Glycan–Lectin Interactions in Cancer and Viral Infections and How to Disrupt Them
Abstract
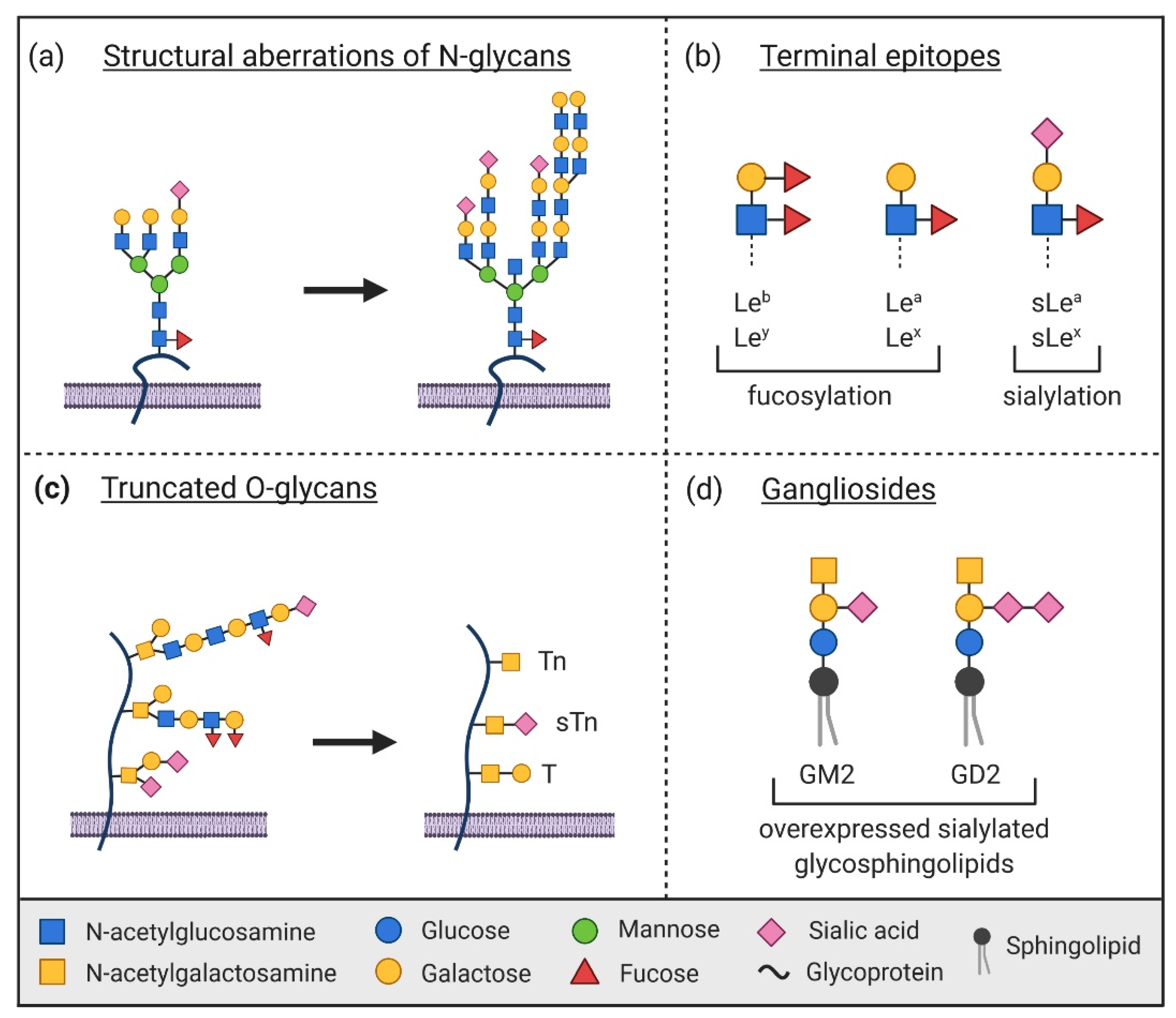
1. Introduction
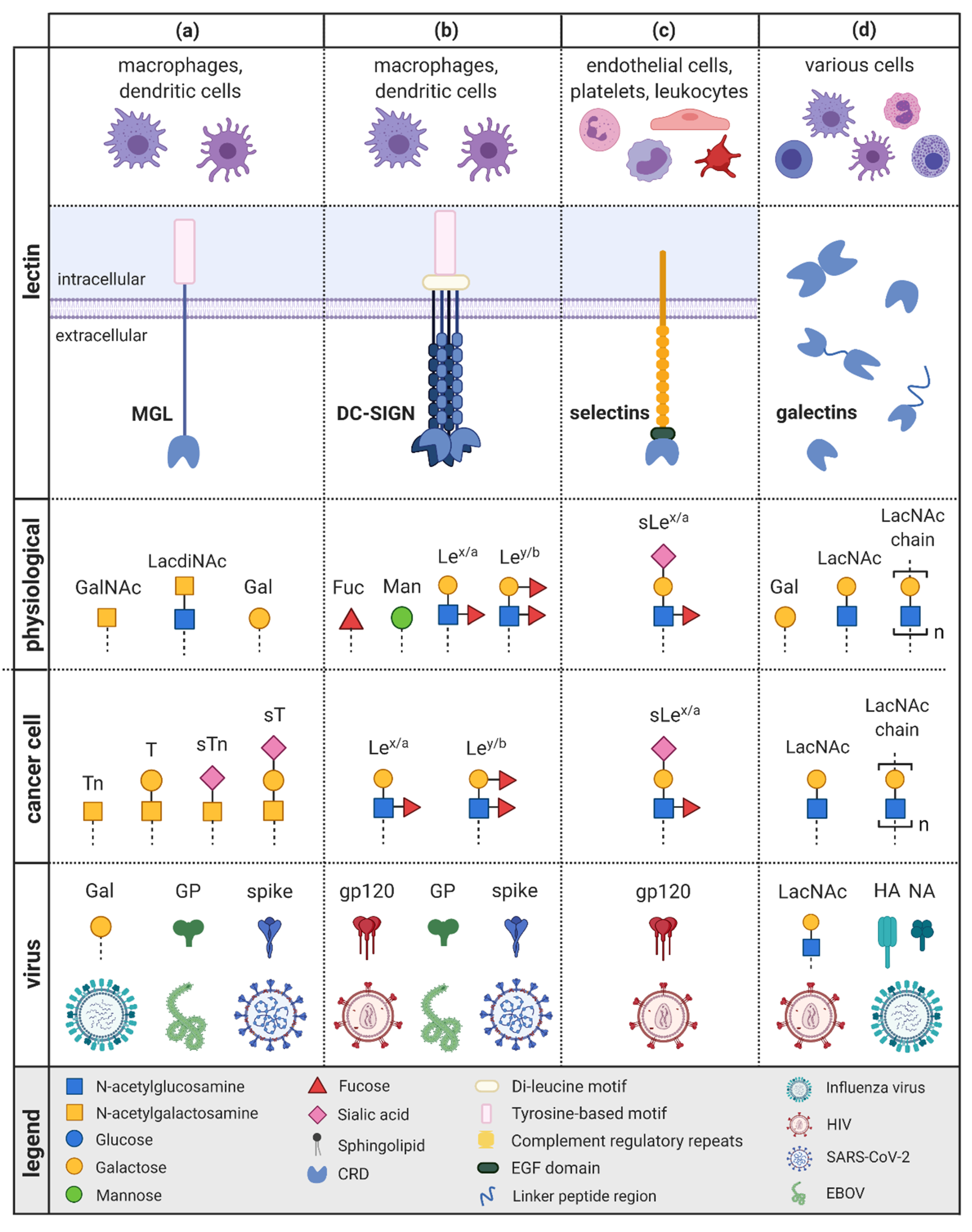
2. Macrophage Galactose-Binding Lectin
2.1. MGL in Cancer
2.2. MGL in Viral Infections
3. Dendritic Cell-Specific ICAM3-Grabbing Non-Integrin
3.1. DC-SIGN in Tumor Escape and Progression
3.2. DC-SIGN in Viral Infections
4. Selectins
4.1. Selectins in Cancer
4.2. Selectins in Viral Infections
5. Galectins
5.1. Galectins in Cancer
5.1.1. Galectin-1 and Galectin-3
5.1.2. Galectin-9
5.2. Galectins in Viral Infections
5.2.1. Galectin-1
5.2.2. Galectin-3
5.2.3. Galectin-9
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | acute myeloid leukemia |
| APC | antigen-presenting cell |
| CAR | chimeric antigen receptor |
| CEA | carcinoembryonic antigen |
| CLR | C-type lectin receptor |
| CML | chronic myeloid leukemia |
| CRD | carbohydrate recognition domain |
| CTL | cytotoxic T-lymphocyte |
| CTLD | C-type lectin-like domain |
| DC | dendritic cell |
| DC-SIGN | DC-specific intracellular adhesion molecule 3-grabbing nonintegrin |
| EBOV | Ebola virus |
| EGF | epidermal growth factor |
| GalNAc | α-N-acetylgalactosamine |
| GP | glycoprotein |
| HIV | human immunodeficiency virus |
| ICAM3 | intracellular adhesion molecule 3 |
| iDC | immature dendritic cell |
| IL | interleukin |
| LacNAc | N-acetyllactosamine |
| Le | Lewis antigen |
| MGL | macrophage galactose-binding lectin |
| MHC | major histocompatibility complex |
| MUC1 | mucin 1 |
| NK cell | natural killer cell |
| PSGL-1 | P-selectin glycoprotein ligand-1 |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SCR | short consensus repeat |
| Th cell | T-helper cell |
| TIM-3 | T cell immunoglobulin and mucin-domain containing 3 |
References
- Alberts, B.; Johnson, A.D.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Varki, A.; Kannagi, R.; Toole, B.; Stanley, P. Glycosylation Changes in Cancer. In Essentials Glycobiology, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2015. [Google Scholar]
- Kaltner, H.; Gabius, H.J. Sensing Glycans as Biochemical Messages by Tissue Lectins: The Sugar Code at Work in Vascular Biology. Thromb. Haemost. 2019, 119, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Crihfield, C.L.; Gattu, S.; Veltri, L.M.; Holland, L.A. Capillary Electrophoresis Separations of Glycans. Chem. Rev. 2018, 118, 7867–7885. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Schnaar, R.L.; Esko, J.D.; Drickamer, K.; Taylor, M.E. Principles of Glycan Recognition, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2015. [Google Scholar]
- Cagnoni, A.J.; Pérez Sáez, J.M.; Rabinovich, G.A.; Mariño, K.V. Turning-Off Signaling by Siglecs, Selectins, and Galectins: Chemical Inhibition of Glycan-Dependent Interactions in Cancer. Front. Oncol. 2016, 6, 109. [Google Scholar] [CrossRef]
- RodrÍguez, E.; Schetters, S.T.T.; van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Van Breedam, W.; Pöhlmann, S.; Favoreel, H.W.; de Groot, R.J.; Nauwynck, H.J. Bitter-sweet symphony: Glycan–lectin interactions in virus biology. FEMS Microbiol. Rev. 2014, 38, 598–632. [Google Scholar] [CrossRef]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- Alley, W.R., Jr.; Novotny, M.V. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J. Proteome Res. 2010, 9, 3062–3072. [Google Scholar] [CrossRef]
- Suenaga, T.; Arase, H. Viral Interactions with Glycans. Glycosci. Biol. Med. 2014, 785–794. [Google Scholar] [CrossRef]
- Bagdonaite, I.; Wandall, H.H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. [Google Scholar] [CrossRef]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, S.J.; Saeland, E.; van Kooyk, Y. Sweet preferences of MGL: Carbohydrate specificity and function. Trends Immunol. 2008, 29, 83–90. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Drickamer, K.; Fadden, A.J. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002, 59–72. [Google Scholar] [CrossRef]
- Drickamer, K. Ca2+-dependent carbohydrate-recognition domains in animal proteins. Curr. Opin. Struct. Biol. 1993, 3, 393–400. [Google Scholar] [CrossRef]
- Zizzari, I.G.; Napoletano, C.; Battisti, F.; Rahimi, H.; Caponnetto, S.; Pierelli, L.; Nuti, M.; Rughetti, A. MGL Receptor and Immunity: When the Ligand Can Make the Difference. J. Immunol. Res. 2015, 2015, 450695. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, S.J.; van Liempt, E.; Saeland, E.; Aarnoudse, C.A.; Appelmelk, B.; Irimura, T.; Geijtenbeek, T.B.; Blixt, O.; Alvarez, R.; van Die, I.; et al. Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 2005, 17, 661–669. [Google Scholar] [CrossRef]
- Van Vliet, S.J.; Gringhuis, S.I.; Geijtenbeek, T.B.; van Kooyk, Y. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 2006, 7, 1200–1208. [Google Scholar] [CrossRef]
- Suzuki, N.; Yamamoto, K.; Toyoshima, S.; Osawa, T.; Irimura, T. Molecular cloning and expression of cDNA encoding human macrophage C-type lectin. Its unique carbohydrate binding specificity for Tn antigen. J. Immunol. 1996, 156, 128–135. [Google Scholar]
- Higashi, N.; Fujioka, K.; Denda-Nagai, K.; Hashimoto, S.; Nagai, S.; Sato, T.; Fujita, Y.; Morikawa, A.; Tsuiji, M.; Miyata-Takeuchi, M.; et al. The macrophage C-type lectin specific for galactose/N-acetylgalactosamine is an endocytic receptor expressed on monocyte-derived immature dendritic cells. J. Biol. Chem. 2002, 277, 20686–20693. [Google Scholar] [CrossRef] [PubMed]
