Tomato Bushy Stunt Virus Nanoparticles as a Platform for Drug Delivery to Shh-Dependent Medulloblastoma
Abstract
:1. Introduction
2. Results
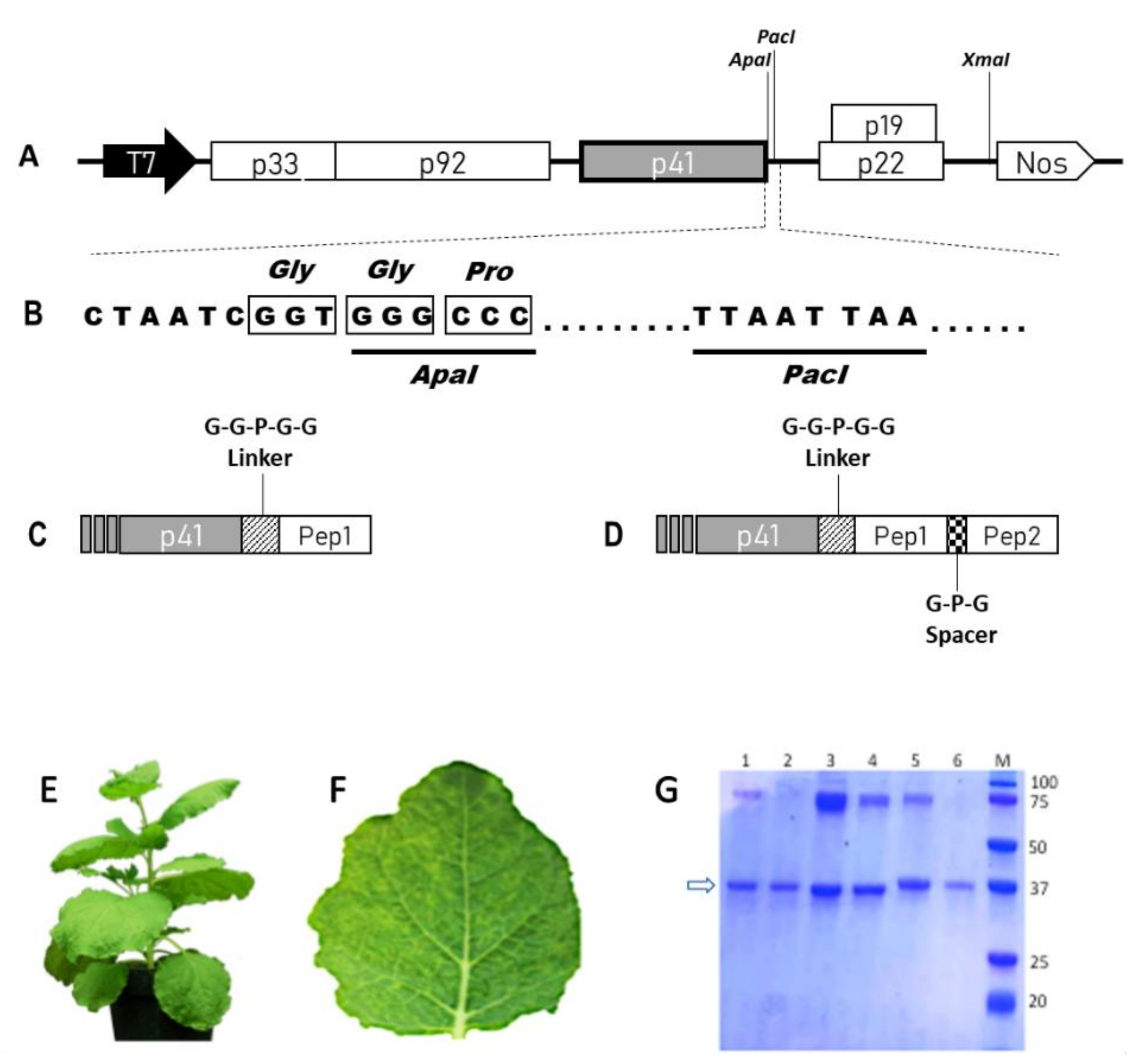
2.1. Construction, Production and Purification of WT and Chimeric TBSV NPs
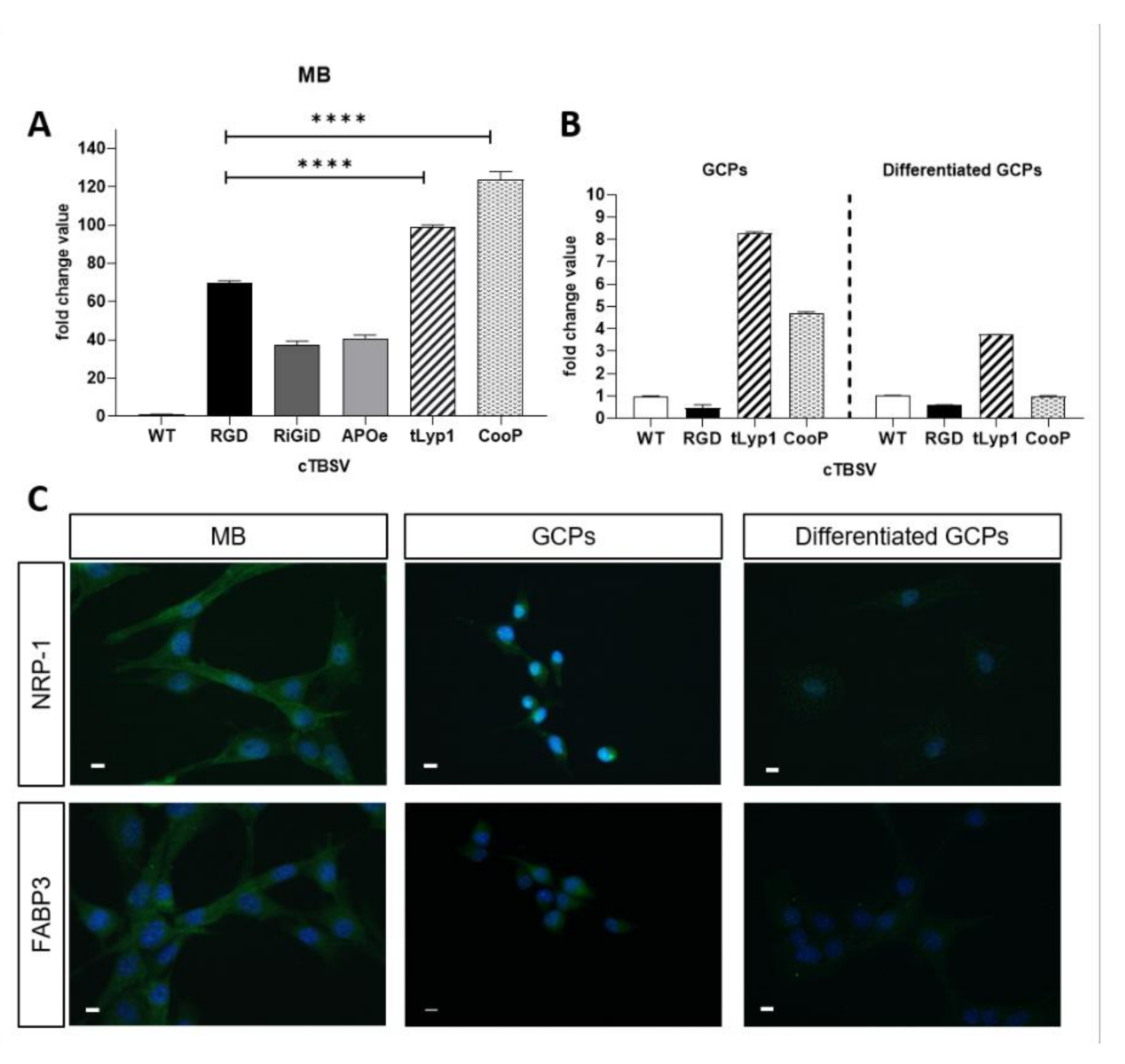
2.2. In Vitro Validation of cTBSV Uptake
2.3. Immunolocalization of the Interaction Partners
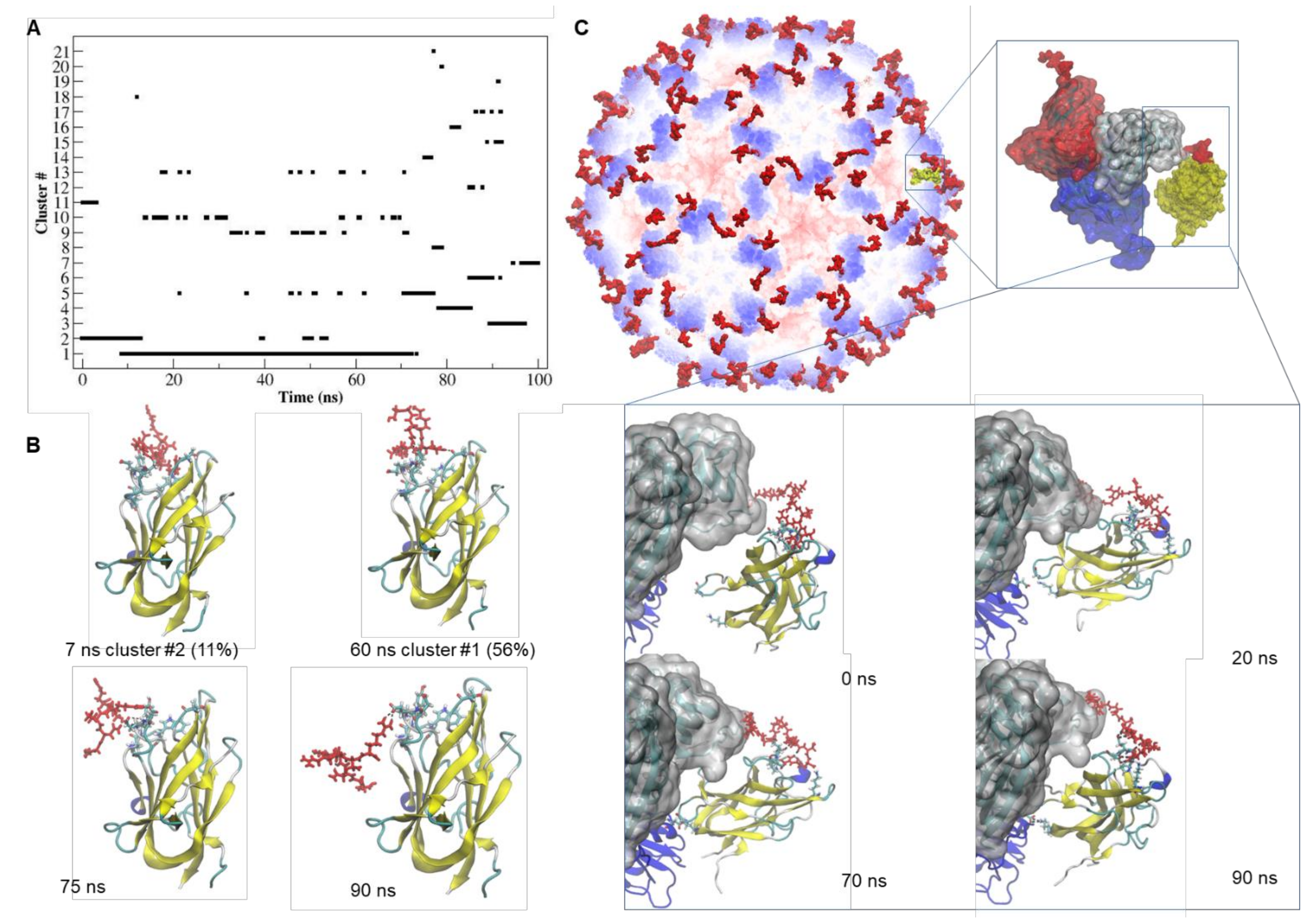
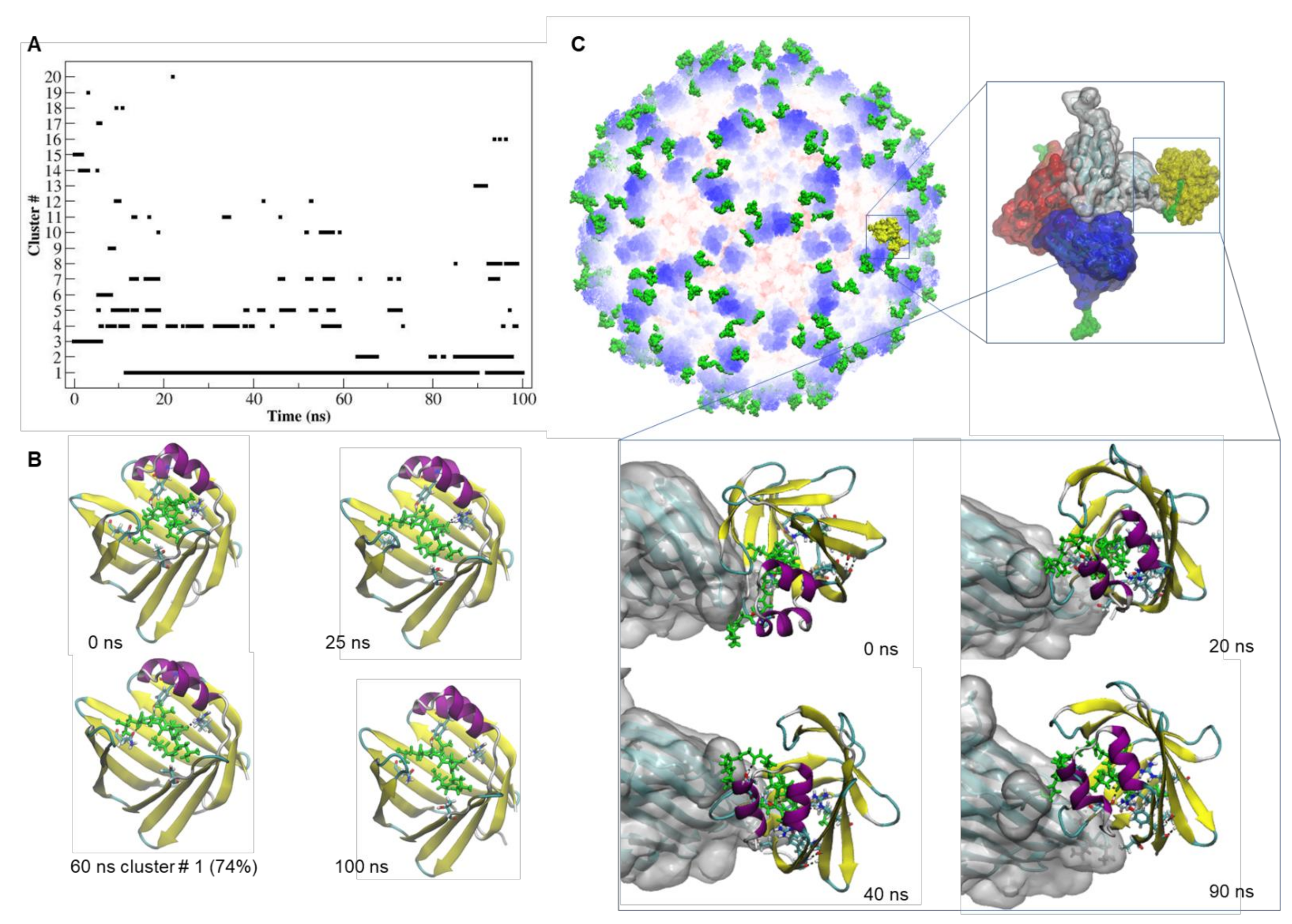
2.4. Docking and MD Simulation of the tLyp1-NRP-1 and CooP-FABP3 Complexes
2.5. TBSV-tLyp1 and TBSV-CooP Characterization and Loading with DOX
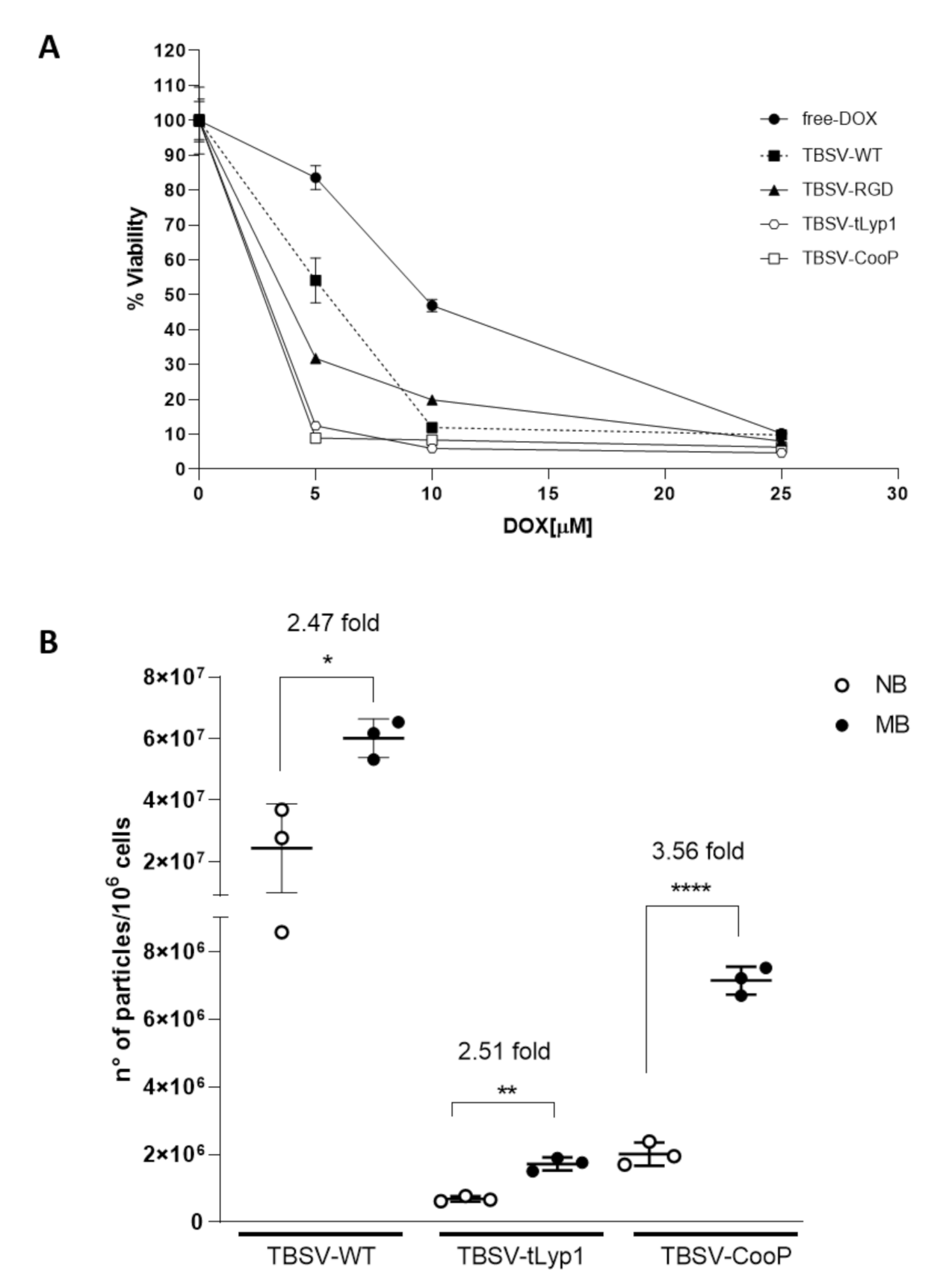
2.6. cTBSV-Mediated Delivery of DOX to Shh-MB Cells
2.7. cTBSV NPs Targeting to Shh-MB In Vivo
3. Discussion
4. Materials and Methods
4.1. TBSV Genetic Engineering, Production in Plants, Purification and Characterization
4.2. Mouse Model
4.3. Primary Cell Cultures and TBSV NPs Internalization
4.4. RNA Isolation and Real-Time qPCR (qPCR) Analysis
4.5. Immunofluorescence Analysis
4.6. Molecular Docking and Molecular Dynamics Simulations
4.7. Transmission Electron Microscopy
4.8. Dynamic Light Scattering and ζ-Potential Analyses
4.9. Production of DOX Loaded TBSV NPs
4.10. Cell Viability Assay
4.11. Absolute Quantification of c-TBSV in Target Tissues after Intravenous Injection
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | blood brain barrier |
| CNS | central nervous system |
| CP | coat protein |
| CPMV | cowpea mosaic virus |
| cTBSV | chimeric TBSV |
| DAPI | 4′,6-diamidino-2-phenylindole |
| d.p.i. | days post infection |
| DLS | dynamic light scattering |
| DOX | doxorubicin |
| EDTA | ethylenediaminetetraacetic acid |
| EE | encapsulation efficiency |
| FABP3 | mammary-derived growth inhibitor (MDGI/H-FABP/FABP3) |
| Gadph | glyceraldehyde-3-phosphate dehydrogenase |
| GBM | glioblastoma |
| GCP | granule cell precursor |
| ELS | electrophoretic light scattering |
