A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening
Abstract
:1. Introduction
2. Results
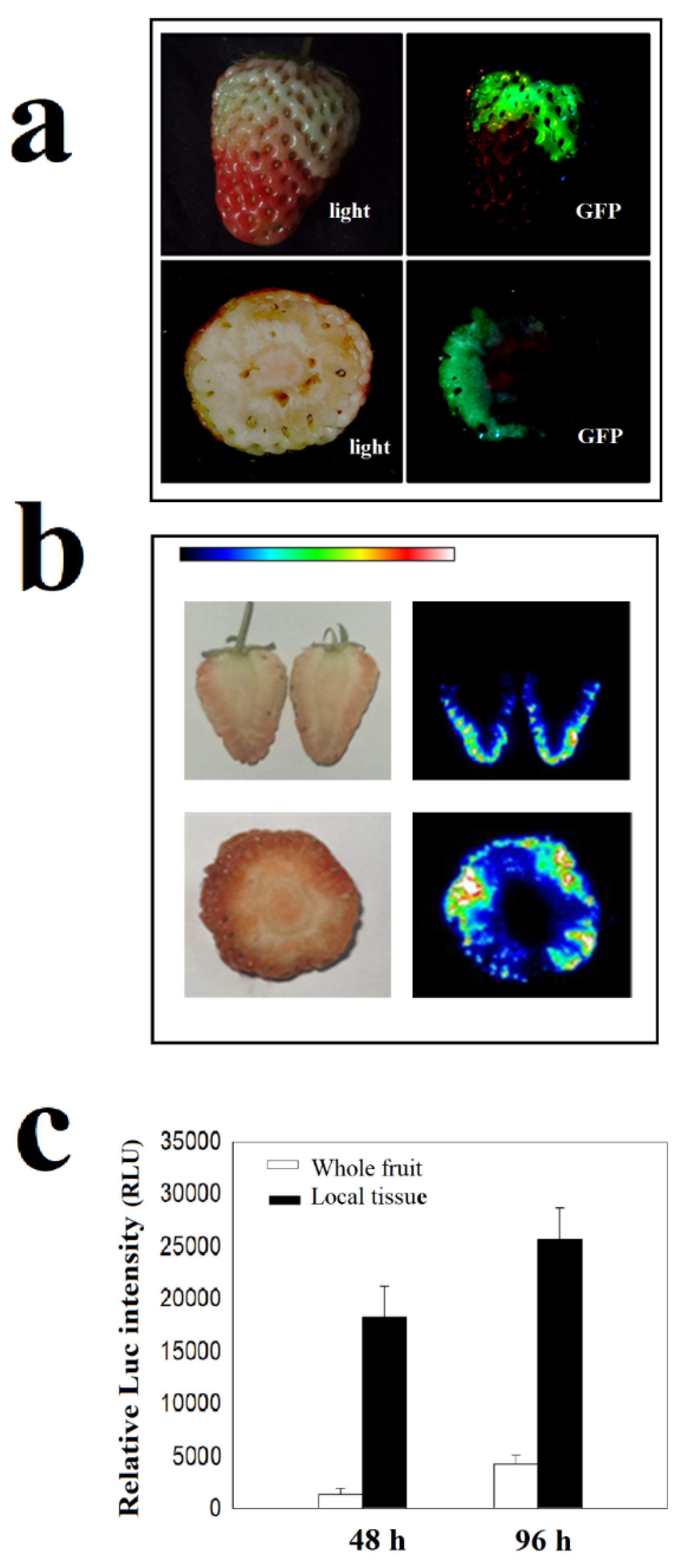
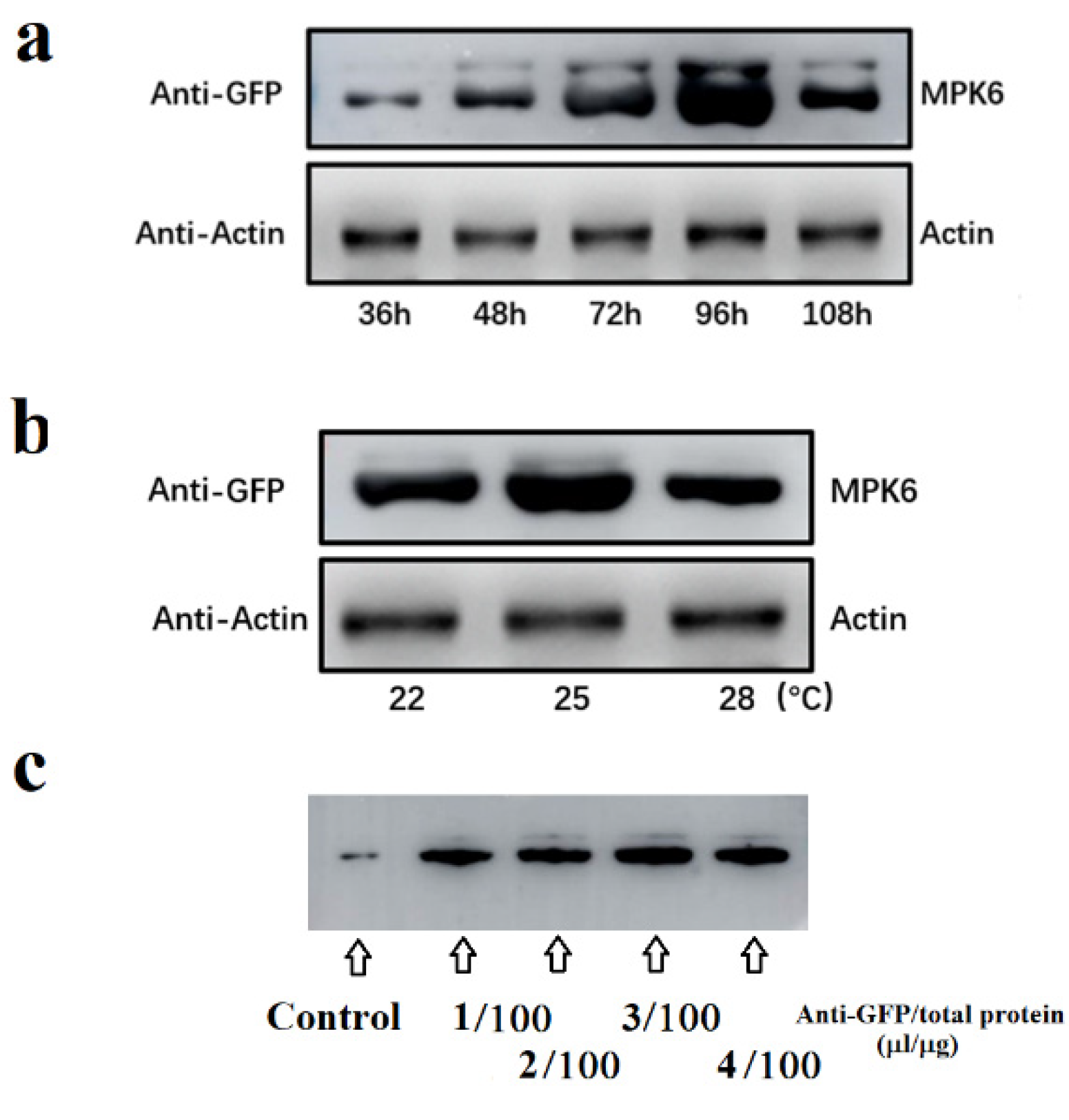
2.1. Optimization of Assays for Protein Kinase Activity In Vivo
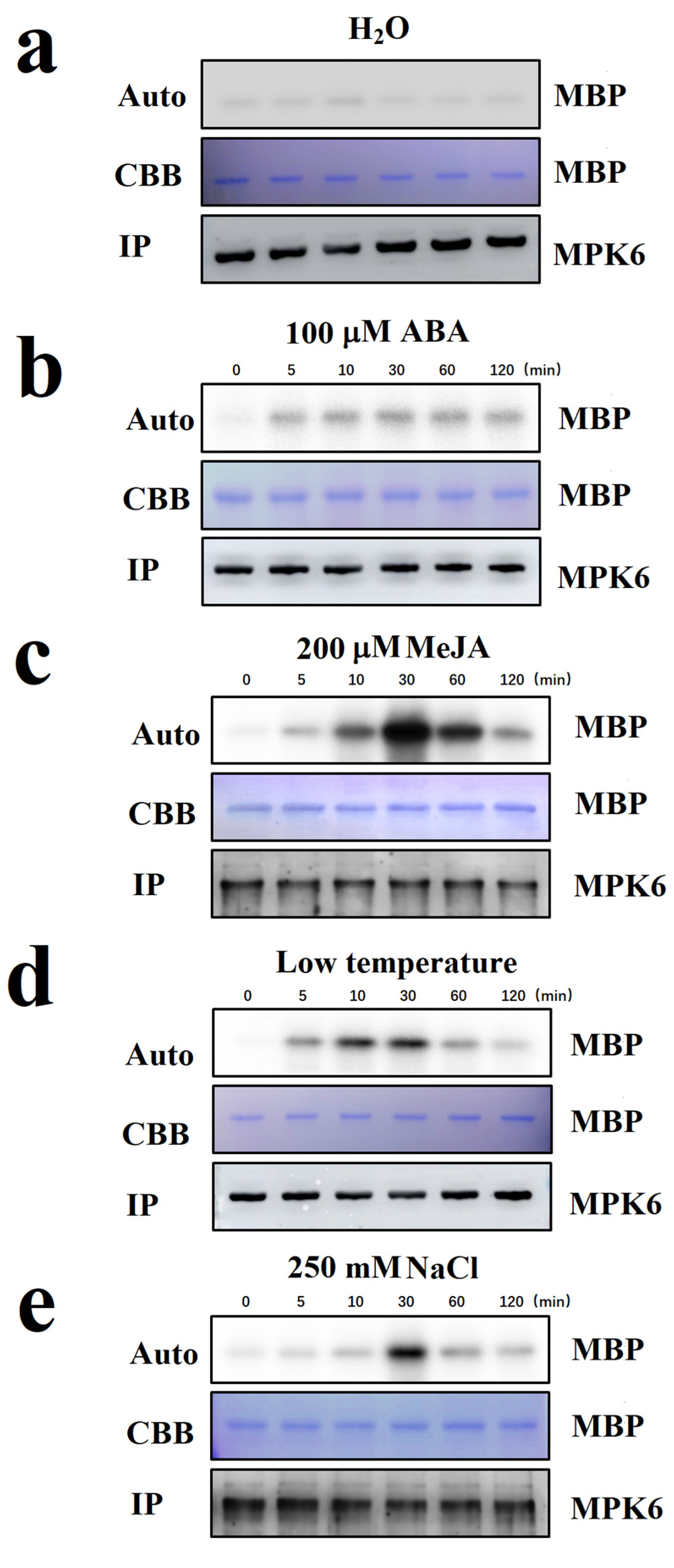
2.2. Screening for Internal and External Signals Upstream of FaMPK6
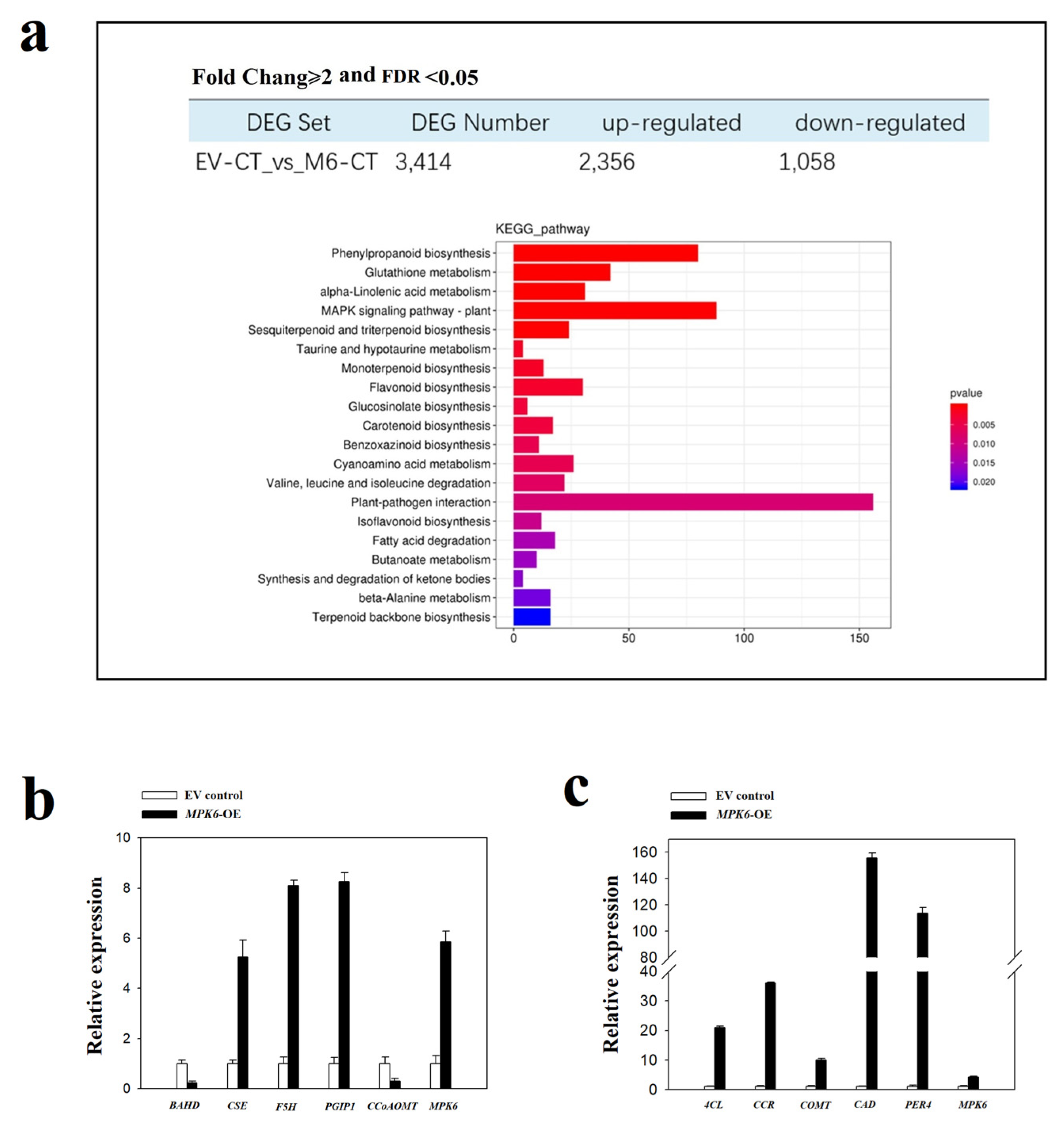
2.3. FaMPK6 Functions in the Regulation of Phenylpropanoid-Associated Metabolism
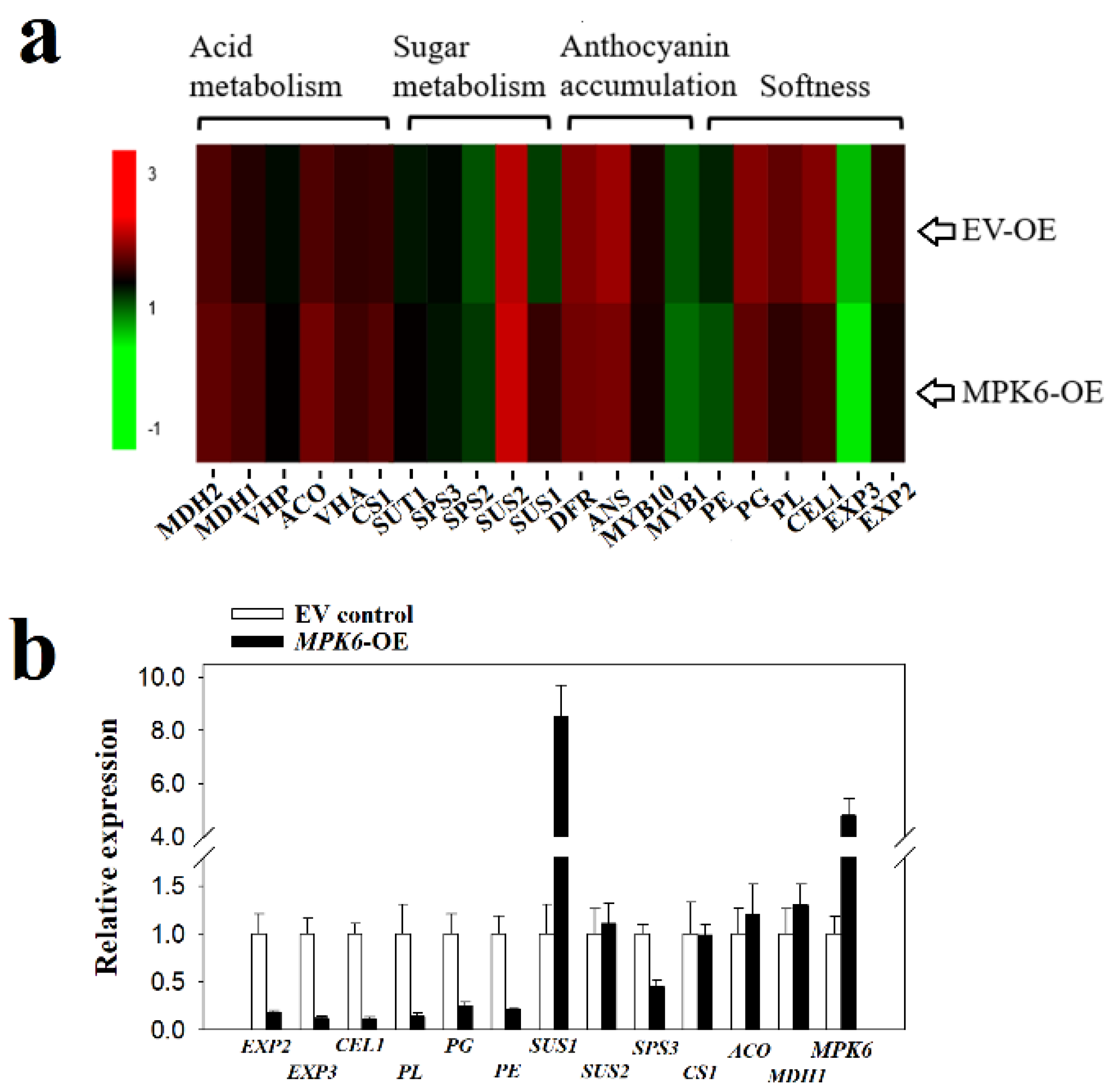
2.4. FaMPK6 Functions in the Regulation of Fruit-Ripening Associated Metabolism
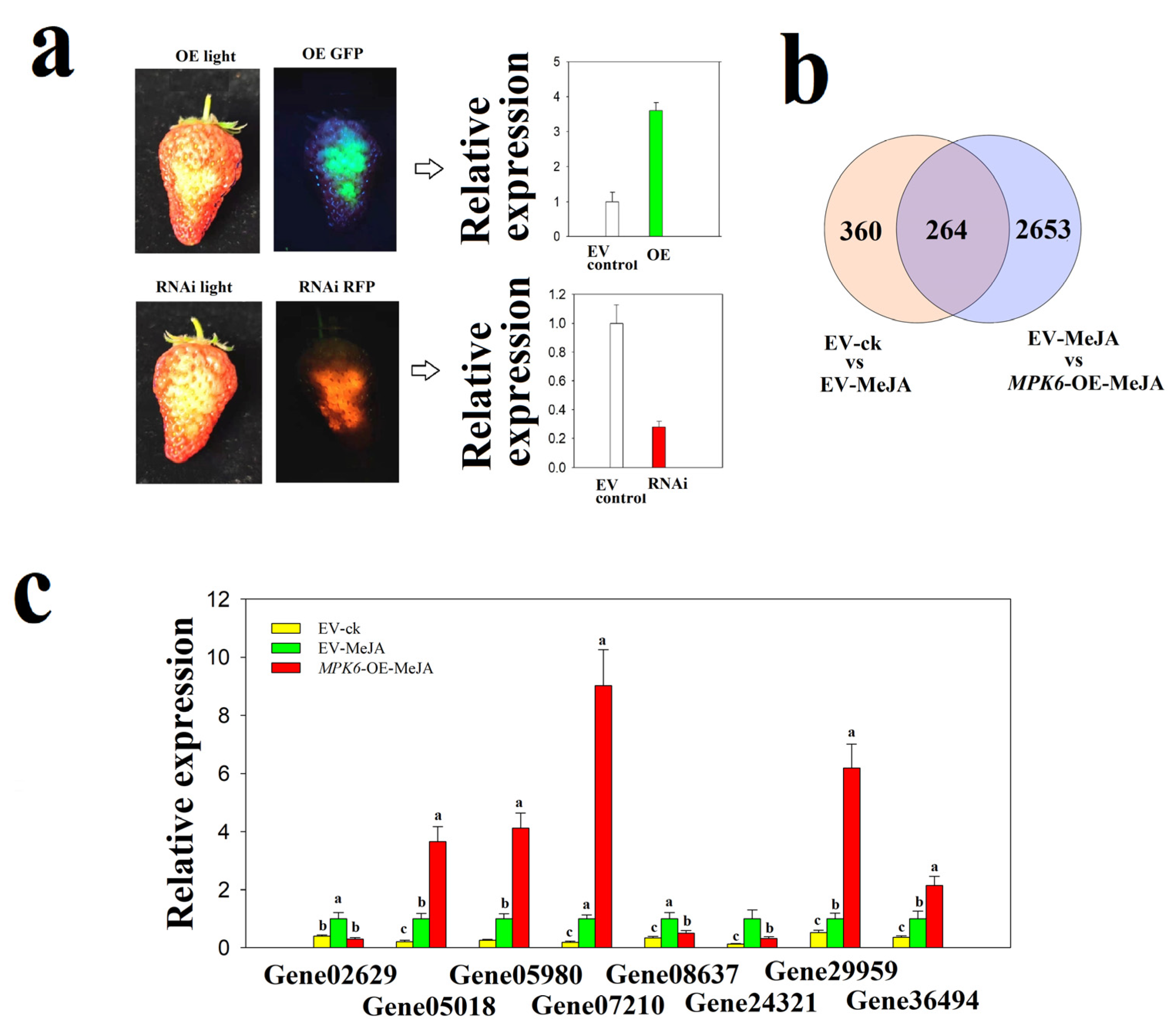
2.5. FaMPK6 Functions in the Mediation of JA-Induced Gene Expression
3. Discussion
3.1. Importance of In Vivo Protein Kinase Activity Assay Method Development
3.2. Broader Use of the In Vivo Protein Kinase Activity Assay
3.3. Key Points for In Vivo Assaying of Protein Kinase Activity
3.4. FaMPK6 Contributes to the Regulation of Secondary Wall Metabolism during Fruit Ripening
3.5. FaMPK6 Mediates Multiple Signaling Cascades
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Given, N.K.; Venis, M.A.; Gierson, D. Hormonal regulation of ripening in the strawberry, A non-climacteric fruit. Planta 1988, 174, 402–426. [Google Scholar] [CrossRef] [PubMed]