- Saeland, E.; van Vliet, S.J.; Bäckström, M.; van den Berg, V.C.; Geijtenbeek, T.B.; Meijer, G.A.; van Kooyk, Y. The C-type lectin MGL expressed by dendritic cells detects glycan changes on MUC1 in colon carcinoma. Cancer Immunol. Immunother. 2007, 56, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.A.; Burchell, J.M.; Taylor-Papadimitriou, J. The polymorphic epithelial mucin: Potential as an immunogen for a cancer vaccine. Cancer Immunol. Immunother. 1996, 42, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zaal, A.; Li, R.J.E.; Lübbers, J.; Bruijns, S.C.M.; Kalay, H.; van Kooyk, Y.; van Vliet, S.J. Activation of the C-Type Lectin MGL by Terminal GalNAc Ligands Reduces the Glycolytic Activity of Human Dendritic Cells. Front. Immunol. 2020, 11, 305. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.A.; Dong, H.F.; Howard, O.M.; Oppenheim, J.J.; Hanisch, F.G.; Finn, O.J. Human tumor antigen MUC1 is chemotactic for immature dendritic cells and elicits maturation but does not promote Th1 type immunity. J. Immunol. 2005, 175, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.H.; Schrama, D.; Thor Straten, P.; Becker, J.C. Cytotoxic T cells. J. Investig. Dermatol. 2006, 126, 32–41. [Google Scholar] [CrossRef]
- Rughetti, A.; Pellicciotta, I.; Biffoni, M.; Bäckström, M.; Link, T.; Bennet, E.P.; Clausen, H.; Noll, T.; Hansson, G.C.; Burchell, J.M.; et al. Recombinant tumor-associated MUC1 glycoprotein impairs the differentiation and function of dendritic cells. J. Immunol. 2005, 174, 7764–7772. [Google Scholar] [CrossRef]
- Kobayashi, M.; Fitz, L.; Ryan, M.; Hewick, R.M.; Clark, S.C.; Chan, S.; Loudon, R.; Sherman, F.; Perussia, B.; Trinchieri, G. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 1989, 170, 827–845. [Google Scholar] [CrossRef]
- Trinchieri, G.; Wysocka, M.; D’Andrea, A.; Rengaraju, M.; Aste-Amezaga, M.; Kubin, M.; Valiante, N.M.; Chehimi, J. Natural killer cell stimulatory factor (NKSF) or interleukin-12 is a key regulator of immune response and inflammation. Prog. Growth Factor Res. 1992, 4, 355–368. [Google Scholar] [CrossRef]
- Posey, A.D., Jr.; Schwab, R.D.; Boesteanu, A.C.; Steentoft, C.; Mandel, U.; Engels, B.; Stone, J.D.; Madsen, T.D.; Schreiber, K.; Haines, K.M.; et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity 2016, 44, 1444–1454. [Google Scholar] [CrossRef]
- Lavrsen, K.; Madsen, C.B.; Rasch, M.G.; Woetmann, A.; Ødum, N.; Mandel, U.; Clausen, H.; Pedersen, A.E.; Wandall, H.H. Aberrantly glycosylated MUC1 is expressed on the surface of breast cancer cells and a target for antibody-dependent cell-mediated cytotoxicity. Glycoconj. J. 2013, 30, 227–236. [Google Scholar] [CrossRef]
- Napoletano, C.; Zizzari, I.G.; Rughetti, A.; Rahimi, H.; Irimura, T.; Clausen, H.; Wandall, H.H.; Belleudi, F.; Bellati, F.; Pierelli, L.; et al. Targeting of macrophage galactose-type C-type lectin (MGL) induces DC signaling and activation. Eur. J. Immunol. 2012, 42, 936–945. [Google Scholar] [CrossRef]
- Pirro, M.; Rombouts, Y.; Stella, A.; Neyrolles, O.; Burlet-Schiltz, O.; van Vliet, S.J.; de Ru, A.H.; Mohammed, Y.; Wuhrer, M.; van Veelen, P.A.; et al. Characterization of Macrophage Galactose-type Lectin (MGL) ligands in colorectal cancer cell lines. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129513. [Google Scholar] [CrossRef]
- Basak, S.; Tomana, M.; Compans, R.W. Sialic acid is incorporated into influenza hemagglutinin glycoproteins in the absence of viral neuraminidase. Virus Res. 1985, 2, 61–68. [Google Scholar] [CrossRef]
- Upham, J.P.; Pickett, D.; Irimura, T.; Anders, E.M.; Reading, P.C. Macrophage receptors for influenza A virus: Role of the macrophage galactose-type lectin and mannose receptor in viral entry. J. Virol. 2010, 84, 3730–3737. [Google Scholar] [CrossRef]
- Ng, W.C.; Liong, S.; Tate, M.D.; Irimura, T.; Denda-Nagai, K.; Brooks, A.G.; Londrigan, S.L.; Reading, P.C. The macrophage galactose-type lectin can function as an attachment and entry receptor for influenza virus. J. Virol. 2014, 88, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Fujioka, K.; Tsuiji, M.; Morikawa, A.; Higashi, N.; Ebihara, H.; Kobasa, D.; Feldmann, H.; Irimura, T.; Kawaoka, Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 2004, 78, 2943–2947. [Google Scholar] [CrossRef] [PubMed]
- Usami, K.; Matsuno, K.; Igarashi, M.; Denda-Nagai, K.; Takada, A.; Irimura, T. Involvement of viral envelope GP2 in Ebola virus entry into cells expressing the macrophage galactose-type C-type lectin. Biochem. Biophys. Res. Commun. 2011, 407, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Fujihira, H.; Usami, K.; Matsuno, K.; Takeuchi, H.; Denda-Nagai, K.; Furukawa, J.-I.; Shinohara, Y.; Takada, A.; Kawaoka, Y.; Irimura, T. A Critical Domain of Ebolavirus Envelope Glycoprotein Determines Glycoform and Infectivity. Sci. Rep. 2018, 8, 5495. [Google Scholar] [CrossRef]
- Sano, Y.; Usami, K.; Izawa, R.; Denda-Nagai, K.; Higashi, N.; Kimura, T.; Suzuki, N.; Irimura, T. Properties of blocking and non-blocking monoclonal antibodies specific for human macrophage galactose-type C-type lectin (MGL/ClecSF10A/CD301). J. Biochem. 2007, 141, 127–136. [Google Scholar] [CrossRef]
- Marzi, A.; Yoshida, R.; Miyamoto, H.; Ishijima, M.; Suzuki, Y.; Higuchi, M.; Matsuyama, Y.; Igarashi, M.; Nakayama, E.; Kuroda, M.; et al. Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PLoS ONE 2012, 7, e36192. [Google Scholar] [CrossRef]
- Corti, D.; Misasi, J.; Mulangu, S.; Stanley, D.A.; Kanekiyo, M.; Wollen, S.; Ploquin, A.; Doria-Rose, N.A.; Staupe, R.P.; Bailey, M.; et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 2016, 351, 1339–1342. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Ebihara, H.; Jones, S.; Feldmann, H.; Kawaoka, Y. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine 2007, 25, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Fernando, L.; Melito, P.L.; Audet, J.; Feldmann, H.; Kobinger, G.; Alimonti, J.B.; Jones, S.M. Ebola GP-specific monoclonal antibodies protect mice and guinea pigs from lethal Ebola virus infection. PLoS Negl. Trop. Dis. 2012, 6, e1575. [Google Scholar] [CrossRef] [PubMed]
- FDA. First FDA-Approved Vaccine for the Prevention of Ebola Virus Disease, Marking a Critical Milestone in Public Health Preparedness and Response. Available online: https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health (accessed on 15 March 2021).
