Discovery and Validation of Lmj_04_BRCT Domain, a Novel Therapeutic Target: Identification of Candidate Drugs for Leishmaniasis
Abstract
1. Introduction
2. Results
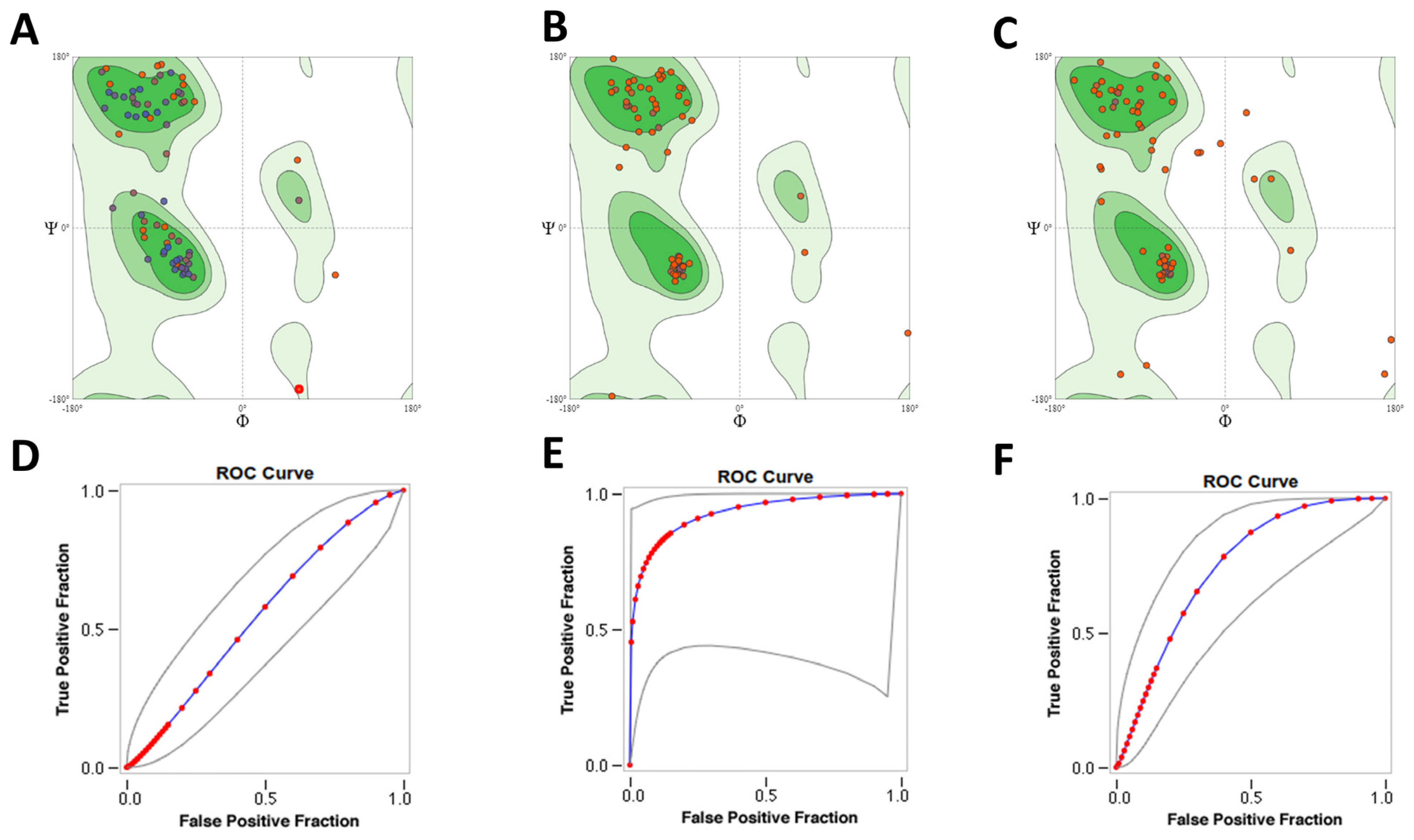
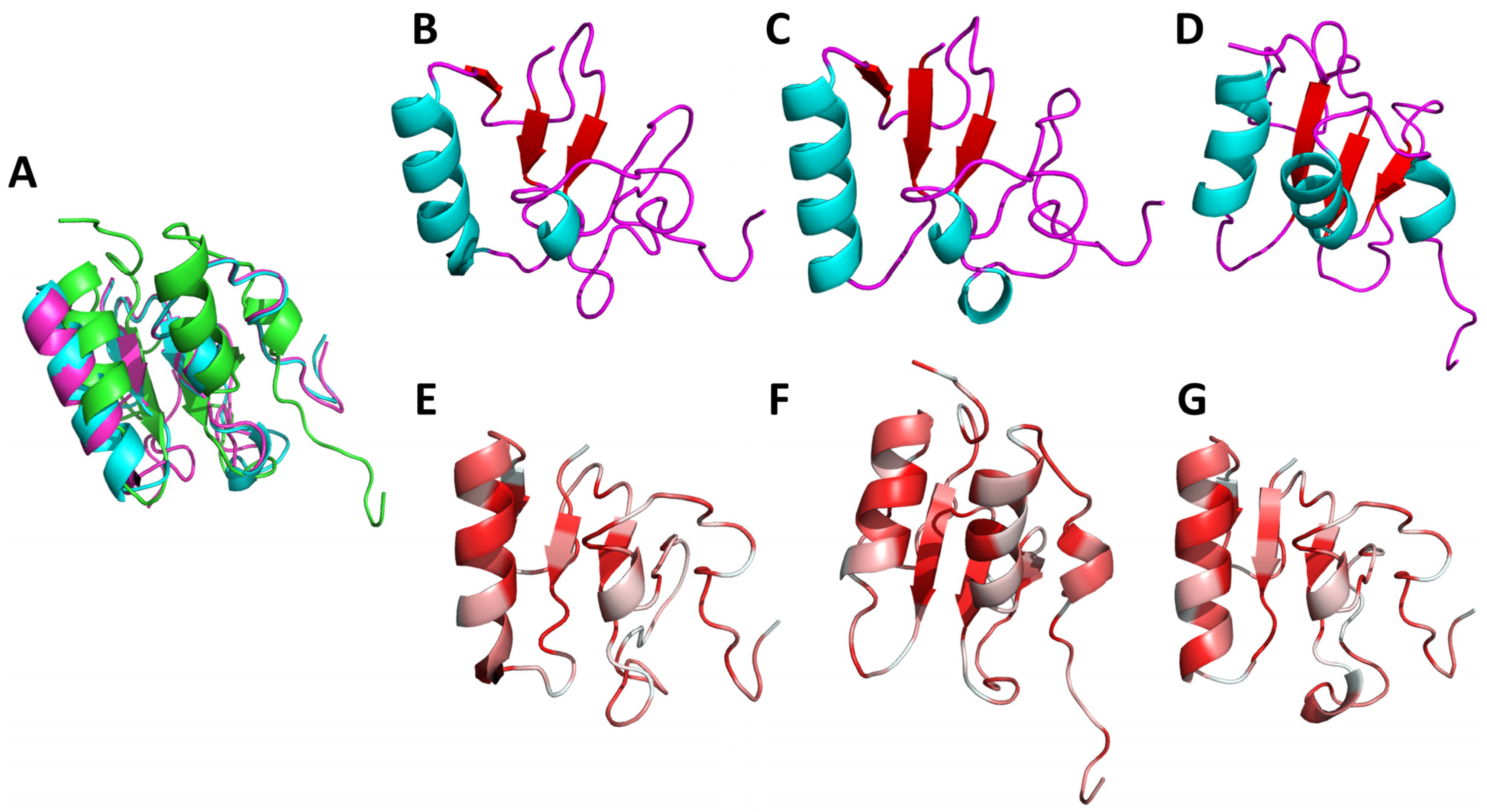