| HDL | high density lipoprotein |
| HPI | shh pathway inhibitor |
| LC | loading capacity |
| MB | medulloblastoma |
| MD | molecular dynamics |
| MM/PBSA | molecular mechanics/Poisson-Boltzmann solvent accessible surface area |
| NB | normal brain |
| NP | nanoparticles |
| NRP-1 | neuropilin-1 |
| OLA | oleic acid |
| PFA | paraformaldehyde |
| PLGA-PEG | poly(lactic-co-glycolic acid) conjugated to polyethylene glycol |
| PME | particle mesh Ewald |
| Ptch1+/- | Patched1 heterozygous mice |
| qPCR | real time quantitative PCR |
| rmsd | root mean square displacement |
| RPL13a | ribosomal protein L13A |
| RPL32 | ribosomal protein L32 |
| RT | room temperature |
| RT-PCR | reverse transcriptase-PCR |
| Shh | sonic hedgehog |
| SMO | smoothened |
| SSEA-1 | stage-specific embryonic antigen 1 |
| TBP | TATA box binding protein |
| TBSV | tomato bushy stunt virus |
| TEM | transmission electron microscopy |
| WNT | wingless |
References
- Senapati, S.; Mahanta, A.K.; Kumar, S.; Maiti, P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct. Target. Ther. 2018, 16, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Lochhead, J.J.; Yang, J.; Ronaldson, P.T.; Davis, T.P. Structure, Function, and Regulation of the Blood-Brain Barrier Tight Junction in Central Nervous System Disorders. Front. Physiol. 2020, 11, 914. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Korshunov, A.; Witt, H.; Hielscher, T.; Eberhart, C.G.; Mack, S.; Bouffet, E.; Clifford, S.C.; Hawkins, C.E.; French, P.; et al. Medulloblastoma Comprises Four Distinct Molecular Variants. J. Clin. Oncol. 2011, 29, 1408–1414. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, S.C.; Doz, F.; Kool, M.; Dufour, C.; Vassal, G.; Milde, T.; et al. Risk stratification of childhood medulloblastoma in the molecular era: The current consensus. Acta Neuropathol. 2016, 131, 821–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.F.; Liu, L.; Xue, X.; Liang, X.J. Nanoparticle-based drug delivery systems: What can they really do in vivo? F1000Research 2017, 6, 681. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Cai, H.; Steinmetz, N.F. Viral nanoparticles for drug delivery, imaging, immunotherapy, and theranostic applications. Adv. Drug Deliv. Rev. 2010, 27, 214–235. [Google Scholar] [CrossRef] [PubMed]
- Grasso, S.; Lico, C.; Imperatori, F.; Santi, L. A plant derived multifunctional tool for nanobiotechnology based on Tomato bushy stunt virus. Transgenic Res. 2013, 22, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Blandino, A.; Lico, C.; Baschieri, S.; Barberini, L.; Cirotto, C.; Blasi, P.; Santi, L. In vitro and in vivo toxicity evaluation of plant virus nanocarriers. Colloids Surf. B Biointerfaces 2015, 129, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Lico, C.; Giardullo, P.; Mancuso, M.; Benvenuto, E.; Santi, L.; Baschieri, S. A biodistribution study of two differently shaped plant virus nanoparticles reveals new peculiar traits. Colloids Surf. B Biointerfaces 2016, 148, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Merk, D.J.; Segal, R.A. Sonic Hedgehog Signaling is Blue: Insights from the Patched Mutant Mice. Trends Neurosci. 2018, 41, 870–872. [Google Scholar] [CrossRef] [PubMed]
- Pazzaglia, S.; Tanori, M.; Mancuso, M.; Gessi, M.; Pasquali, E.; Leonardi, S.; Oliva, M.A.; Rebessi, S.; Di Majo, V.; Covelli, V.; et al. Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene 2006, 25, 5575–5580. [Google Scholar] [CrossRef] [Green Version]
- Mancuso, M.; Pasquali, E.; Leonardi, S.; Tanori, M.; Rebessi, S.; Di Majo, V.; Pazzaglia, S.; Toni, M.P.; Pimpinella, M.; Covelli, V.; et al. Oncogenic bystander radiation effects in Patched heterozygous mouse cerebellum. Proc. Natl. Acad. Sci. USA 2008, 105, 12445–12450. [Google Scholar] [CrossRef] [Green Version]