- Poetsch, J.H.; Dahlke, C.; Zinser, M.E.; Kasonta, R.; Lunemann, S.; Rechtien, A.; Ly, M.L.; Stubbe, H.C.; Krähling, V.; Biedenkopf, N.; et al. Detectable Vesicular Stomatitis Virus (VSV)-Specific Humoral and Cellular Immune Responses Following VSV-Ebola Virus Vaccination in Humans. J. Infect. Dis. 2019, 219, 556–561. [Google Scholar] [CrossRef]
- Suder, E.; Furuyama, W.; Feldmann, H.; Marzi, A.; de Wit, E. The vesicular stomatitis virus-based Ebola virus vaccine: From concept to clinical trials. Hum. Vaccin. Immunother. 2018, 14, 2107–2113. [Google Scholar] [CrossRef]
- EMA. New Vaccine for Prevention of Ebola Virus Disease Recommended for Approval in the European Union. Available online: https://www.ema.europa.eu/en/news/new-vaccine-prevention-ebola-virus-disease-recommended-approval-european-union (accessed on 15 March 2021).
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gao, C.; Zeng, J.; Jia, N.; Stavenhagen, K.; Matsumoto, Y.; Zhang, H.; Li, J.; Hume, A.J.; Mühlberger, E.; van Die, I.; et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Pöhlmann, S.; Baribaud, F.; Doms, R. DC-SIGN and DC-SIGNR: Helping hands for HIV. Trends Immunol. 2001, 22, 643–646. [Google Scholar] [CrossRef]
- Curtis, B.M.; Scharnowske, S.; Watson, A.J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 1992, 89, 8356–8360. [Google Scholar] [CrossRef]
- Figdor, C.G.; van Kooyk, Y.; Adema, G.J. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2002, 2, 77–84. [Google Scholar] [CrossRef]
- Van Vliet, S.J.; García-Vallejo, J.J.; van Kooyk, Y. Dendritic cells and C-type lectin receptors: Coupling innate to adaptive immune responses. Immunol. Cell Biol. 2008, 86, 580–587. [Google Scholar] [CrossRef]
- Feinberg, H.; Mitchell, D.A.; Drickamer, K.; Weis, W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 2001, 294, 2163–2166. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- Pederson, K.; Mitchell, D.A.; Prestegard, J.H. Structural Characterization of the DC-SIGN–LewisX Complex. Biochemistry 2014, 53, 5700–5709. [Google Scholar] [CrossRef] [PubMed]
- Van Gisbergen, K.P.; Aarnoudse, C.A.; Meijer, G.A.; Geijtenbeek, T.B.; van Kooyk, Y. Dendritic cells recognize tumor-specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin. Cancer Res. 2005, 65, 5935–5944. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Van Kooyk, Y.; Rabinovich, G.A. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 2008, 9, 593–601. [Google Scholar] [CrossRef]
- Guo, Y.; Feinberg, H.; Conroy, E.; Mitchell, D.A.; Alvarez, R.; Blixt, O.; Taylor, M.E.; Weis, W.I.; Drickamer, K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct Mol. Biol. 2004, 11, 591–598. [Google Scholar] [CrossRef]
- Blanas, A.; Sahasrabudhe, N.M.; Rodríguez, E.; van Kooyk, Y.; van Vliet, S.J. Fucosylated Antigens in Cancer: An Alliance toward Tumor Progression, Metastasis, and Resistance to Chemotherapy. Front. Oncol. 2018, 8, 39. [Google Scholar] [CrossRef]
- Nonaka, M.; Ma, B.Y.; Murai, R.; Nakamura, N.; Baba, M.; Kawasaki, N.; Hodohara, K.; Asano, S.; Kawasaki, T. Glycosylation-dependent interactions of C-type lectin DC-SIGN with colorectal tumor-associated Lewis glycans impair the function and differentiation of monocyte-derived dendritic cells. J. Immunol. 2008, 180, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Jonuleit, H.; Schmitt, E.; Schuler, G.; Knop, J.; Enk, A.H. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J. Exp. Med. 2000, 192, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Schlapbach, C.; Ochsenbein, A.; Kaelin, U.; Hassan, A.S.; Hunger, R.E.; Yawalkar, N. High numbers of DC-SIGN+ dendritic cells in lesional skin of cutaneous T-cell lymphoma. J. Am. Acad. Dermatol. 2010, 62, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Mourcin, F.; Uhel, F.; Pangault, C.; Ruminy, P.; Dupré, L.; Guirriec, M.; Marchand, T.; Fest, T.; Lamy, T.; et al. DC-SIGN-expressing macrophages trigger activation of mannosylated IgM B-cell receptor in follicular lymphoma. Blood 2015, 126, 1911–1920. [Google Scholar] [CrossRef]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Sabatte, J.; Faigle, W.; Ceballos, A.; Morelle, W.; Rodríguez Rodrígues, C.; Remes Lenicov, F.; Thépaut, M.; Fieschi, F.; Malchiodi, E.; Fernández, M.; et al. Semen Clusterin Is a Novel DC-SIGN Ligand. J. Immunol. 2011, 187, 5299–5309. [Google Scholar] [CrossRef] [PubMed]
- Merlotti, A.; Malizia, A.L.; Michea, P.; Bonte, P.-E.; Goudot, C.; Carregal, M.S.; Nuñez, N.; Sedlik, C.; Ceballos, A.; Soumelis, V.; et al. Aberrant fucosylation enables breast cancer clusterin to interact with dendritic cell-specific ICAM-grabbing non-integrin (DC-SIGN). Oncoimmunology 2019, 8, e1629257. [Google Scholar] [CrossRef] [PubMed]
- Tellez, T.; Garcia-Aranda, M.; Redondo, M. The Role of Clusterin in Carcinogenesis and its Potential Utility as Therapeutic Target. Curr. Med. Chem. 2016, 23, 4297–4308. [Google Scholar] [CrossRef]
- Kretz-Rommel, A.; Qin, F.; Dakappagari, N.; Torensma, R.; Faas, S.; Wu, D.; Bowdish, K.S. In vivo targeting of antigens to human dendritic cells through DC-SIGN elicits stimulatory immune responses and inhibits tumor growth in grafted mouse models. J. Immunother. 2007, 30, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.F.; Torensma, R.; Hebeda, K.; Kretz-Rommel, A.; Faas, S.J.; Figdor, C.G.; Adema, G.J. In vivo targeting of DC-SIGN-positive antigen-presenting cells in a nonhuman primate model. J. Immunother. 2007, 30, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Tacken, P.J.; de Vries, I.J.M.; Gijzen, K.; Joosten, B.; Wu, D.; Rother, R.P.; Faas, S.J.; Punt, C.J.A.; Torensma, R.; Adema, G.J.; et al. Effective induction of naive and recall T-cell responses by targeting antigen to human dendritic cells via a humanized anti–DC-SIGN antibody. Blood 2005, 106, 1278–1285. [Google Scholar] [CrossRef]
- Unger, W.W.; Mayer, C.T.; Engels, S.; Hesse, C.; Perdicchio, M.; Puttur, F.; Streng-Ouwehand, I.; Litjens, M.; Kalay, H.; Berod, L.; et al. Antigen targeting to dendritic cells combined with transient regulatory T cell inhibition results in long-term tumor regression. Oncoimmunology 2015, 4, e970462. [Google Scholar] [CrossRef]
- Hu, B.; Wang, Z.; Zeng, H.; Qi, Y.; Chen, Y.; Wang, T.; Wang, J.; Chang, Y.; Bai, Q.; Xia, Y.; et al. Blockade of DC-SIGN+ Tumor-Associated Macrophages Reactivates Antitumor Immunity and Improves Immunotherapy in Muscle-Invasive Bladder Cancer. Cancer Res. 2020, 80, 1707–1719. [Google Scholar] [CrossRef]
- Boghaert, E.R.; Sridharan, L.; Armellino, D.C.; Khandke, K.M.; DiJoseph, J.F.; Kunz, A.; Dougher, M.M.; Jiang, F.; Kalyandrug, L.B.; Hamann, P.R.; et al. Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-acetyl gamma calicheamicin dimethyl hydrazide targets Lewisy and eliminates Lewisy-positive human carcinoma cells and xenografts. Clin. Cancer Res. 2004, 10, 4538–4549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krug, L.M.; Milton, D.T.; Jungbluth, A.A.; Chen, L.-C.; Quaia, E.; Pandit-Taskar, N.; Nagel, A.; Jones, J.; Kris, M.G.; Finn, R.; et al. Targeting Lewis Y (Ley) in Small Cell Lung Cancer with a Humanized Monoclonal Antibody, hu3S193: A Pilot Trial Testing Two Dose Levels. J. Thorac. Oncol. 2007, 2, 947–952. [Google Scholar] [CrossRef]
- Scott, A.M.; Tebbutt, N.; Lee, F.-T.; Cavicchiolo, T.; Liu, Z.; Gill, S.; Poon, A.M.T.; Hopkins, W.; Smyth, F.E.; Murone, C.; et al. A Phase I Biodistribution and Pharmacokinetic Trial of Humanized Monoclonal Antibody Hu3s193 in Patients with Advanced Epithelial Cancers that Express the Lewis-Y Antigen. Clin. Cancer Res. 2007, 13, 3286–3292. [Google Scholar] [CrossRef] [PubMed]
- Smaletz, O.; Diz, M.D.; do Carmo, C.C.; Sabbaga, J.; Cunha-Junior, G.F.; Azevedo, S.J.; Maluf, F.C.; Barrios, C.H.; Costa, R.L.; Fontana, A.G.; et al. A phase II trial with anti-Lewis-Y monoclonal antibody (hu3S193) for the treatment of platinum resistant/refractory ovarian, fallopian tube and primary peritoneal carcinoma. Gynecol. Oncol. 2015, 138, 272–277. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Middel, J.; Cornelissen, I.L.; Nottet, H.S.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef]
- Alvarez, C.P.; Lasala, F.; Carrillo, J.; Muñiz, O.; Corbí, A.L.; Delgado, R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002, 76, 6841–6844. [Google Scholar] [CrossRef]
- Marzi, A.; Gramberg, T.; Simmons, G.; Möller, P.; Rennekamp, A.J.; Krumbiegel, M.; Geier, M.; Eisemann, J.; Turza, N.; Saunier, B.; et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 12090–12095. [Google Scholar] [CrossRef] [PubMed]