- Jang, R.; Loewus, F.A.; Seegmiller, C.G. The conversion of C14-labeled sugars to L-ascorbic acid in ripening strawberries. J. Biol. Chem. 1956, 222, 649–664. [Google Scholar] [PubMed]
- Loewus, F.A.; Kelly, S. The metabolism of p-galacturonic acid and its methyl ester in the detached ripening strawberry. Arch. Biochem. Biophys. 1961, 95, 483–493. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.) Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef] [PubMed]
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folta, K.M.; Barbey, C.R. The strawberry genome: A complicated past and promising future. Hortic. Res. 2019, 6, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Ma, H.; Jung, S.; Main, D.; Guo, L. Developmental Mechanisms of Fleshy Fruit Diversity in Rosaceae. Annu. Rev. Plant Biol. 2020, 71, 547–573. [Google Scholar] [CrossRef] [PubMed]
- Martín-Pizarro, C.; Vallarino, J.G.; Osorio, S.; Meco, V.; Urrutia, M.; Pillet, J.; Casañal, A.; Merchante, C.; Amaya, I.; Willmitzer, L. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 2021. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Osorio, S.; Bombarely, A.; Casañal, A.; Cruz-Rus, E.; Sánchez-Sevilla, J.F.; Amaya, I.; Giavalisco, P.; Fernie, A.R.; Botella, M.A. Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 2015, 208, 482–496. [Google Scholar] [CrossRef]