- Alipour, M.; Baneshi, M.; Hosseinkhani, S.; Mahmoudi, R.; Jabari Arabzadeh, A.; Akrami, M.; Mehrzad, J.; Bardania, H. Recent progress in biomedical applications of RGD-based ligand: From precise cancer theranostics to biomaterial engineering: A systematic review. J. Biomed. Mater. Res. A 2020, 108, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Agemy, L.; Kotamraju, V.R.; Braun, G.; Teesalu, T.; Sugahara, K.N.; Hamzah, J.; Ruoslahti, E. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene 2012, 31, 3754–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Gao, X.; Kang, T.; Jiang, D.; Yao, J.; Jing, Y.; Song, Q.; Jiang, X.; Liang, J.; Chen, J. Mammary-Derived Growth Inhibitor Targeting Peptide-Modified PEG-PLA Nanoparticles for Enhanced Targeted Glioblastoma Therapy. Bioconjug. Chem. 2015, 2015 26, 1850–1861. [Google Scholar] [CrossRef]
- Wang, Q.; Kumar, V.; Lin, F.; Sethi, B.; Coulter, D.W.; McGuire, T.R.; Mahato, R.I. ApoE mimetic peptide targeted nanoparticles carrying a BRD4 inhibitor for treating Medulloblastoma in mice. J. Control. Release 2020, 323, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Eberhart, C.G. Medulloblastoma stem cells. J. Clin. Oncol. 2008, 26, 2821–2827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyvönen, M.; Enbäck, J.; Huhtala, T.; Lammi, J.; Sihto, H.; Weisell, J.; Joensuu, H.; Rosenthal-Aizman, K.; El-Andaloussi, S.; Langel, U.; et al. Novel target for peptide-based imaging and treatment of brain tumors. Mol. Cancer Ther. 2014, 13, 996–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatr. 2019, 24, 353–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kool, M.; Jones, D.T.W.; Jäger, N.; Northcott, P.A.; Pugh, T.J.; Hovestadt, V. Genome Sequencing of Shh Medulloblastoma Predicts Genotype-Related Response to Smoothened Inhibition. Cancer Cell 2014, 25, 393–405. [Google Scholar] [CrossRef] [Green Version]
- Zottel, A.; Videtič Paska, A.; Jovčevska, I. Nanotechnology Meets Oncology: Nanomaterials in Brain Cancer Research, Diagnosis and Therapy. Materials 2019, 12, 1588. [Google Scholar] [CrossRef] [Green Version]
