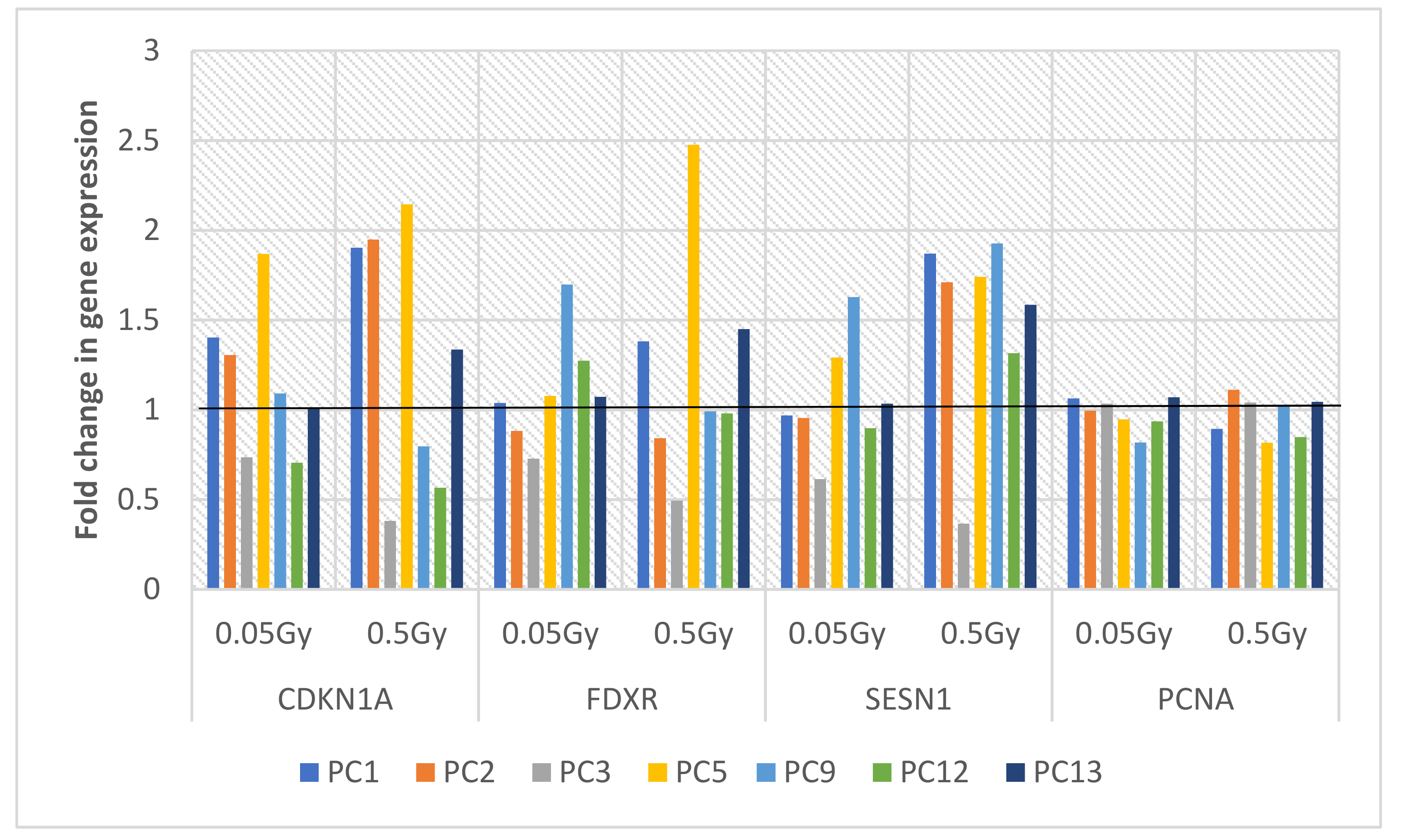
2.2. Relative Gene Expression of 4-Gene Panel
2.2.1. Healthy Control Donors
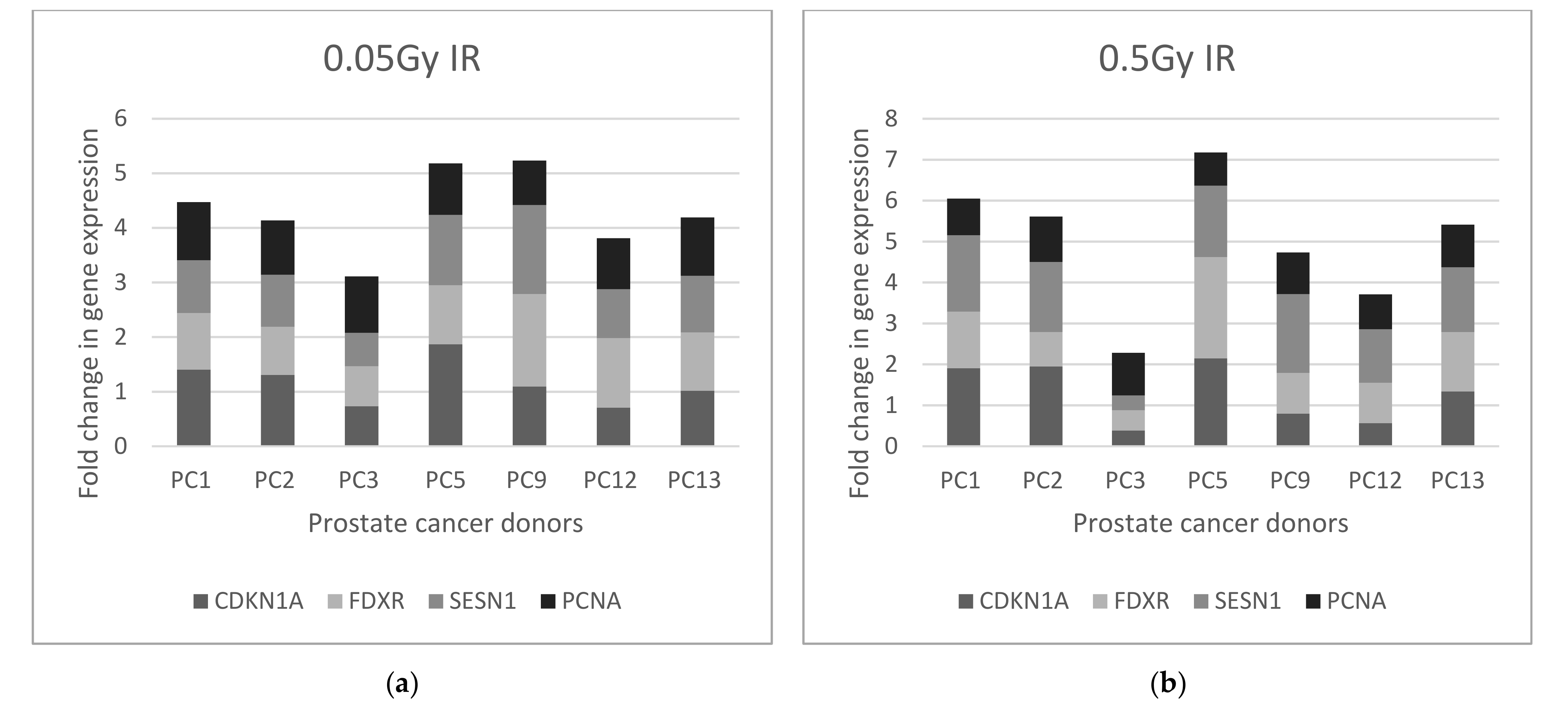
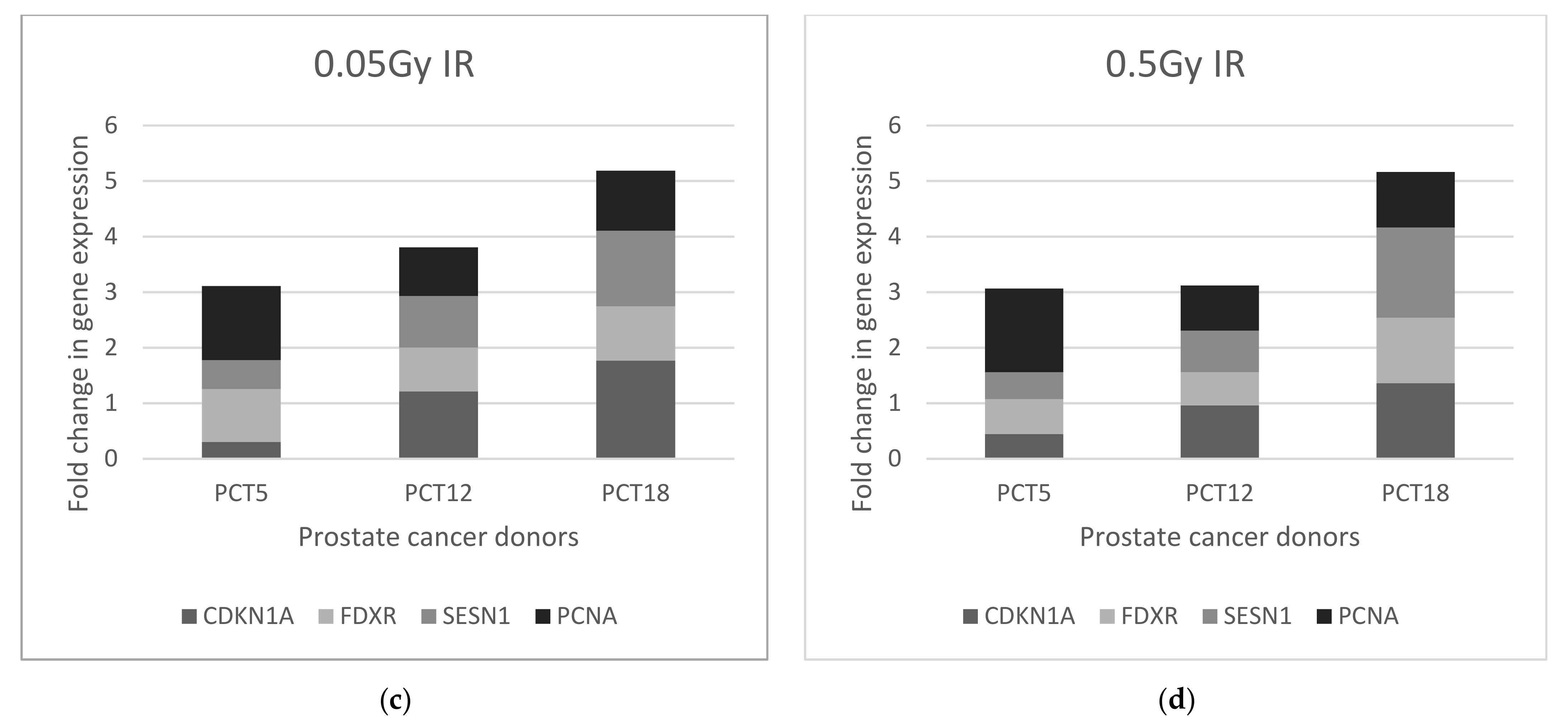
2.2.2. Prostate Cancer Donors
2.2.3. Biomarker Correlations
3. Discussion
4. Materials and Methods
4.1. Blood Samples and Donors
4.2. Whole Blood Cultures and Irradiation
4.3. G2 Chromosomal Radiosensitivity Assay
4.4. RNA Isolation and cDNA Synthesis
4.5. Multiplex Quantitative Real Time PCR (MQRT-PCR)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salomaa, S.; Averbeck, D.; Ottolenghi, A.; Sabatier, L.; Bouffler, S.; Atkinson, M.; Jourdain, J.-R. European low-dose radiation risk research strategy: Future of research on biological effects at low doses. Radiat. Prot. Dosim. 2015, 164, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Kulka, U.; Abend, M.; Ainsbury, E.; Badie, C.; Barquinero, J.F.; Barrios, L.; Beinke, C.; Bortolin, E.; Cucu, A.; De Amicis, A.; et al. RENEB—Running the European Network of biological dosimetry and physical retrospective dosimetry. Int. J. Radiat. Biol. 2017, 93, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Jeggo, P.A.; West, C.; Gomolka, M.; Quintens, R.; Badie, C.; Laurent, O.; Aerts, A.; Anastasov, N.; Azimzadeh, O.; et al. Ionizing radiation biomarkers in epidemiological studies—An update. Mutat. Res. Mutat. Res. 2017, 771, 59–84. [Google Scholar] [CrossRef] [PubMed]
- Pernot, E.; Hall, J.; Baatout, S.; Benotmane, M.A.; Blanchardon, E.; Bouffler, S.; El Saghire, H.; Gomolka, M.; Guertler, A.; Harms-Ringdahl, M.; et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat. Res. Rev. Mutat. Res. 2012, 751, 258–286. [Google Scholar] [CrossRef] [PubMed]
- Gomolka, M.; Blyth, B.; Bourguignon, M.; Badie, C.; Schmitz, A.; Talbot, C.; Hoeschen, C.; Salomaa, S. Potential screening assays for individual radiation sensitivity and susceptibility and their current validation state. Int. J. Radiat. Biol. 2020, 96, 280–296. [Google Scholar] [CrossRef]
- Abend, M.; Badie, C.; Quintens, R.; Kriehuber, R.; Manning, G.; Macaeva, E.; Njima, M.; Oskamp, D.; Strunz, S.; Moertl, S.; et al. Examining radiation-induced in vivo and in vitro gene expression changes of the peripheral blood in different laboratories for biodosimetry purposes: First RENEB Gene Expression Study. Radiat. Res. 2017, 185, 109–123. [Google Scholar] [CrossRef]