- Hodges, A.; Sharrocks, K.; Edelmann, M.; Baban, D.; Moris, A.; Schwartz, O.; Drakesmith, H.; Davies, K.; Kessler, B.; McMichael, A.; et al. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat. Immunol. 2007, 8, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Shan, M.; Klasse, P.J.; Banerjee, K.; Dey, A.K.; Iyer, S.P.; Dionisio, R.; Charles, D.; Campbell-Gardener, L.; Olson, W.C.; Sanders, R.W.; et al. HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog. 2007, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Venturi, M.; Schiffner, L.; Kalyanaraman, R.; Katinger, H.; Lloyd, K.O.; Kwong, P.D.; Moore, J.P. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 2002, 76, 7293–7305. [Google Scholar] [CrossRef]
- Garcia-Vallejo, J.J.; Koning, N.; Ambrosini, M.; Kalay, H.; Vuist, I.; Sarrami-Forooshani, R.; Geijtenbeek, T.B.; van Kooyk, Y. Glycodendrimers prevent HIV transmission via DC-SIGN on dendritic cells. Int. Immunol. 2013, 25, 221–233. [Google Scholar] [CrossRef]
- Martínez-Avila, O.; Bedoya, L.M.; Marradi, M.; Clavel, C.; Alcamí, J.; Penadés, S. Multivalent manno-glyconanoparticles inhibit DC-SIGN-mediated HIV-1 trans-infection of human T cells. Chembiochem 2009, 10, 1806–1809. [Google Scholar] [CrossRef]
- Sattin, S.; Daghetti, A.; Thépaut, M.; Berzi, A.; Sánchez-Navarro, M.; Tabarani, G.; Rojo, J.; Fieschi, F.; Clerici, M.; Bernardi, A. Inhibition of DC-SIGN-Mediated HIV Infection by a Linear Trimannoside Mimic in a Tetravalent Presentation. ACS Chem. Biol. 2010, 5, 301–312. [Google Scholar] [CrossRef]
- Varga, N.; Sutkeviciute, I.; Ribeiro-Viana, R.; Berzi, A.; Ramdasi, R.; Daghetti, A.; Vettoretti, G.; Amara, A.; Clerici, M.; Rojo, J.; et al. A multivalent inhibitor of the DC-SIGN dependent uptake of HIV-1 and Dengue virus. Biomaterials 2014, 35, 4175–4184. [Google Scholar] [CrossRef]
- Berzi, A.; Ordanini, S.; Joosten, B.; Trabattoni, D.; Cambi, A.; Bernardi, A.; Clerici, M. Pseudo-Mannosylated DC-SIGN Ligands as Immunomodulants. Sci. Rep. 2016, 6, 35373. [Google Scholar] [CrossRef]
- Naarding, M.A.; Baan, E.; Pollakis, G.; Paxton, W.A. Effect of chloroquine on reducing HIV-1 replication in vitro and the DC-SIGN mediated transfer of virus to CD4+T-lymphocytes. Retrovirology 2007, 4, 6. [Google Scholar] [CrossRef]
- Pustylnikov, S.; Dave, R.S.; Khan, Z.K.; Porkolab, V.; Rashad, A.A.; Hutchinson, M.; Fieschi, F.; Chaiken, I.; Jain, P. Short Communication: Inhibition of DC-SIGN-Mediated HIV-1 Infection by Complementary Actions of Dendritic Cell Receptor Antagonists and Env-Targeting Virus Inactivators. AIDS Res. Hum. Retrovir. 2016, 32, 93–100. [Google Scholar] [CrossRef]
- Nair, M.P.; Reynolds, J.L.; Mahajan, S.D.; Schwartz, S.A.; Aalinkeel, R.; Bindukumar, B.; Sykes, D. RNAi-directed inhibition of DC-SIGN by dendritic cells: Prospects for HIV-1 therapy. Aaps. J. 2005, 7, E572–E578. [Google Scholar] [CrossRef][Green Version]
- Geyer, H.; Will, C.; Feldmann, H.; Klenk, H.D.; Geyer, R. Carbohydrate structure of Marburg virus glycoprotein. Glycobiology 1992, 2, 299–312. [Google Scholar] [CrossRef]
- Simmons, G.; Reeves, J.D.; Grogan, C.C.; Vandenberghe, L.H.; Baribaud, F.; Whitbeck, J.C.; Burke, E.; Buchmeier, M.J.; Soilleux, E.J.; Riley, J.L.; et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 2003, 305, 115–123. [Google Scholar] [CrossRef]
- Francica, J.R.; Varela-Rohena, A.; Medvec, A.; Plesa, G.; Riley, J.L.; Bates, P. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 2010, 6, e1001098. [Google Scholar] [CrossRef]
- Lennemann, N.J.; Rhein, B.A.; Ndungo, E.; Chandran, K.; Qiu, X.; Maury, W. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 2014, 5, e00862-13. [Google Scholar] [CrossRef]
- Marzi, A.; Möller, P.; Hanna, S.L.; Harrer, T.; Eisemann, J.; Steinkasserer, A.; Becker, S.; Baribaud, F.; Pöhlmann, S. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J. Infect. Dis. 2007, 196 (Suppl. 2), S237–S246. [Google Scholar] [CrossRef]
- Lasala, F.; Arce, E.; Otero, J.R.; Rojo, J.; Delgado, R. Mannosyl glycodendritic structure inhibits DC-SIGN-mediated Ebola virus infection in cis and in trans. Antimicrob. Agents Chemother 2003, 47, 3970–3972. [Google Scholar] [CrossRef]
- Ribeiro-Viana, R.; Sánchez-Navarro, M.; Luczkowiak, J.; Koeppe, J.R.; Delgado, R.; Rojo, J.; Davis, B.G. Virus-like glycodendrinanoparticles displaying quasi-equivalent nested polyvalency upon glycoprotein platforms potently block viral infection. Nat. Commun. 2012, 3, 1303. [Google Scholar] [CrossRef]
- Thépaut, M.; Luczkowiak, J.; Vivès, C.; Labiod, N.; Bally, I.; Lasala, F.; Grimoire, Y.; Fenel, D.; Sattin, S.; Thielens, N.; et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cummings, R.D.; McEver, R.P. C-type Lectins. In Essentials of Glycobiology; Harbor Laboratory (NY): New York, NY, USA, 2009. [Google Scholar]
- Kansas, G.S. Selectins and their ligands: Current concepts and controversies. Blood 1996, 88, 3259–3287. [Google Scholar] [CrossRef]
- Vestweber, D.; Blanks, J.E. Mechanisms that regulate the function of the selectins and their ligands. Physiol. Rev. 1999, 79, 181–213. [Google Scholar] [CrossRef]
- Foxall, C.; Watson, S.R.; Dowbenko, D.; Fennie, C.; Lasky, L.A.; Kiso, M.; Hasegawa, A.; Asa, D.; Brandley, B.K. The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewis(x) oligosaccharide. J. Cell Biol. 1992, 117, 895–902. [Google Scholar] [CrossRef]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology 2017, 6, 16. [Google Scholar] [CrossRef]
- Somers, W.S.; Tang, J.; Shaw, G.D.; Camphausen, R.T. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 2000, 103, 467–479. [Google Scholar] [CrossRef]
- Eppihimer, M.J.; Wolitzky, B.; Anderson, D.C.; Labow, M.A.; Granger, D.N. Heterogeneity of expression of E- and P-selectins in vivo. Circ. Res. 1996, 79, 560–569. [Google Scholar] [CrossRef]
- Ivetic, A.; Hoskins Green, H.L.; Hart, S.J. L-selectin: A Major Regulator of Leukocyte Adhesion, Migration and Signaling. Front. Immunol. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Peschon, J.J.; Slack, J.L.; Reddy, P.; Stocking, K.L.; Sunnarborg, S.W.; Lee, D.C.; Russell, W.E.; Castner, B.J.; Johnson, R.S.; Fitzner, J.N.; et al. An essential role for ectodomain shedding in mammalian development. Science 1998, 282, 1281–1284. [Google Scholar] [CrossRef]
- McEver, R.P. Selectin-carbohydrate interactions during inflammation and metastasis. Glycoconj. J. 1997, 14, 585–591. [Google Scholar] [CrossRef]
- Bullard, D.C.; Kunkel, E.J.; Kubo, H.; Hicks, M.J.; Lorenzo, I.; Doyle, N.A.; Doerschuk, C.M.; Ley, K.; Beaudet, A.L. Infectious susceptibility and severe deficiency of leukocyte rolling and recruitment in E-selectin and P-selectin double mutant mice. J. Exp. Med. 1996, 183, 2329–2336. [Google Scholar] [CrossRef]
- Chen, S.; Alon, R.; Fuhlbrigge, R.C.; Springer, T.A. Rolling and transient tethering of leukocytes on antibodies reveal specializations of selectins. Proc. Natl. Acad. Sci. USA 1997, 94, 3172–3177. [Google Scholar] [CrossRef] [PubMed]
- Mayadas, T.N.; Johnson, R.C.; Rayburn, H.; Hynes, R.O.; Wagner, D.D. Leukocyte rolling and extravasation are severely compromised in P selectin-deficient mice. Cell 1993, 74, 541–554. [Google Scholar] [CrossRef]
- Luscinskas, F.W.; Kansas, G.S.; Ding, H.; Pizcueta, P.; Schleiffenbaum, B.E.; Tedder, T.F.; Gimbrone, M.A., Jr. Monocyte rolling, arrest and spreading on IL-4-activated vascular endothelium under flow is mediated via sequential action of L-selectin, beta 1-integrins, and beta 2-integrins. J. Cell Biol. 1994, 125, 1417–1427. [Google Scholar] [CrossRef]
- Bargatze, R.F.; Kurk, S.; Butcher, E.C.; Jutila, M.A. Neutrophils roll on adherent neutrophils bound to cytokine-induced endothelial cells via L-selectin on the rolling cells. J. Exp. Med. 1994, 180, 1785–1792. [Google Scholar] [CrossRef]
- Wang, H.B.; Wang, J.T.; Zhang, L.; Geng, Z.H.; Xu, W.L.; Xu, T.; Huo, Y.; Zhu, X.; Plow, E.F.; Chen, M.; et al. P-selectin primes leukocyte integrin activation during inflammation. Nat. Immunol. 2007, 8, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Zarbock, A.; Abram, C.L.; Hundt, M.; Altman, A.; Lowell, C.A.; Ley, K. PSGL-1 engagement by E-selectin signals through Src kinase Fgr and ITAM adapters DAP12 and FcR gamma to induce slow leukocyte rolling. J. Exp. Med. 2008, 205, 2339–2347. [Google Scholar] [CrossRef] [PubMed]
- Weyrich, A.S.; McIntyre, T.M.; McEver, R.P.; Prescott, S.M.; Zimmerman, G.A. Monocyte tethering by P-selectin regulates monocyte chemotactic protein-1 and tumor necrosis factor-alpha secretion. Signal integration and NF-kappa B translocation. J. Clin. Investig. 1995, 95, 2297–2303. [Google Scholar] [CrossRef]
- Crockett-Torabi, E.; Sulenbarger, B.; Smith, C.W.; Fantone, J.C. Activation of human neutrophils through L-selectin and Mac-1 molecules. J. Immunol. 1995, 154, 2291–2302. [Google Scholar]
- Palabrica, T.; Lobb, R.; Furie, B.C.; Aronovitz, M.; Benjamin, C.; Hsu, Y.M.; Sajer, S.A.; Furie, B. Leukocyte accumulation promoting fibrin deposition is mediated in vivo by P-selectin on adherent platelets. Nature 1992, 359, 848–851. [Google Scholar] [CrossRef]
- Polgar, J.; Matuskova, J.; Wagner, D.D. The P-selectin, tissue factor, coagulation triad. J. Thromb. Haemost. 2005, 3, 1590–1596. [Google Scholar] [CrossRef]