- Catanzaro, G.; Curcio, M.; Cirillo, G.; Spizzirri, U.G.; Besharat, Z.M.; Abballe, L.; Vacca, A.; Iemma, F.; Picci, N.; Ferretti, E. Albumin nanoparticles for glutathione-responsive release of cisplatin: New opportunities for medulloblastoma. Int. J. Pharm. 2017, 517, 168–174. [Google Scholar] [CrossRef]
- Chenna, V.; Hu, C.; Pramanik, D.; Aftab, B.T.; Karikari, C.; Campbell, N.R.; Hong, S.M.; Zhao, M.; Rudek, M.A.; Khan, S.R.; et al. A polymeric nanoparticle encapsulated small-molecule inhibitor of Hedgehog signaling (NanoHHI) bypasses secondary mutational resistance to Smoothened antagonists. Mol. Cancer Ther. 2012, 11, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Bell, J.B.; Rink, J.S.; Eckerdt, F.; Clymer, J.; Goldman, S.; Thaxton, C.S.; Platanias, L.C. HDL nanoparticles targeting sonic hedgehog subtype medulloblastoma. Sci. Rep. 2018, 8, 1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Dey, A.; Malhotra, A.; Liu, J.; Ahn, S.I.; Sei, Y.J.; Kenney, A.M.; MacDonald, T.J.; Kim, Y. Engineered biomimetic nanoparticle for dual targeting of the cancer stem-like cell population in sonic hedgehog medulloblastoma. Proc. Natl. Acad. Sci. USA 2020, 117, 24205–24212. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Dismuke, T.; Tikunov, A.; Rosen, E.P.; Kagel, J.R.; Ramsey, J.D.; Lim, C.; Zamboni, W.; Kabanov, A.V.; Gershon, T.R.; et al. Poly(2-oxazoline) nanoparticle delivery enhances the therapeutic potential of vismodegib for medulloblastoma by improving CNS pharmacokinetics and reducing systemic toxicity. Nanomedicine 2021, 32, 102345. [Google Scholar] [CrossRef] [PubMed]
- Chariou, P.L.; Ortega-Rivera, O.A.; Steinmetz, N.F. Nanocarriers for the delivery of medical, veterinary, and agricultural active ingredients. ACS Nano 2020, 14, 2678–2701. [Google Scholar] [CrossRef]