- Whitaker, V.M. A roadmap for research in octoploid strawberry. Hortic. Res. 2020, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Zorrilla-Fontanesi, Y.; Rambla, J.; Cabeza, A.; Medina, J.J.; Sánchez-Sevilla, J.F.; Valpuesta, V.; Botella, M.A.; Granell, A.; Amaya, I. Genetic analysis of strawberry fruit aroma and identification of O-methyltransferase FaOMT as the locus controlling natural variation in mesifurane content. Plant Physiol. 2012, 159, 851–870. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.G.; Doke, N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Jiang, M.; Zhang, J.; Tan, M.; Hu, X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006, 141, 475–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, S.; Ohta, K.; Yoshioka, H. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 2008, 20, 1390–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaspers, P.; Kangasjärvi, J. Reactive oxygen species in abiotic stress signaling. Physiol. Plant. 2010, 138, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Burnett, E.C.; Desikan, R.; Moser, R.C.; Neill, S.J. ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 2000, 51, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.-F.; Andreoli, S.; Tiriac, H.; Alonso, J.; Harper, J.F.; Ecker, J.; et al. CDPKs CPK6 and CPK3 Function in ABA Regulation of Guard Cell S-Type Anion- and Ca2+-Permeable Channels and Stomatal Closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [Green Version]
- Gudesblat, G.E.; Iusem, N.D.; Morris, P.C. Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 2007, 173, 713–721. [Google Scholar] [CrossRef]
- Jammes, F.; Song, C.; Shin, D.; Munemasa, S.; Takeda, K.; Gu, D.; Cho, D.; Lee, S.; Giordo, R.; Sritubtim, S.; et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20520–20525. [Google Scholar] [CrossRef] [Green Version]
- Wurzinger, B.; Mair, A.; Pfister, B.; Teige, M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 2011, 6, 8–12. [Google Scholar] [CrossRef]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A Unique Short-Chain Dehydrogenase/Reductase in Arabidopsis Glucose Signaling and Abscisic Acid Biosynthesis and Functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- Koornneef, M.; Bentsink, L.; Hilhorst, H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002, 5, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Han, M.H.; Guevara-Garcia, A.; Fedoroff, N.V. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 2002, 99, 15812–15817. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Jia, W.; Zhang, J. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007, 58, 2969–2981. [Google Scholar] [CrossRef]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. Cell Mol. Biol. 2008, 54, 440–451. [Google Scholar] [CrossRef]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol. Biol. 2009, 70, 725–736. [Google Scholar] [CrossRef]
- Ludwików, A.; Cieśla, A.; Kasprowicz-Maluśki, A.; Mituła, F.; Tajdel, M.; Gałgański, Ł.; Ziółkowski, P.A.; Kubiak, P.; Małecka, A.; Piechalak, A.; et al. Arabidopsis Protein Phosphatase 2C ABI1 Interacts with Type I ACC Synthases and Is Involved in the Regulation of Ozone-Induced Ethylene Biosynthesis. Mol. Plant 2014, 7, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, Y.M.; Jia, H.F.; Li, C.L.; Dong, Q.H.; Shen, Y.Y. FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 2011, 62, 5079–5089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Symons, G.M.; Chua, Y.-J.; Ross, J.J.; Quittenden, L.; Davies, N.; Reid, J.B. Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 2012, 63, 4741–4750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Wang, Y.; Sun, M.; Li, B.; Han, Y.; Zhao, Y.; Li, X.; Ding, N.; Li, C.; Ji, W.; et al. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013, 198, 453–465. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.G.; Hong, Y.; Lee, E.J. Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J. Plant Physiol. 2019, 239, 52–60. [Google Scholar] [CrossRef]