- Subramaniam, M.; Frenette, P.S.; Saffaripour, S.; Johnson, R.C.; Hynes, R.O.; Wagner, D.D. Defects in hemostasis in P-selectin-deficient mice. Blood 1996, 87, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Arbonés, M.L.; Ord, D.C.; Ley, K.; Ratech, H.; Maynard-Curry, C.; Otten, G.; Capon, D.J.; Tedder, T.F. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity 1994, 1, 247–260. [Google Scholar] [CrossRef]
- Läubli, H.; Borsig, L. Selectins as mediators of lung metastasis. Cancer Microenviron. Off. J. Int. Cancer Microenviron. Soc. 2010, 3, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Khatib, A.M.; Kontogiannea, M.; Fallavollita, L.; Jamison, B.; Meterissian, S.; Brodt, P. Rapid induction of cytokine and E-selectin expression in the liver in response to metastatic tumor cells. Cancer Res. 1999, 59, 1356–1361. [Google Scholar] [PubMed]
- Barbier, V.; Erbani, J.; Fiveash, C.; Davies, J.M.; Tay, J.; Tallack, M.R.; Lowe, J.; Magnani, J.L.; Pattabiraman, D.R.; Perkins, A.C.; et al. Endothelial E-selectin inhibition improves acute myeloid leukaemia therapy by disrupting vascular niche-mediated chemoresistance. Nat. Commun. 2020, 11, 2042. [Google Scholar] [CrossRef]
- Muz, B.; Abdelghafer, A.; Markovic, M.; Yavner, J.; Melam, A.; Salama, N.N.; Azab, A.K. Targeting E-selectin to Tackle Cancer Using Uproleselan. Cancers 2021, 13, 335. [Google Scholar] [CrossRef] [PubMed]
- Kageshita, T.; Hirai, S.; Kimura, T.; Hanai, N.; Ohta, S.; Ono, T. Association between sialyl Lewis(a) expression and tumor progression in melanoma. Cancer Res. 1995, 55, 1748–1751. [Google Scholar] [CrossRef]
- Numahata, K.; Satoh, M.; Handa, K.; Saito, S.; Ohyama, C.; Ito, A.; Takahashi, T.; Hoshi, S.; Orikasa, S.; Hakomori, S.I. Sialosyl-Le(x) expression defines invasive and metastatic properties of bladder carcinoma. Cancer 2002, 94, 673–685. [Google Scholar] [CrossRef]
- Villar-Portela, S.; Martin, C.; Muinelo-Romay, L.; Cuevas, E.; Gil Martín, E.; Fernández, A. sLe a and sLe x expression in colorectal cancer: Implications for tumourigenesis and disease prognosis. Histol. Histopathol. 2011, 26, 1305–1316. [Google Scholar] [CrossRef]
- Zhong, L.; Simoneau, B.; Tremblay, P.-L.; Gout, S.; Simard, M.J.; Huot, J. E-Selectin-mediated adhesion and extravasation in cancer. Encycl. Cancer 2017, 1618–1624. [Google Scholar]
- DeAngelo, D.J.; Jonas, B.A.; Liesveld, J.L.; Bixby, D.L.; Advani, A.S.; Marlton, P.; O’Dwyer, M.E.; Fogler, W.E.; Magnani, J.L.; Chen, M.M.; et al. High E-Selectin Ligand Expression Contributes to Chemotherapy-Resistance in Poor Risk Relapsed and Refractory (R/R) Acute Myeloid Leukemia Patients and Can be Overcome with the Addition of Uproleselan. Blood 2019, 134, 2690. [Google Scholar] [CrossRef]
- Winkler, I.G.; Barbier, V.; Nowlan, B.; Jacobsen, R.N.; Forristal, C.E.; Patton, J.T.; Magnani, J.L.; Lévesque, J.P. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 2012, 18, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Mondal, N.; Greco, T.M.; Wei, Y.; Spadazzi, C.; Lin, S.C.; Zheng, H.; Cheung, C.; Magnani, J.L.; Lin, S.H.; et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 2019, 21, 627–639. [Google Scholar] [CrossRef]
- Chen, M.; Geng, J.G. P-selectin mediates adhesion of leukocytes, platelets, and cancer cells in inflammation, thrombosis, and cancer growth and metastasis. Arch. Immunol. Ther. Exp. 2006, 54, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Ernst, B.; Magnani, J.L. From carbohydrate leads to glycomimetic drugs. Nat. Reviews Drug Discov. 2009, 8, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Scott, E. Targeting Aberrant Sialylation to Treat Cancer. Medicines 2019, 6, 102. [Google Scholar] [CrossRef]
- DeAngelo, D.J.; Jonas, B.A.; Liesveld, J.L.; Bixby, D.L.; Advani, A.S.; Marlton, P.; O’Dwyer, M.E.; Fogler, W.E.; Wolfgang, C.D.; Magnani, J.L.; et al. Uproleselan (GMI-1271), an E-Selectin Antagonist, Improves the Efficacy and Safety of Chemotherapy in Relapsed/Refractory (R/R) and Newly Diagnosed Older Patients with Acute Myeloid Leukemia: Final, Correlative, and Subgroup Analyses. Blood 2018, 132, 331. [Google Scholar] [CrossRef]
- DeAngelo, D.J.; Erba, H.P.; Jonas, B.A.; O’Dwyer, M.; Marlton, P.; Huls, G.A.; Liesveld, J.; Cooper, B.W.; Bhatnagar, B.; Armstrong, M.; et al. A phase III trial to evaluate the efficacy of uproleselan (GMI-1271) with chemotherapy in patients with relapsed/refractory acute myeloid leukemia. J. Clin. Oncol. 2019, 37, TPS7066. [Google Scholar] [CrossRef]
- Muz, B.; Azab, F.; Fiala, M.; King, J.; Kohnen, D.; Fogler, W.E.; Smith, T.; Magnani, J.L.; Vij, R.; Azab, A.K. Inhibition of E-Selectin (GMI-1271) or E-selectin together with CXCR4 (GMI-1359) re-sensitizes multiple myeloma to therapy. Blood Cancer J. 2019, 9, 68. [Google Scholar] [CrossRef]
- Kononchik, J.; Ireland, J.; Zou, Z.; Segura, J.; Holzapfel, G.; Chastain, A.; Wang, R.; Spencer, M.; He, B.; Stutzman, N.; et al. HIV-1 targets L-selectin for adhesion and induces its shedding for viral release. Nat. Commun. 2018, 9, 2825. [Google Scholar] [CrossRef]
- Giuliani, E.; Vassena, L.; Galardi, S.; Michienzi, A.; Desimio, M.G.; Doria, M. Dual regulation of L-selectin (CD62L) by HIV-1: Enhanced expression by Vpr in contrast with cell-surface down-modulation by Nef and Vpu. Virology 2018, 523, 121–128. [Google Scholar] [CrossRef]
- Vassena, L.; Giuliani, E.; Koppensteiner, H.; Bolduan, S.; Schindler, M.; Doria, M. HIV-1 Nef and Vpu Interfere with L-Selectin (CD62L) Cell Surface Expression To Inhibit Adhesion and Signaling in Infected CD4+ T Lymphocytes. J. Virol. 2015, 89, 5687–5700. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, R.N.; Watson, H.A.; Vigar, M.; Ohme, J.; Thomson, A.; Humphreys, I.R.; Ager, A. L-selectin Is Essential for Delivery of Activated CD8(+) T Cells to Virus-Infected Organs for Protective Immunity. Cell Rep. 2016, 14, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, F.; Wang, Q.J.; Rosenberg, S.A.; Morgan, R.A. The shedding of CD62L (L-selectin) regulates the acquisition of lytic activity in human tumor reactive T lymphocytes. PLoS ONE 2011, 6, e22560. [Google Scholar] [CrossRef]
- Hoffman, M.; Ipp, H.; Phatlhane, D.V.; Erasmus, R.T.; Zemlin, A.E. E-Selectin and markers of HIV disease severity, inflammation and coagulation in HIV-infected treatment-naïve individuals. Afr. Health Sci. 2018, 18, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Cota-Gomez, A.; Flores, N.C.; Cruz, C.; Casullo, A.; Aw, T.Y.; Ichikawa, H.; Schaack, J.; Scheinman, R.; Flores, S.C. The human immunodeficiency virus-1 Tat protein activates human umbilical vein endothelial cell E-selectin expression via an NF-kappa B-dependent mechanism. J. Biol. Chem. 2002, 277, 14390–14399. [Google Scholar] [CrossRef] [PubMed]
- Colomb, F.; Giron, L.B.; Kuri-Cervantes, L.; Adeniji, O.S.; Ma, T.; Dweep, H.; Battivelli, E.; Verdin, E.; Palmer, C.S.; Tateno, H.; et al. Sialyl-Lewis(X) Glycoantigen Is Enriched on Cells with Persistent HIV Transcription during Therapy. Cell Rep. 2020, 32, 107991. [Google Scholar] [CrossRef]
- Fu, Y.; He, S.; Waheed, A.A.; Dabbagh, D.; Zhou, Z.; Trinité, B.; Wang, Z.; Yu, J.; Wang, D.; Li, F.; et al. PSGL-1 restricts HIV-1 infectivity by blocking virus particle attachment to target cells. Proc. Natl. Acad. Sci. USA 2020, 117, 9537–9545. [Google Scholar] [CrossRef]
- He, S.; Waheed, A.A.; Hetrick, B.; Dabbagh, D.; Akhrymuk, I.V.; Kehn-Hall, K.; Freed, E.O.; Wu, Y. PSGL-1 Inhibits the Incorporation of SARS-CoV and SARS-CoV-2 Spike Glycoproteins into Pseudovirions and Impairs Pseudovirus Attachment and Infectivity. Viruses 2021, 13, 46. [Google Scholar] [CrossRef]
- Barondes, S.H.; Castronovo, V.; Cooper, D.N.; Cummings, R.D.; Drickamer, K.; Feizi, T.; Gitt, M.A.; Hirabayashi, J.; Hughes, C.; Kasai, K.; et al. Galectins: A family of animal beta-galactoside-binding lectins. Cell 1994, 76, 597–598. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2009. [Google Scholar]
- Cummings, R.D.; Liu, F.T.; Vasta, G.R. Galectins. In Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2017. [Google Scholar]
- Rabinovich, G.A.; Liu, F.T.; Hirashima, M.; Anderson, A. An emerging role for galectins in tuning the immune response: Lessons from experimental models of inflammatory disease, autoimmunity and cancer. Scand. J. Immunol. 2007, 66, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Kasai, K. The family of metazoan metal-independent beta-galactoside-binding lectins: Structure, function and molecular evolution. Glycobiology 1993, 3, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Salatino, M.; Dalotto-Moreno, T.; Rabinovich, G.A. Sweetening the hallmarks of cancer: Galectins as multifunctional mediators of tumor progression. J. Exp. Med. 2020, 217, e20182041. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Lin, C.Y.; Chang, M.R.; Urbina, A.N.; Assavalapsakul, W.; Thitithanyanont, A.; Chen, Y.H.; Liu, F.T.; Wang, S.F. The role of galectins in virus infection—A systemic literature review. J. Microbiol. Immunol. Infect. 2020, 53, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Leffler, H.; Barondes, S.H. Specificity of binding of three soluble rat lung lectins to substituted and unsubstituted mammalian beta-galactosides. J. Biol. Chem. 1986, 261, 10119–10126. [Google Scholar] [CrossRef]