- Manning, G.; Macaeva, E.; Majewski, M.; Kriehuber, R.; Brzóska, K.; Abend, M.; Doucha-Senf, S.; Oskamp, D.; Strunz, S.; Quintens, R.; et al. Comparable dose estimates of blinded whole blood samples are obtained independently of culture conditions and analytical approaches. Second RENEB gene expression study. Int. J. Radiat. Biol. 2017, 93, 87–98. [Google Scholar] [CrossRef]
- Badie, C.; Dziwura, S.; Raffy, C.; Tsigani, T.; Alsbeih, G.; Moody, J.; Finnon, P.; Levine, E.; Scott, D.; Bouffler, S. Aberrant CDKN1A transcriptional response associates with abnormal sensitivity to radiation treatment. Br. J. Cancer 2008, 98, 1845–1851. [Google Scholar] [CrossRef]
- El-Deiry, W.S. p21(WAF1) Mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016, 76, 5189–5191. [Google Scholar] [CrossRef]
- Manning, G.; Rothkamm, K. Deoxyribonucleic acid damage-associated biomarkers of ionising radiation: Current status and future relevance for radiology and radiotherapy. Br. J. Radiol. 2013, 86, 20130173. [Google Scholar] [CrossRef]
- Cruz-Garcia, L.; O’Brien, G.; Sipos, B.; Mayes, S.; Love, M.I.; Turner, D.J.; Badie, C. Generation of a transcriptional radiation exposure signature in human blood using long-read nanopore sequencing. Radiat. Res. 2020, 193, 143–154. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, G.; Cruz-Garcia, L.; Majewski, M.; Grepl, J.; Abend, M.; Port, M.; Tichý, A.; Sirak, I.; Malkova, A.; Donovan, E.; et al. FDXR is a biomarker of radiation exposure in vivo. Sci. Rep. 2018, 8, 684. [Google Scholar] [CrossRef]
- Kabacik, S.; Manning, G.; Raffy, C.; Bouffler, S.; Badie, C. Time, Dose and Ataxia Telangiectasia Mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiat. Res. 2015, 183, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Kabacik, S.; Mackay, A.; Tamber, N.; Manning, G.; Finnon, P.; Paillier, F.; Ashworth, A.; Bouffler, S.; Badie, C. Gene expression following ionising radiation: Identification of biomarkers for dose estimation and prediction of individual response. Int. J. Radiat. Biol. 2011, 87, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Buckbinder, L.; Talbott, R.; Seizinger, B.R.; Kley, N. Gene regulation by temperature-sensitive p53 mutants: Identification of p53 response genes. Proc. Natl. Acad. Sci. USA 1994, 91, 10640–10644. [Google Scholar] [CrossRef]