- Zhao, Y.; Mao, W.; Chen, Y.; Wang, W.; Dai, Z.; Dou, Z.; Zhang, K.; Wei, L.; Li, T.; Zeng, B.; et al. Optimization and standardization of transient expression assays for gene functional analyses in strawberry fruits. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hardie, D.G. Plant Protein Serine/Threonine Kinases: Classification and Functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 97–131. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.C. Exploring the role of protein phosphorylation in plants: From signalling to metabolism. Biochem. Soc. Trans. 2007, 35, 28–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, S. Protein phosphatases in plants. Annu. Rev. Plant Biol. 2003, 54, 63–92. [Google Scholar] [CrossRef] [PubMed]
- MacKintosh, C. Regulation of cytosolic enzymes in primary metabolism by reversible protein phosphorylation. Curr. Opin. Plant Biol. 1998, 1, 224–229. [Google Scholar] [CrossRef]
- Smith, R.D.; Walker, J.C. PLANT PROTEIN PHOSPHATASES. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 101–125. [Google Scholar] [CrossRef] [Green Version]
- Zeng, B.; Li, T.; Wang, W.; Dai, Z.; Li, J.; Xi, Z.; Jia, K.; Xing, Y.; Li, B.; Yan, J.; et al. An effector–reporter system to study cellular signal transduction in strawberry fruit (Fragaria ananassa). Hortic. Res. 2021, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000, 24, 655–665. [Google Scholar] [CrossRef]
- Kishi-Kaboshi, M.; Okada, K.; Kurimoto, L.; Murakami, S.; Umezawa, T.; Shibuya, N.; Yamane, H.; Miyao, A.; Takatsuji, H.; Takahashi, A.; et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabo-lite synthesis. Plant J. 2010, 63, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spolaore, S.; Trainotti, L.; Casadoro, G. A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 2001, 52, 845–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, T.; Kalinowski, G.; Schwab, W. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: A rapid assay for gene function analysis. Plant J. Cell Mol. Biol. 2006, 48, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Concha, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.J.; Schwab, W.; Figueroa, C. Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W. Changes in the cell wall during fruit development and ripening in Fragaria vesca. Plant Physiol. Biochem. 2020, 154, 54–65. [Google Scholar] [CrossRef]