- Zhou, Q.; Cummings, R.D. The S-type lectin from calf heart tissue binds selectively to the carbohydrate chains of laminin. Arch. Biochem. Biophys. 1990, 281, 27–35. [Google Scholar] [CrossRef]
- Bourne, Y.; Bolgiano, B.; Liao, D.-I.; Strecker, G.; Cantau, P.; Herzberg, O.; Feizi, T.; Cambillau, C. Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat. Struct. Biol. 1994, 1, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Brewer, C.F.; Miceli, M.C.; Baum, L.G. Clusters, bundles, arrays and lattices: Novel mechanisms for lectin–saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 2002, 12, 616–623. [Google Scholar] [CrossRef]
- Sharon, N. When lectin meets oligosaccharide. Nat. Struct. Biol. 1994, 1, 843–845. [Google Scholar] [CrossRef]
- Ahmad, N.; Gabius, H.J.; Kaltner, H.; André, S.; Kuwabara, I.; Liu, F.-T.; Oscarson, S.; Norberg, T.; Brewer, C.F. Thermodynamic binding studies of cell surface carbohydrate epitopes to galectins-1, -3, and -7: Evidence for differential binding specificities. Can. J. Chem. 2002, 80, 1096–1104. [Google Scholar] [CrossRef]
- Hirashima, M.; Kashio, Y.; Nishi, N.; Yamauchi, A.; Imaizumi, T.A.; Kageshita, T.; Saita, N.; Nakamura, T. Galectin-9 in physiological and pathological conditions. Glycoconj. J. 2002, 19, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Gabius, H.J.; André, S.; Kaltner, H.; Sabesan, S.; Roy, R.; Liu, B.; Macaluso, F.; Brewer, C.F. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J. Biol. Chem. 2004, 279, 10841–10847. [Google Scholar] [CrossRef]
- Ideo, H.; Seko, A.; Yamashita, K. Recognition mechanism of galectin-4 for cholesterol 3-sulfate. J. Biol. Chem. 2007, 282, 21081–21089. [Google Scholar] [CrossRef] [PubMed]
- Fermin Lee, A.; Chen, H.Y.; Wan, L.; Wu, S.Y.; Yu, J.S.; Huang, A.C.; Miaw, S.C.; Hsu, D.K.; Wu-Hsieh, B.A.; Liu, F.T. Galectin-3 modulates Th17 responses by regulating dendritic cell cytokines. Am. J. Pathol. 2013, 183, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, G.A.; Iglesias, M.M.; Modesti, N.M.; Castagna, L.F.; Wolfenstein-Todel, C.; Riera, C.M.; Sotomayor, C.E. Activated rat macrophages produce a galectin-1-like protein that induces apoptosis of T cells: Biochemical and functional characterization. J. Immunol. 1998, 160, 4831–4840. [Google Scholar] [PubMed]
- Frigeri, L.G.; Liu, F.T. Surface expression of functional IgE binding protein, an endogenous lectin, on mast cells and macrophages. J. Immunol. 1992, 148, 861–867. [Google Scholar]
- Ge, X.N.; Ha, S.G.; Liu, F.-T.; Rao, S.P.; Sriramarao, P. Eosinophil-expressed galectin-3 regulates cell trafficking and migration. Front. Pharm. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lingblom, C.; Andersson, K.; Wennerås, C. Kinetic studies of galectin-10 release from eosinophils exposed to proliferating T cells. Clin. Exp. Immunol. 2021, 203, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Truong, M.J.; Gruart, V.; Kusnierz, J.P.; Papin, J.P.; Loiseau, S.; Capron, A.; Capron, M. Human neutrophils express immunoglobulin E (IgE)-binding proteins (Mac-2/epsilon BP) of the S-type lectin family: Role in IgE-dependent activation. J. Exp. Med. 1993, 177, 243–248. [Google Scholar] [CrossRef]
- Koopman, L.A.; Kopcow, H.D.; Rybalov, B.; Boyson, J.E.; Orange, J.S.; Schatz, F.; Masch, R.; Lockwood, C.J.; Schachter, A.D.; Park, P.J.; et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 2003, 198, 1201–1212. [Google Scholar] [CrossRef]
- Blaser, C.; Kaufmann, M.; Müller, C.; Zimmermann, C.; Wells, V.; Mallucci, L.; Pircher, H. Beta-galactoside-binding protein secreted by activated T cells inhibits antigen-induced proliferation of T cells. Eur. J. Immunol. 1998, 28, 2311–2319. [Google Scholar] [CrossRef]
- Meissner, N.; Radke, J.; Hedges, J.F.; White, M.; Behnke, M.; Bertolino, S.; Abrahamsen, M.; Jutila, M.A. Serial Analysis of Gene Expression in Circulating γδ T Cell Subsets Defines Distinct Immunoregulatory Phenotypes and Unexpected Gene Expression Profiles. J. Immunol. 2003, 170, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.L.; Chammas, R.; Ricon, L.; Fermino, M.L.; Bernardes, E.S.; Hsu, D.K.; Liu, F.-T.; Borojevic, R.; El-Cheikh, M.C. Galectin-3 regulates peritoneal B1-cell differentiation into plasma cells. Glycobiology 2009, 19, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Zuñiga, E.; Rabinovich, G.A.; Iglesias, M.M.; Gruppi, A. Regulated expression of galectin-1 during B-cell activation and implications for T-cell apoptosis. J. Leukoc. Biol. 2001, 70, 73–79. [Google Scholar]
- Hernandez, J.D.; Baum, L.G. Ah, sweet mystery of death! Galectins and control of cell fate. Glycobiology 2002, 12, 127R–136R. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Patterson, R.J.; Wang, J.L. Intracellular functions of galectins. Biochim. Biophys. Acta 2002, 1572, 263–273. [Google Scholar] [CrossRef]
- Elola, M.T.; Wolfenstein-Todel, C.; Troncoso, M.F.; Vasta, G.R.; Rabinovich, G.A. Galectins: Matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell Mol. Life Sci. 2007, 64, 1679–1700. [Google Scholar] [CrossRef]
- Garner, O.B.; Baum, L.G. Galectin-glycan lattices regulate cell-surface glycoprotein organization and signalling. Biochem. Soc. Trans. 2008, 36, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Baum, L.G. Galectin interactions with extracellular matrix and effects on cellular function. Methods Enzym. 2006, 417, 247–256. [Google Scholar] [CrossRef]
- Nabi, I.R.; Shankar, J.; Dennis, J.W. The galectin lattice at a glance. J. Cell Sci. 2015, 128, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Perillo, N.L.; Marcus, M.E.; Baum, L.G. Galectins: Versatile modulators of cell adhesion, cell proliferation, and cell death. J. Mol. Med. (Berl) 1998, 76, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N.; Barondes, S.H. God must love galectins; he made so many of them. Glycobiology 1999, 9, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Rabinovich, G.A. Galectins as modulators of tumour progression. Nat. Rev. Cancer 2005, 5, 29–41. [Google Scholar] [CrossRef]
- Wdowiak, K.; Francuz, T.; Gallego-Colon, E.; Ruiz-Agamez, N.; Kubeczko, M.; Grochoła, I.; Wojnar, J. Galectin Targeted Therapy in Oncology: Current Knowledge and Perspectives. Int. J. Mol. Sci. 2018, 19, 210. [Google Scholar] [CrossRef]
- Thijssen, V.L.; Heusschen, R.; Caers, J.; Griffioen, A.W. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim. Biophys. Acta (BBA) Rev. Cancer 2015, 1855, 235–247. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.G.; et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim. Biophys. Acta (BBA) Gen. Subj. 2002, 1572, 232–254. [Google Scholar] [CrossRef]
- Lau, K.S.; Partridge, E.A.; Grigorian, A.; Silvescu, C.I.; Reinhold, V.N.; Demetriou, M.; Dennis, J.W. Complex N-Glycan Number and Degree of Branching Cooperate to Regulate Cell Proliferation and Differentiation. Cell 2007, 129, 123–134. [Google Scholar] [CrossRef]
- Stanley, P. A method to the madness of N-glycan complexity? Cell 2007, 129, 27–29. [Google Scholar] [CrossRef]
- Camby, I.; Decaestecker, C.; Lefranc, F.; Kaltner, H.; Gabius, H.J.; Kiss, R. Galectin-1 knocking down in human U87 glioblastoma cells alters their gene expression pattern. Biochem. Biophys. Res. Commun. 2005, 335, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Puchades, M.; Nilsson, C.L.; Emmett, M.R.; Aldape, K.D.; Ji, Y.; Lang, F.F.; Liu, T.J.; Conrad, C.A. Proteomic investigation of glioblastoma cell lines treated with wild-type p53 and cytotoxic chemotherapy demonstrates an association between galectin-1 and p53 expression. J. Proteome Res. 2007, 6, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Raimond, J.; Rouleux, F.; Monsigny, M.; Legrand, A. The second intron of the human galectin-3 gene has a strong promoter activity down-regulated by p53. FEBS Lett. 1995, 363, 165–169. [Google Scholar] [CrossRef]
- Potikha, T.; Pappo, O.; Mizrahi, L.; Olam, D.; Maller, S.M.; Rabinovich, G.A.; Galun, E.; Goldenberg, D.S. Lack of galectin-1 exacerbates chronic hepatitis, liver fibrosis, and carcinogenesis in murine hepatocellular carcinoma model. FASEB J. 2019, 33, 7995–8007. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, M.R.; Stephen, T.L.; Svoronos, N.; Allegrezza, M.J.; Tesone, A.J.; Perales-Puchalt, A.; Brencicova, E.; Escovar-Fadul, X.; Nguyen, J.M.; Cadungog, M.G.; et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 2015, 27, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.S.; Fernandes, V.C.; Nepomuceno, T.C.; Rodrigues, D.C.; Woods, N.T.; Suarez-Kurtz, G.; Chammas, R.; Monteiro, A.N.; Carvalho, M.A. Characterization of LGALS3 (galectin-3) as a player in DNA damage response. Cancer Biol. Ther. 2014, 15, 840–850. [Google Scholar] [CrossRef]