- Budanov, A.V.; Shoshani, T.; Faerman, A.; Zelin, E.; Kamer, I.; Kalinski, H.; Gorodin, S.; Fishman, A.; Chajut, A.; Einat, P.; et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene 2002, 21, 6017–6031. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Tan, C.K.; Castillo, C.; So, A.G.; Downey, K.M. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J. Biol. Chem. 1986, 261, 12310–12316. [Google Scholar] [CrossRef]
- Prelich, G.; Tan, C.-K.; Kostura, M.; Mathews, M.B.; So, A.G.; Downey, K.M.; Stillman, B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-δ auxiliary protein. Nature 1987, 326, 517–520. [Google Scholar] [CrossRef]
- Cazzalini, O.; Sommatis, S.; Tillhon, M.; Dutto, I.; Bachi, A.; Rapp, A.; Nardo, T.; Scovassi, A.I.; Necchi, D.; Cardoso, M.C.; et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res. 2014, 42, 8433–8448. [Google Scholar] [CrossRef]
- Mailand, N.; Gibbs-Seymour, I.; Bekker-Jensen, S. Regulation of PCNA–protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 2013, 14, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Kawabe, T.; Ohara, H.; Ducommun, B.; Itoh, M.; Okamoto, T. Involvement of the Interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J. Biol. Chem. 2001, 276, 42971–42977. [Google Scholar] [CrossRef] [PubMed]
- Cazzalini, O.; Perucca, P.; Savio, M.; Necchi, D.; Bianchi, L.; Stivala, L.A.; Ducommun, B.; Scovassi, A.I.; Prosperi, E. Interaction of p21 CDKN1A with PCNA regulates the histone acetyltransferase activity of p300 in nucleotide excision repair. Nucleic Acids Res. 2008, 36, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Parshad, R.; Sanford, K.K.; Jones, G.M. Chromatid damage after G2 phase x-irradiation of cells from cancer-prone individuals implicates deficiency in DNA repair. Proc. Natl. Acad. Sci. USA 1983, 80, 5612–5616. [Google Scholar] [CrossRef] [PubMed]
- Parshad, R.; Sanford, K.K.; Jones, G.M. Chromosomal radiosensitivity during the G2 cell-cycle period of skin fibroblasts from individuals with familial cancer. Proc. Natl. Acad. Sci. USA 1985, 82, 5400–5403. [Google Scholar] [CrossRef]