- Koudelka, K.J.; Destito, G.; Plummer, E.M.; Trauger, S.A.; Siuzdak, G.; Manchester, M. Endothelial Targeting of Cowpea Mosaic Virus (CPMV) via Surface Vimentin. PLoS Pathog. 2009, 5, e1000417. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, L.; Dong, X.; Liu, L.; Huo, L.; Chen, H. High Expression of Vimentin is Associated With Progression and a Poor Outcome in Glioblastoma. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 337–344. [Google Scholar] [CrossRef]
- Nkanga, C.I.; Steinmetz, N.F. The pharmacology of plant virus nanoparticles. Virology 2021, 556, 39–61. [Google Scholar] [CrossRef] [PubMed]
- Arcangeli, C.; Lico, C.; Baschieri, S.; Mancuso, M. Characterization Of Blood-Brain Barrier Crossing And Tumor Homing Peptides By Molecular Dynamics Simulations. Int. J. Nanomed. 2019, 14, 10123–10136. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, L.; Nandy, D.; Zhang, Y.; Basu, A.; Radisky, D.; Mukhopadhyay, D. Neuropilin-1 upholds dedifferentiation and propagation phenotypes of renal cell carcinoma cells by activating Akt and sonic hedgehog axes. Cancer Res. 2008, 68, 8667–8672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osada, H.; Tokunaga, T.; Nishi, M.; Hatanaka, H.; Abe, Y.; Tsugu, A.; Kijima, H.; Yamazaki, H.; Ueyama, Y.; Nakamura, M. Overexpression of the neuropilin 1 (NRP1) gene correlated with poor prognosis in human glioma. Anticancer Res. 2004, 24, 547–552. [Google Scholar] [PubMed]
- Snuderl, M.; Batista, A.; Kirkpatrick, N.D.; Ruiz de Almodovar, C.; Riedemann, L.; Walsh, E.C.; Anolik, R.; Huang, Y.; Martin, J.D.; Kamoun, W.; et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell 2013, 152, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinskey, J.M.; Franks, N.E.; McMellen, A.N.; Giger, R.J.; Allen, B.L. Neuropilin-1 promotes Hedgehog signaling through a novel cytoplasmic motif. J. Biol. Chem. 2017, 292, 15192–15204. [Google Scholar] [CrossRef] [Green Version]
- Smits, M.; van Rijn, S.; Hulleman, E.; Biesmans, D.; van Vuurden, D.G.; Kool, M.; Haberler, C.; Aronica, E.; Vandertop, W.P.; Noske, D.P.; et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin. Cancer Res. 2012, 18, 4048–4058. [Google Scholar] [CrossRef] [Green Version]