- Gebert, J.; Kloor, M.; Lee, J.; Lohr, M.; André, S.; Wagner, R.; Kopitz, J.; Gabius, H.J. Colonic carcinogenesis along different genetic routes: Glycophenotyping of tumor cases separated by microsatellite instability/stability. Histochem. Cell Biol. 2012, 138, 339–350. [Google Scholar] [CrossRef]
- Chen, C.; Duckworth, C.A.; Zhao, Q.; Pritchard, D.M.; Rhodes, J.M.; Yu, L.G. Increased circulation of galectin-3 in cancer induces secretion of metastasis-promoting cytokines from blood vascular endothelium. Clin. Cancer Res. 2013, 19, 1693–1704. [Google Scholar] [CrossRef]
- Lee, Y.K.; Lin, T.H.; Chang, C.F.; Lo, Y.L. Galectin-3 silencing inhibits epirubicin-induced ATP binding cassette transporters and activates the mitochondrial apoptosis pathway via β-catenin/GSK-3β modulation in colorectal carcinoma. PLoS ONE 2013, 8, e82478. [Google Scholar] [CrossRef]
- Chauhan, D.; Li, G.; Podar, K.; Hideshima, T.; Neri, P.; He, D.; Mitsiades, N.; Richardson, P.; Chang, Y.; Schindler, J.; et al. A novel carbohydrate-based therapeutic GCS-100 overcomes bortezomib resistance and enhances dexamethasone-induced apoptosis in multiple myeloma cells. Cancer Res. 2005, 65, 8350–8358. [Google Scholar] [CrossRef]
- Fukumori, T.; Kanayama, H.O.; Raz, A. The role of galectin-3 in cancer drug resistance. Drug Resist. Updat. 2007, 10, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, H.; Yu, X.; Collins, P.M.; Bum-Erdene, K. Galectin-3 inhibitors: A patent review (2008-present). Expert. Opin. Ther. Pat. 2014, 24, 1053–1065. [Google Scholar] [CrossRef]
- Chou, F.C.; Chen, H.Y.; Kuo, C.C.; Sytwu, H.K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef]
- Goud, N.S.; Soukya, P.S.L.; Ghouse, M.; Komal, D.; Alvala, R.; Alvala, M. Human Galectin-1 and Its Inhibitors: Privileged Target for Cancer and HIV. Mini. Rev. Med. Chem. 2019, 19, 1369–1378. [Google Scholar] [CrossRef]
- Miller, M.C.; Klyosov, A.; Mayo, K.H. The alpha-galactomannan Davanat binds galectin-1 at a site different from the conventional galectin carbohydrate binding domain. Glycobiology 2009, 19, 1034–1045. [Google Scholar] [CrossRef]
- Oberg, C.T.; Leffler, H.; Nilsson, U.J. Inhibition of galectins with small molecules. Chimia 2011, 65, 18–23. [Google Scholar] [CrossRef]
- Klyosov, A.; Zomer, E.; Platt, D. DAVANAT® (GM-CT-01) and Colon Cancer: Preclinical and Clinical (Phase I and II) Studies. In Glycobiology and Drug Design; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2012; Volume 1102, pp. 89–130. [Google Scholar]
- Cada, Z.; Smetana, K., Jr.; Lacina, L.; Plzáková, Z.; Stork, J.; Kaltner, H.; Russwurm, R.; Lensch, M.; André, S.; Gabius, H.J. Immunohistochemical fingerprinting of the network of seven adhesion/growth-regulatory lectins in human skin and detection of distinct tumour-associated alterations. Folia. Biol. 2009, 55, 145–152. [Google Scholar]
- Kageshita, T.; Kashio, Y.; Yamauchi, A.; Seki, M.; Abedin, M.J.; Nishi, N.; Shoji, H.; Nakamura, T.; Ono, T.; Hirashima, M. Possible role of galectin-9 in cell aggregation and apoptosis of human melanoma cell lines and its clinical significance. Int. J. Cancer 2002, 99, 809–816. [Google Scholar] [CrossRef]
- Kasamatsu, A.; Uzawa, K.; Nakashima, D.; Koike, H.; Shiiba, M.; Bukawa, H.; Yokoe, H.; Tanzawa, H. Galectin-9 as a regulator of cellular adhesion in human oral squamous cell carcinoma cell lines. Int. J. Mol. Med. 2005, 16, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Ueno, M.; Oomizu, S.; Arikawa, T.; Shinonaga, R.; Zhang, S.; Yamauchi, A.; Hirashima, M. Galectin-9 expression links to malignant potential of cervical squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2008, 134, 899–907. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Dong, J.H.; Chen, Y.W.; Wang, X.Q.; Li, C.H.; Wang, J.; Wang, G.Q.; Li, H.L.; Wang, X.D. Galectin-9 acts as a prognostic factor with antimetastatic potential in hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 2503–2509. [Google Scholar] [CrossRef]
- Chan, S.W.; Kallarakkal, T.G.; Abraham, M.T. Changed expression of E-cadherin and galectin-9 in oral squamous cell carcinomas but lack of potential as prognostic markers. Asian Pac. J. Cancer Prev. 2014, 15, 2145–2152. [Google Scholar] [CrossRef]
- Terris, B.; Blaveri, E.; Crnogorac-Jurcevic, T.; Jones, M.; Missiaglia, E.; Ruszniewski, P.; Sauvanet, A.; Lemoine, N.R. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am. J. Pathol. 2002, 160, 1745–1754. [Google Scholar] [CrossRef]
- Türeci, Ö.; Schmitt, H.; Fadle, N.; Pfreundschuh, M.; Sahin, U. Molecular Definition of a Novel Human Galectin Which Is Immunogenic in Patients with Hodgkin’s Disease *. J. Biol. Chem. 1997, 272, 6416–6422. [Google Scholar] [CrossRef]
- Zhang, C.-X.; Huang, D.-J.; Baloche, V.; Zhang, L.; Xu, J.-X.; Li, B.-W.; Zhao, X.-R.; He, J.; Mai, H.-Q.; Chen, Q.-Y.; et al. Galectin-9 promotes a suppressive microenvironment in human cancer by enhancing STING degradation. Oncogenesis 2020, 9, 65. [Google Scholar] [CrossRef]
- Gleason, M.K.; Lenvik, T.R.; McCullar, V.; Felices, M.; O’Brien, M.S.; Cooley, S.A.; Verneris, M.R.; Cichocki, F.; Holman, C.J.; Panoskaltsis-Mortari, A.; et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012, 119, 3064–3072. [Google Scholar] [CrossRef]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Le Moulec, S.; Guigay, J.; Hirashima, M.; Guemira, F.; et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef]
- Li, H.; Wu, K.; Tao, K.; Chen, L.; Zheng, Q.; Lu, X.; Liu, J.; Shi, L.; Liu, C.; Wang, G.; et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2012, 56, 1342–1351. [Google Scholar] [CrossRef] [PubMed]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef]
- John, S.; Mishra, R. Galectin-9: From cell biology to complex disease dynamics. J. Biosci. 2016, 41, 507–534. [Google Scholar] [CrossRef]
- Gonçalves Silva, I.; Yasinska, I.M.; Sakhnevych, S.S.; Fiedler, W.; Wellbrock, J.; Bardelli, M.; Varani, L.; Hussain, R.; Siligardi, G.; Ceccone, G.; et al. The Tim-3-galectin-9 Secretory Pathway is Involved in the Immune Escape of Human Acute Myeloid Leukemia Cells. EBioMedicine 2017, 22, 44–57. [Google Scholar] [CrossRef]
- Irie, A.; Yamauchi, A.; Kontani, K.; Kihara, M.; Liu, D.; Shirato, Y.; Seki, M.; Nishi, N.; Nakamura, T.; Yokomise, H.; et al. Galectin-9 as a prognostic factor with antimetastatic potential in breast cancer. Clin. Cancer Res. 2005, 11, 2962–2968. [Google Scholar] [CrossRef] [PubMed]
- Akashi, E.; Fujihara, S.; Morishita, A.; Tadokoro, T.; Chiyo, T.; Fujikawa, K.; Kobara, H.; Mori, H.; Iwama, H.; Okano, K.; et al. Effects of galectin-9 on apoptosis, cell cycle and autophagy in human esophageal adenocarcinoma cells. Oncol. Rep. 2017, 38, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Chiyo, T.; Fujita, K.; Iwama, H.; Fujihara, S.; Tadokoro, T.; Ohura, K.; Matsui, T.; Goda, Y.; Kobayashi, N.; Nishiyama, N.; et al. Galectin-9 Induces Mitochondria-Mediated Apoptosis of Esophageal Cancer In Vitro and In Vivo in a Xenograft Mouse Model. Int. J. Mol. Sci. 2019, 20, 2634. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Iwama, H.; Sakamoto, T.; Okura, R.; Kobayashi, K.; Takano, J.; Katsura, A.; Tatsuta, M.; Maeda, E.; Mimura, S.; et al. Galectin-9 suppresses the growth of hepatocellular carcinoma via apoptosis in vitro and in vivo. Int. J. Oncol. 2015, 46, 2419–2430. [Google Scholar] [CrossRef]
- Itoh, A.; Nonaka, Y.; Ogawa, T.; Nakamura, T.; Nishi, N. Galectin-9 induces atypical ubiquitination leading to cell death in PC-3 prostate cancer cells. Glycobiology 2019, 29, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Jafari, S.M.; Nazri, A.; Shabani, M.; Balajam, N.Z.; Aghaei, M. Galectin-9 induces apoptosis in OVCAR-3 ovarian cancer cell through mitochondrial pathway. Res. Pharm. Sci. 2018, 13, 557–565. [Google Scholar] [CrossRef]
- Kobayashi, K.; Morishita, A.; Iwama, H.; Fujita, K.; Okura, R.; Fujihara, S.; Yamashita, T.; Fujimori, T.; Kato, K.; Kamada, H.; et al. Galectin-9 suppresses cholangiocarcinoma cell proliferation by inducing apoptosis but not cell cycle arrest. Oncol. Rep. 2015, 34, 1761–1770. [Google Scholar] [CrossRef]
- Kuroda, J.; Yamamoto, M.; Nagoshi, H.; Kobayashi, T.; Sasaki, N.; Shimura, Y.; Horiike, S.; Kimura, S.; Yamauchi, A.; Hirashima, M.; et al. Targeting Activating Transcription Factor 3 by Galectin-9 Induces Apoptosis and Overcomes Various Types of Treatment Resistance in Chronic Myelogenous Leukemia. Mol. Cancer Res. 2010, 8, 994–1001. [Google Scholar] [CrossRef]
- Tadokoro, T.; Morishita, A.; Fujihara, S.; Iwama, H.; Niki, T.; Fujita, K.; Akashi, E.; Mimura, S.; Oura, K.; Sakamoto, T.; et al. Galectin-9: An anticancer molecule for gallbladder carcinoma. Int. J. Oncol. 2016, 48, 1165–1174. [Google Scholar] [CrossRef]