- Haskins, J.S.; Kato, T.A. G2 chromosomal radiosensitivity assay for testing individual radiation sensitivity. Adv. Struct. Saf. Stud. 2019, 1984, 39–45. [Google Scholar] [CrossRef]
- White, L.; Shields, L.; Howe, O.; Vega Carrascal, I.; Maguire, A.; Meade, A.; McClean, B.; Lyng, F. A comparison of radiobiological response in cells exposed to low LET radiation with different beam energies. Radiat. Environ. Med. 2020, 9, 1–6. [Google Scholar]
- Meade, A.D.; Maguire, A.; Bryant, J.; Cullen, D.; Medipally, D.; White, L.; McClean, B.; Shields, L.; Armstrong, J.; Dunne, M.; et al. Prediction of DNA damage and G2 chromosomal radio-sensitivity ex vivo in peripheral blood mononuclear cells with label-free Raman micro-spectroscopy. Int. J. Radiat. Biol. 2019, 95, 44–53. [Google Scholar] [CrossRef]
- Howe, O.; O’Sullivan, J.; Nolan, B.; Vaughan, J.; Gorman, S.; Clarke, C.; McClean, B.; Lyng, F.M. Do radiation-induced bystander effects correlate to the intrinsic radiosensitivity of individuals and have clinical significance? Radiat. Res. 2009, 171, 521–529. [Google Scholar] [CrossRef]
- Howe, O.; O’Malley, K.; Lavin, M.; Gardner, R.A.; Seymour, C.; Lyng, F.; Mulvin, D.; Quinlan, D.M.; Mothersill, C. Cell death mechanisms associated with G2 radiosensitivity in patients with prostate cancer and benign prostatic hyperplasia. Radiat. Res. 2005, 164, 627–634. [Google Scholar] [CrossRef]
- Howe, O.L.; Daly, P.A.; Seymour, C.; Ormiston, W.; Nolan, C.; Mothersill, C. Elevated G2 chromosomal radiosensitivity in Irish breast cancer patients: A comparison with other studies. Int. J. Radiat. Biol. 2005, 81, 373–378. [Google Scholar] [CrossRef]
- Borgmann, K.; Raabe, A.; Reuther, S.; Szymczak, S.; Schlomm, T.; Isbarn, H.; Gomolka, M.; Busjahn, A.; Bonin, M.; Ziegler, A.; et al. The potential role of G2- but not of G0-radiosensitivity for predisposition of prostate cancer. Radiother. Oncol. 2010, 96, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Marples, B.; Wouters, B.G.; Collis, S.J.; Chalmers, A.J.; Joiner, M.C. Low-dose hyper-radiosensitivity: A consequence of ineffective cell cycle arrest of radiation-damaged G2-phase cells. Radiat. Res. 2004, 161, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Marples, B.; Collis, S.J. Low-dose hyper-radiosensitivity: Past, present, and future. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, D.G.; Diehn, M.; Kesarwala, A.; Maity, A.; Morgan, M.A.; Schwarz, J.K.; Bristow, R.; DeMaria, S.; Eke, I.; Griffin, R.J.; et al. The future of radiobiology. J. Natl. Cancer Inst. 2018, 110, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Badie, C.; Hess, J.; Zitzelsberger, H.; Kulka, U. Established and emerging biomarkers of radiation exposure. Clin. Oncol. 2016, 28, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Vinnikov, V.; Hande, M.P.; Wilkins, R.; Wojcik, A.; Zubizarreta, E.; Belyakov, O. Prediction of the acute or late radiation toxicity effects in radiotherapy patients using ex vivo induced biodosimetric markers: A review. J. Pers. Med. 2020, 10, 285. [Google Scholar] [CrossRef]
- Crawford, E.D.; Heidenreich, A.; Lawrentschuk, N.; Tombal, B.; Pompeo, A.C.L.; Mendoza-Valdes, A.; Miller, K.; Debruyne, F.M.J.; Klotz, L. Androgen-targeted therapy in men with prostate cancer: Evolving practice and future considerations. Prostate Cancer Prostatic Dis. 2019, 22, 24–38. [Google Scholar] [CrossRef]
- Siddiqui, Z.A.; Krauss, D.J. Adjuvant androgen deprivation therapy for prostate cancer treated with radiation therapy. Transl. Androl. Urol. 2018, 7, 378–389. [Google Scholar] [CrossRef]
- Spratt, D.E.; Evans, M.J.; Davis, B.J.; Doran, M.G.; Lee, M.X.; Shah, N.; Wongvipat, J.; Carnazza, K.E.; Klee, G.G.; Polkinghorn, W.; et al. Androgen receptor upregulation mediates radioresistance after ionizing radiation. Cancer Res. 2015, 75, 4688–4696. [Google Scholar] [CrossRef]
- Curwen, G.B.; Cadwell, K.K.; Tawn, E.J.; Winther, J.F.; Boice, J.D. Intra-individual variation in G2 chromosomal radiosensitivity. Mutagenesis 2012, 27, 471–475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vral, A.; Thierens, H.; Baeyens, A.; De Ridder, L. Chromosomal aberrations and in vitro radiosensitivity: Intra-individual versus inter-individual variability. Toxicol. Lett. 2004, 149, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Badie, C.; Kabacik, S.; Balagurunathan, Y.; Bernard, N.; Brengues, M.; Faggioni, G.; Greither, R.; Lista, F.; Peinnequin, A.; Poyot, T.; et al. Laboratory intercomparison of gene expression assays. Radiat. Res. 2013, 180, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Zyla, J.; Kabacik, S.; O’Brien, G.; Wakil, S.; Al-Harbi, N.; Kaprio, J.; Badie, C.; Polanska, J.; Alsbeih, G. Combining CDKN1A gene expression and genome-wide SNPs in a twin cohort to gain insight into the heritability of individual radiosensitivity. Funct. Integr. Genom. 