- Katzen, F. Gateway(®) recombinational cloning: A biological operating system. Expert Opin. Drug Discov. 2007, 2, 571–589. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3797–3800. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Dai, Z.; Li, J.; Ouyang, J.; Li, T.; Zeng, B.; Kang, L.; Jia, K.; Xi, Z.; Jia, W. A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening. Int. J. Mol. Sci. 2021, 22, 10495. https://doi.org/10.3390/ijms221910495
Wang W, Dai Z, Li J, Ouyang J, Li T, Zeng B, Kang L, Jia K, Xi Z, Jia W. A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening. International Journal of Molecular Sciences. 2021; 22(19):10495. https://doi.org/10.3390/ijms221910495
Chicago/Turabian StyleWang, Wei, Zhengrong Dai, Jie Li, Jinyao Ouyang, Tianyu Li, Baozhen Zeng, Li Kang, Kenan Jia, Zhiyuan Xi, and Wensuo Jia. 2021. "A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening" International Journal of Molecular Sciences 22, no. 19: 10495. https://doi.org/10.3390/ijms221910495
APA StyleWang, W., Dai, Z., Li, J., Ouyang, J., Li, T., Zeng, B., Kang, L., Jia, K., Xi, Z., & Jia, W. (2021). A Method for Assaying of Protein Kinase Activity In Vivo and Its Use in Studies of Signal Transduction in Strawberry Fruit Ripening. International Journal of Molecular Sciences, 22(19), 10495. https://doi.org/10.3390/ijms221910495