- Tanno, B.; Leonardi, S.; Babini, G.; Giardullo, P.; De Stefano, I.; Pasquali, E.; Saran, A.; Mancuso, M. Nanog-driven cell-reprogramming and self-renewal maintenance in Ptch1 +/- granule cell precursors after radiation injury. Sci. Rep. 2017, 7, 14238. [Google Scholar] [CrossRef] [Green Version]
- Mota, F.; Fotinou, C.; Rana, R.R.; Chan, A.W.E.; Yelland, T.; Arooz, M.T.; O’Leary, A.P.; Hutton, J.; Frankel, P.; Zachary, I.; et al. Architecture and hydration of the arginine-binding site of neuropilin-1. FEBS J. 2018, 285, 1290–1304. [Google Scholar] [CrossRef] [Green Version]
- Howard, E.I.; Guillot, B.; Blakeley, M.P.; Haertlein, M.; Moulin, M.; Mitschler, A.; Cousido-Siah, A.; Fadel, F.; Valsecchi, W.M.; Tomizaki, T.; et al. High-resolution neutron and X-ray diffraction room-temperature studies of an H-FABP-oleic acid complex: Study of the internal water cluster and ligand binding by a transferred multipolar electron-density distribution. IUCrJ 2016, 3 Pt 2, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Fiser, A.; Sali, A. Modeller: Generation and refinement of homology-based protein structure models. Meth. Enzymol. 2003, 374, 461–491. [Google Scholar]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins 2017, 85, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Spoel, D.V.D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comp. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Alemzadeh, E.; Izadpanah, K.; Ahmadi, F. Generation of recombinant protein shells of Johnson grass chlorotic stripe mosaic virus in tobacco plants and their use as drug carrier. J. Virol. Methods 2017, 248, 148–153. [Google Scholar] [CrossRef]
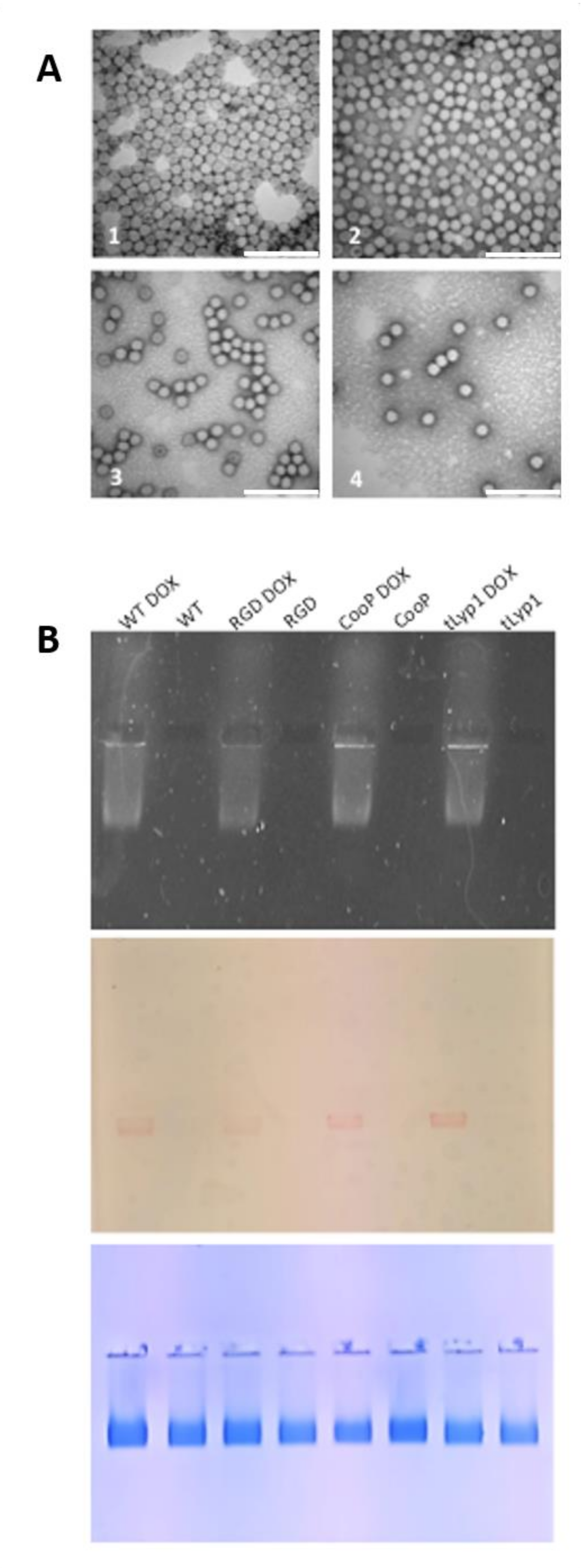
| NP | Size * (d.nm) by TEM | Size § (d.nm) by DLS | PDI | ζ-Potential (mV) | DOX Molecules/Particle § | Ng DOX/ µg Virus | [µM] | EE § (%) | LC § (%) |