- Takano, J.; Morishita, A.; Fujihara, S.; Iwama, H.; Kokado, F.; Fujikawa, K.; Fujita, K.; Chiyo, T.; Tadokoro, T.; Sakamoto, T.; et al. Galectin-9 suppresses the proliferation of gastric cancer cells in vitro. Oncol. Rep. 2016, 35, 851–860. [Google Scholar] [CrossRef]
- Wiersma, V.R.; de Bruyn, M.; van Ginkel, R.J.; Sigar, E.; Hirashima, M.; Niki, T.; Nishi, N.; Samplonius, D.F.; Helfrich, W.; Bremer, E. The glycan-binding protein galectin-9 has direct apoptotic activity toward melanoma cells. J. Investig. Dermatol. 2012, 132, 2302–2305. [Google Scholar] [CrossRef]
- Yang, R.; Sun, L.; Li, C.F.; Wang, Y.H.; Yao, J.; Li, H.; Yan, M.; Chang, W.C.; Hsu, J.M.; Cha, J.H.; et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun. 2021, 12, 832. [Google Scholar] [CrossRef] [PubMed]
- Baum, L.G.; Pang, M.; Perillo, N.L.; Wu, T.; Delegeane, A.; Uittenbogaart, C.H.; Fukuda, M.; Seilhamer, J.J. Human thymic epithelial cells express an endogenous lectin, galectin-1, which binds to core 2 O-glycans on thymocytes and T lymphoblastoid cells. J. Exp. Med. 1995, 181, 877–887. [Google Scholar] [CrossRef]
- Perillo, N.L.; Uittenbogaart, C.H.; Nguyen, J.T.; Baum, L.G. Galectin-1, an endogenous lectin produced by thymic epithelial cells, induces apoptosis of human thymocytes. J. Exp. Med. 1997, 185, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, M.; Mercier, S.; Pelletier, I.; Bounou, S.; Roy, J.; Hirabayashi, J.; Sato, S.; Tremblay, M.J. Galectin-1 Acts as a Soluble Host Factor That Promotes HIV-1 Infectivity through Stabilization of Virus Attachment to Host Cells. J. Immunol. 2005, 174, 4120–4126. [Google Scholar] [CrossRef] [PubMed]
- Mercier, S.; St-Pierre, C.; Pelletier, I.; Ouellet, M.; Tremblay, M.J.; Sato, S. Galectin-1 promotes HIV-1 infectivity in macrophages through stabilization of viral adsorption. Virology 2008, 371, 121–129. [Google Scholar] [CrossRef]
- Lantéri, M.; Giordanengo, V.; Hiraoka, N.; Fuzibet, J.G.; Auberger, P.; Fukuda, M.; Baum, L.G.; Lefebvre, J.C. Altered T cell surface glycosylation in HIV-1 infection results in increased susceptibility to galectin-1-induced cell death. Glycobiology 2003, 13, 909–918. [Google Scholar] [CrossRef]
- St-Pierre, C.; Ouellet, M.; Giguère, D.; Ohtake, R.; Roy, R.; Sato, S.; Tremblay, M.J. Galectin-1-specific inhibitors as a new class of compounds to treat HIV-1 infection. Antimicrob. Agents Chemother. 2012, 56, 154–162. [Google Scholar] [CrossRef]
- Yang, M.L.; Chen, Y.H.; Wang, S.W.; Huang, Y.J.; Leu, C.H.; Yeh, N.C.; Chu, C.Y.; Lin, C.C.; Shieh, G.S.; Chen, Y.L.; et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011, 85, 10010–10020. [Google Scholar] [CrossRef] [PubMed]
- Fogel, S.; Guittaut, M.; Legrand, A.; Monsigny, M.; Hébert, E. The Tat protein of HIV-1 induces galectin-3 expression. Glycobiology 1999, 9, 383–387. [Google Scholar] [CrossRef]
- Jones, K.A. Tat and the HIV-1 promoter. Curr. Opin. Cell Biol. 1993, 5, 461–468. [Google Scholar] [CrossRef]
- Wang, S.F.; Tsao, C.H.; Lin, Y.T.; Hsu, D.K.; Chiang, M.L.; Lo, C.H.; Chien, F.C.; Chen, P.; Arthur Chen, Y.M.; Chen, H.Y.; et al. Galectin-3 promotes HIV-1 budding via association with Alix and Gag p6. Glycobiology 2014, 24, 1022–1035. [Google Scholar] [CrossRef]
- Xue, J.; Fu, C.; Cong, Z.; Peng, L.; Peng, Z.; Chen, T.; Wang, W.; Jiang, H.; Wei, Q.; Qin, C. Galectin-3 promotes caspase-independent cell death of HIV-1-infected macrophages. FEBS J. 2017, 284, 97–113. [Google Scholar] [CrossRef]
- Barnard, D.L.; Huffman, J.H.; Morris, J.L.; Wood, S.G.; Hughes, B.G.; Sidwell, R.W. Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antiviral. Res. 1992, 17, 63–77. [Google Scholar] [CrossRef]
- Lin, C.-W.; Wu, C.-F.; Hsiao, N.-W.; Chang, C.-Y.; Li, S.-W.; Wan, L.; Lin, Y.-J.; Lin, W.-Y. Aloe-emodin is an interferon-inducing agent with antiviral activity against Japanese encephalitis virus and enterovirus 71. Int. J. Antimicrob. Agents 2008, 32, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Shuangsuo, D.; Zhengguo, Z.; Yunru, C.; Xin, Z.; Baofeng, W.; Lichao, Y.; Yan’an, C. Inhibition of the replication of hepatitis B virus in vitro by emodin. Med. Sci. Monit. 2006, 12, Br302–Br306. [Google Scholar]
- Sydiskis, R.J.; Owen, D.G.; Lohr, J.L.; Rosler, K.H.; Blomster, R.N. Inactivation of enveloped viruses by anthraquinones extracted from plants. Antimicrob. Agents Chemother. 1991, 35, 2463–2466. [Google Scholar] [CrossRef]
- Li, S.W.; Yang, T.C.; Lai, C.C.; Huang, S.H.; Liao, J.M.; Wan, L.; Lin, Y.J.; Lin, C.W. Antiviral activity of aloe-emodin against influenza A virus via galectin-3 up-regulation. Eur. J. Pharmacol. 2014, 738, 125–132. [Google Scholar] [CrossRef]
- Li, F. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Caniglia, J.L.; Guda, M.R.; Asuthkar, S.; Tsung, A.J.; Velpula, K.K. A potential role for Galectin-3 inhibitors in the treatment of COVID-19. PeerJ 2020, 8, e9392. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.S.; Sun, L.W.; Brickner, H.; Sun, P.Q. Downregulating galectin-3 inhibits proinflammatory cytokine production by human monocyte-derived dendritic cells via RNA interference. Cell Immunol. 2015, 294, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Liang, W.; Sheng, J.; Xun, C.; Xu, T.; Cao, R.; Sheng, W. Gal-3 is a potential biomarker for spinal cord injury and Gal-3 deficiency attenuates neuroinflammation through ROS/TXNIP/NLRP3 signaling pathway. Biosci. Rep. 2019, 39, BSR20192368. [Google Scholar] [CrossRef]
- Sethi, A.; Sanam, S.; Munagalasetty, S.; Jayanthi, S.; Alvala, M. Understanding the role of galectin inhibitors as potential candidates for SARS-CoV-2 spike protein: In silico studies. RSC Advances 2020, 10, 29873–29884. [Google Scholar] [CrossRef]
- Bi, S.; Hong, P.W.; Lee, B.; Baum, L.G. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. USA 2011, 108, 10650–10655. [Google Scholar] [CrossRef]
- Colomb, F.; Giron, L.B.; Premeaux, T.A.; Mitchell, B.I.; Niki, T.; Papasavvas, E.; Montaner, L.J.; Ndhlovu, L.C.; Abdel-Mohsen, M. Galectin-9 Mediates HIV Transcription by Inducing TCR-Dependent ERK Signaling. Front. Immunol. 2019, 10, 267. [Google Scholar] [CrossRef]
- Jost, S.; Moreno-Nieves, U.Y.; Garcia-Beltran, W.F.; Rands, K.; Reardon, J.; Toth, I.; Piechocka-Trocha, A.; Altfeld, M.; Addo, M.M. Dysregulated Tim-3 expression on natural killer cells is associated with increased Galectin-9 levels in HIV-1 infection. Retrovirology 2013, 10, 74. [Google Scholar] [CrossRef]
- Elahi, S.; Niki, T.; Hirashima, M.; Horton, H. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood 2012, 119, 4192–4204. [Google Scholar] [CrossRef]
- Abdel-Mohsen, M.; Wang, C.; Strain, M.C.; Lada, S.M.; Deng, X.; Cockerham, L.R.; Pilcher, C.D.; Hecht, F.M.; Liegler, T.; Richman, D.D.; et al. Select host restriction factors are associated with HIV persistence during antiretroviral therapy. Aids 2015, 29, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mohsen, M.; Chavez, L.; Tandon, R.; Chew, G.M.; Deng, X.; Danesh, A.; Keating, S.; Lanteri, M.; Samuels, M.L.; Hoh, R.; et al. Human Galectin-9 Is a Potent Mediator of HIV Transcription and Reactivation. PLoS Pathog. 2016, 12, e1005677. [Google Scholar] [CrossRef]
- Katoh, S.; Ikeda, M.; Shimizu, H.; Mouri, K.; Obase, Y.; Kobashi, Y.; Fukushima, K.; Hirashima, M.; Oka, M. Increased levels of plasma galectin-9 in patients with influenza virus infection. Tohoku J. Exp. Med. 2014, 232, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sundararajan, A.; Suryawanshi, A.; Kumar, N.; Veiga-Parga, T.; Kuchroo, V.K.; Thomas, P.G.; Sangster, M.Y.; Rouse, B.T. T cell immunoglobulin and mucin protein-3 (Tim-3)/Galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc. Natl. Acad. Sci. USA 2011, 22, 108. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kremsreiter, S.M.; Kroell, A.-S.H.; Weinberger, K.; Boehm, H. Glycan–Lectin Interactions in Cancer and Viral Infections and How to Disrupt Them. Int. J. Mol. Sci. 2021, 22, 10577. https://doi.org/10.3390/ijms221910577
Kremsreiter SM, Kroell A-SH, Weinberger K, Boehm H. Glycan–Lectin Interactions in Cancer and Viral Infections and How to Disrupt Them. International Journal of Molecular Sciences. 2021; 22(19):10577. https://doi.org/10.3390/ijms221910577
Chicago/Turabian StyleKremsreiter, Stefanie Maria, Ann-Sophie Helene Kroell, Katharina Weinberger, and Heike Boehm. 2021. "Glycan–Lectin Interactions in Cancer and Viral Infections and How to Disrupt Them" International Journal of Molecular Sciences 22, no. 19: 10577. https://doi.org/10.3390/ijms221910577
APA StyleKremsreiter, S. M., Kroell, A.-S. H., Weinberger, K., & Boehm, H. (2021). Glycan–Lectin Interactions in Cancer and Viral Infections and How to Disrupt Them. International Journal of Molecular Sciences, 22(19), 10577. https://doi.org/10.3390/ijms221910577