2019, 19, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Garcia, L.; O’Brien, G.; Sipos, B.; Mayes, S.; Tichý, A.; Sirák, I.; Davídková, M.; Marková, M.; Turner, D.J.; Badie, C. In vivo validation of alternative FDXR transcripts in human blood in response to ionizing radiation. Int. J. Mol. Sci. 2020, 21, 7851. [Google Scholar] [CrossRef]
- Manning, G.; Tichý, A.; Sirak, I.; Badie, C. Radiotherapy-associated long-term modification of expression of the inflammatory biomarker genes ARG1, BCL2L1, and MYC. Front. Immunol. 2017, 8, 412. [Google Scholar] [CrossRef] [PubMed]
- Port, M.; Ostheim, P.; Majewski, M.; Voss, T.; Haupt, J.; Lamkowski, A.; Abend, M. Rapid high-throughput diagnostic triage after a mass radiation exposure event using early gene expression changes. Radiat. Res. 2019, 192, 208–218. [Google Scholar] [CrossRef]
- Port, M.; Hérodin, F.; Valente, M.; Drouet, M.; Ostheim, P.; Majewski, M.; Abend, M. Persistent mRNA and miRNA expression changes in irradiated baboons. Sci. Rep. 2018, 8, 15353. [Google Scholar] [CrossRef]
- Gao, F.; Liu, P.; Narayanan, J.; Yang, M.; Fish, B.L.; Liu, Y.; Liang, M.; Jacobs, E.R.; Medhora, M. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci. Rep. 2017, 7, 44132. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Omaruddin, R.A.; Brumbaugh, C.D.; Tariq, M.A.; Pourmand, N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 2013, 54, 808–822. [Google Scholar] [CrossRef]
- Bryant, J.; White, L.; Coen, N.; Shields, L.; McClean, B.; Meade, A.D.; Lyng, F.M.; Howe, O. MicroRNA analysis of ATM-deficient cells indicate PTEN and CCDN1 as potential biomarkers of radiation response. Radiat. Res. 2020, 193, 520. [Google Scholar] [CrossRef]
- Labbé, M.; Hoey, C.; Ray, J.; Potiron, V.; Supiot, S.; Liu, S.K.; Fradin, D. microRNAs identified in prostate cancer: Correlative studies on response to ionizing radiation. Mol. Cancer 2020, 19, 63. [Google Scholar] [CrossRef]