|---|---|---|---|---|---|---|---|---|---|
| TBSV-WT | 32.7 ± 1.1 | 50.8 ± 18.4 | 0.429 | −5.73 | 2009 ± 73 | 130.2 ± 4.7 | 207 | 38 ± 6 | 12 ± 2 |
| TBSV-RGD | 32.7 ± 0.9 | 48.5 ± 14.2 | 0.489 | −5.95 | 1911 ± 394 | 136.4 ± 21.2 | 155 | 34 ± 1 | 11 ± 1 |
| TBSV-CooP | 32.7 ± 0.8 | 68.1 ± 17.7 | 0.588 | −3.98 | 2174 ± 326 | 151.9 ± 15.9 | 379 | 47 ± 5 | 15 ± 2 |
| TBSV-tLyp1 | 32.7 ± 0.9 | 66.5 ± 18.8 | 0.419 | −3.61 | 2498 ± 749 | 174.1 ± 32.1 | 207 | 54 ± 10 | 17 ± 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lico, C.; Tanno, B.; Marchetti, L.; Novelli, F.; Giardullo, P.; Arcangeli, C.; Pazzaglia, S.; Podda, M.S.; Santi, L.; Bernini, R.; et al. Tomato Bushy Stunt Virus Nanoparticles as a Platform for Drug Delivery to Shh-Dependent Medulloblastoma. Int. J. Mol. Sci. 2021, 22, 10523. https://doi.org/10.3390/ijms221910523
Lico C, Tanno B, Marchetti L, Novelli F, Giardullo P, Arcangeli C, Pazzaglia S, Podda MS, Santi L, Bernini R, et al. Tomato Bushy Stunt Virus Nanoparticles as a Platform for Drug Delivery to Shh-Dependent Medulloblastoma. International Journal of Molecular Sciences. 2021; 22(19):10523. https://doi.org/10.3390/ijms221910523
Chicago/Turabian StyleLico, Chiara, Barbara Tanno, Luca Marchetti, Flavia Novelli, Paola Giardullo, Caterina Arcangeli, Simonetta Pazzaglia, Maurizio S. Podda, Luca Santi, Roberta Bernini, and et al. 2021. "Tomato Bushy Stunt Virus Nanoparticles as a Platform for Drug Delivery to Shh-Dependent Medulloblastoma" International Journal of Molecular Sciences 22, no. 19: 10523. https://doi.org/10.3390/ijms221910523
APA StyleLico, C., Tanno, B., Marchetti, L., Novelli, F., Giardullo, P., Arcangeli, C., Pazzaglia, S., Podda, M. S., Santi, L., Bernini, R., Baschieri, S., & Mancuso, M. (2021). Tomato Bushy Stunt Virus Nanoparticles as a Platform for Drug Delivery to Shh-Dependent Medulloblastoma. International Journal of Molecular Sciences, 22(19), 10523. https://doi.org/10.3390/ijms221910523