- Malachowska, B.; Tomasik, B.; Stawiski, K.; Kulkarni, S.; Guha, C.; Chowdhury, D.; Fendler, W. Circulating microRNAs as biomarkers of radiation exposure: A systematic review and meta-analysis. Int. J. Radiat. Oncol. Biol Phys. 2020, 106, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Enelund, L.; Nielsen, L.N.; Cirera, S. Evaluation of microRNA stability in plasma and serum from healthy dogs. MicroRNA 2017, 6, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Balzano, F.; Deiana, M.; Giudici, S.D.; Oggiano, A.; Baralla, A.; Pasella, S.; Mannu, A.; Pescatori, M.; Porcu, B.; Fanciulli, G.; et al. miRNA stability in frozen plasma samples. Molecules 2015, 20, 19030–19040. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, V.; Karunasinghe, N.; Jabed, A.; Pallati, R.; Kao, C.H.-J.; Wang, A.; Marlow, G.; Ferguson, L.R. Prostate cancer: Is it a battle lost to age? Geriatrics 2016, 1, 27. [Google Scholar] [CrossRef]
- Furlong, H.; Mothersill, C.; Lyng, F.M.; Howe, O. Apoptosis is signalled early by low doses of ionising radiation in a radiation-induced bystander effect. Mutat. Res. Mol. Mech. Mutagen. 2013, 741–742, 35–43. [Google Scholar] [CrossRef]
- Bryant, P.E.; Gray, L.; Riches, A.C.; Steel, C.M.; Finnon, P.; Howe, O.; Kesterton, I.; Vral, A.; Curwen, G.B.; Smart, V.; et al. The G2 chromosomal radiosensitivity assay. Int. J. Radiat. Biol. 2002, 78, 863–866. [Google Scholar] [CrossRef]
- Scott, D.; Barber, J.B.; Spreadborough, A.R.; Burrill, W.; Roberts, S.A. Increased chromosomal radiosensitivity in breast cancer patients: A comparison of two assays. Int. J. Radiat. Biol. 1999, 75, 1–10. [Google Scholar] [CrossRef]
- Lillicrap, S.C.; Owen, B.; Williams, J.R.; Williams, P.C. Code of Practice for high-energy photon therapy dosimetry based on the NPL absorbed dose calibration service. Phys. Med. Biol. 1990, 35, 1355–1360. [Google Scholar] [CrossRef]
- Lewis, D.; Micke, A.; Yu, X.; Chan, M.F. An efficient protocol for radiochromic film dosimetry combining calibration and measurement in a single scan. Med. Phys. 2012, 39, 6339–6350. [Google Scholar] [CrossRef] [PubMed]
- Fiandra, C.; Ricardi, U.; Ragona, R.; Anglesio, S.; Giglioli, F.R.; Calamia, E.; Lucio, F. Clinical use of EBT model Gafchromic film in radiotherapy. Med. Phys. 2006, 33, 4314–4319. [Google Scholar] [CrossRef] [PubMed]
- Micke, A.; Lewis, D.F.; Yu, X. Multichannel film dosimetry with nonuniformity correction. Med. Phys. 2011, 38, 2523–2534. [Google Scholar] [CrossRef] [PubMed]

| Statistic | 0.05 Gy | 0.5 Gy |
|---|---|---|
| Mean | 33.7 * | 122.6 * |
| Standard deviation | 12.7 | 27.3 |
| Coefficient of variation | 37.9 | 22.3 |
| 90th percentile | 50 * | 152 * |
| Gene | Primers (5′-3′) | Probes (5′-3′) |
|---|---|---|
| * HPRT1 | F-TCAGGCAGTATAATCCAAAGATGG | CGCAAGCTTGCTGGTGAAAAGGACCC |
| CDKN1A | R-AGTCTGGCTTATATCCAACACTTCGT F-GCAGACCAGCATGACAG R-TAGGGCTTCCTCTTGGA | TTTCTACCACTCCAAACGCCGGCT |
| FDXR | F-GTACAACGGGCTTCCTGAGA R-CTCAGGTGGGGTCAGTAGGA | CGGGCCACGTCCAGAGCCA |
| SESN1 | F-GCTGTCTTGTGCATTACTTGTG R-CTGCGCAGCAGTCTACAG | ACATGTCCCACAACTTTGGTGCTGG |
| PCNA | F-CTCAAGGACCTCATCAACGA R-GGACATACTGGTGAGGTTCA | CCGCTGCGACCGCAACCTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howe, O.; White, L.; Cullen, D.; O’Brien, G.; Shields, L.; Bryant, J.; Noone, E.; Bradshaw, S.; Finn, M.; Dunne, M.; et al. A 4-Gene Signature of CDKN1, FDXR, SESN1 and PCNA Radiation Biomarkers for Prediction of Patient Radiosensitivity. Int. J. Mol. Sci. 2021, 22, 10607. https://doi.org/10.3390/ijms221910607
Howe O, White L, Cullen D, O’Brien G, Shields L, Bryant J, Noone E, Bradshaw S, Finn M, Dunne M, et al. A 4-Gene Signature of CDKN1, FDXR, SESN1 and PCNA Radiation Biomarkers for Prediction of Patient Radiosensitivity. International Journal of Molecular Sciences. 2021; 22(19):10607. https://doi.org/10.3390/ijms221910607
Chicago/Turabian StyleHowe, Orla, Lisa White, Daniel Cullen, Grainne O’Brien, Laura Shields, Jane Bryant, Emma Noone, Shirley Bradshaw, Marie Finn, Mary Dunne, and et al. 2021. "A 4-Gene Signature of CDKN1, FDXR, SESN1 and PCNA Radiation Biomarkers for Prediction of Patient Radiosensitivity" International Journal of Molecular Sciences 22, no. 19: 10607. https://doi.org/10.3390/ijms221910607
APA StyleHowe, O., White, L., Cullen, D., O’Brien, G., Shields, L., Bryant, J., Noone, E., Bradshaw, S., Finn, M., Dunne, M., Shannon, A. M., Armstrong, J., McClean, B., Meade, A., Badie, C., & Lyng, F. M. (2021). A 4-Gene Signature of CDKN1, FDXR, SESN1 and PCNA Radiation Biomarkers for Prediction of Patient Radiosensitivity. International Journal of Molecular Sciences, 22(19), 10607. https://doi.org/10.3390/ijms221910607