2.1. Model Generation and Evaluation
2.2. Sequence Analysis
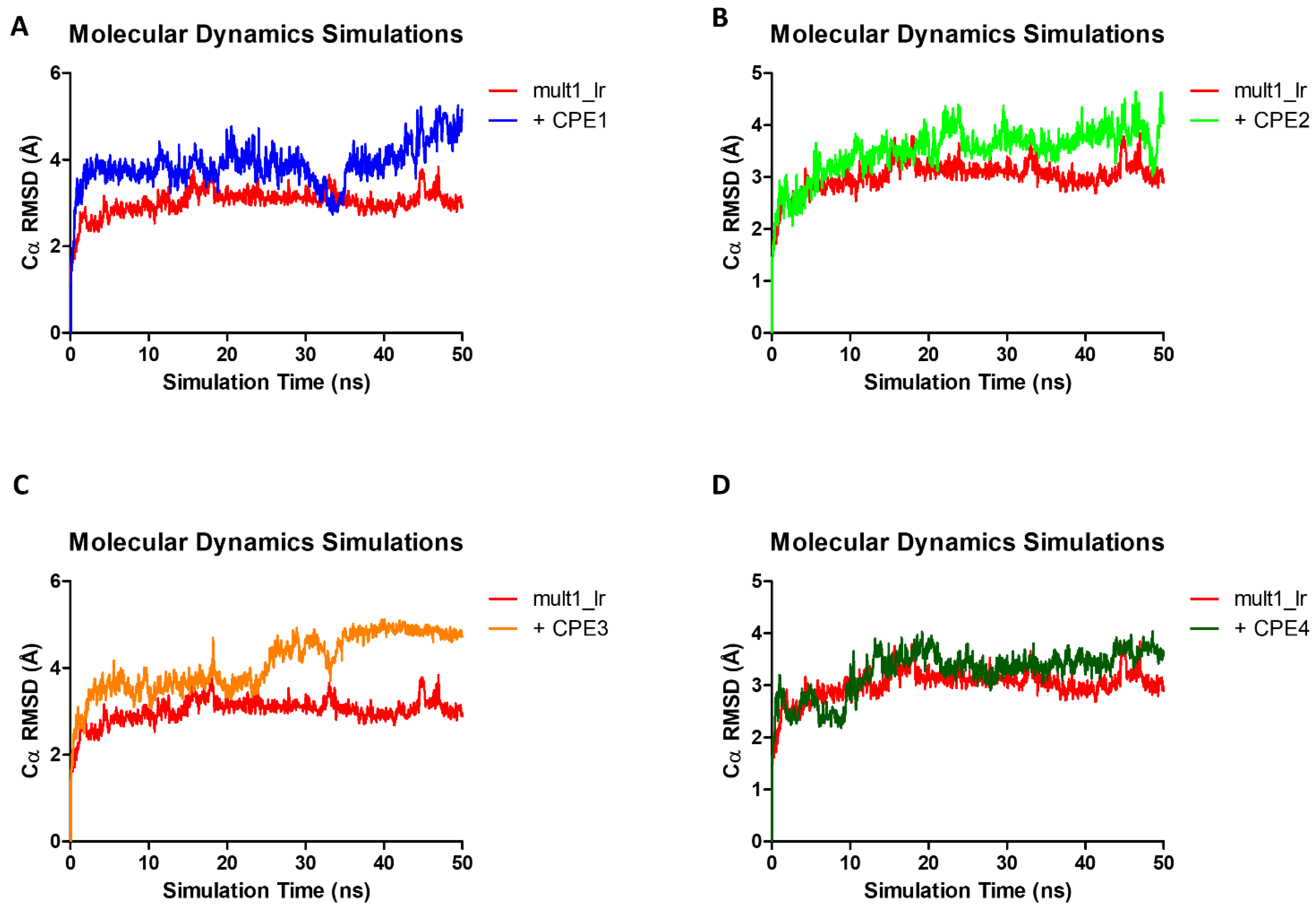
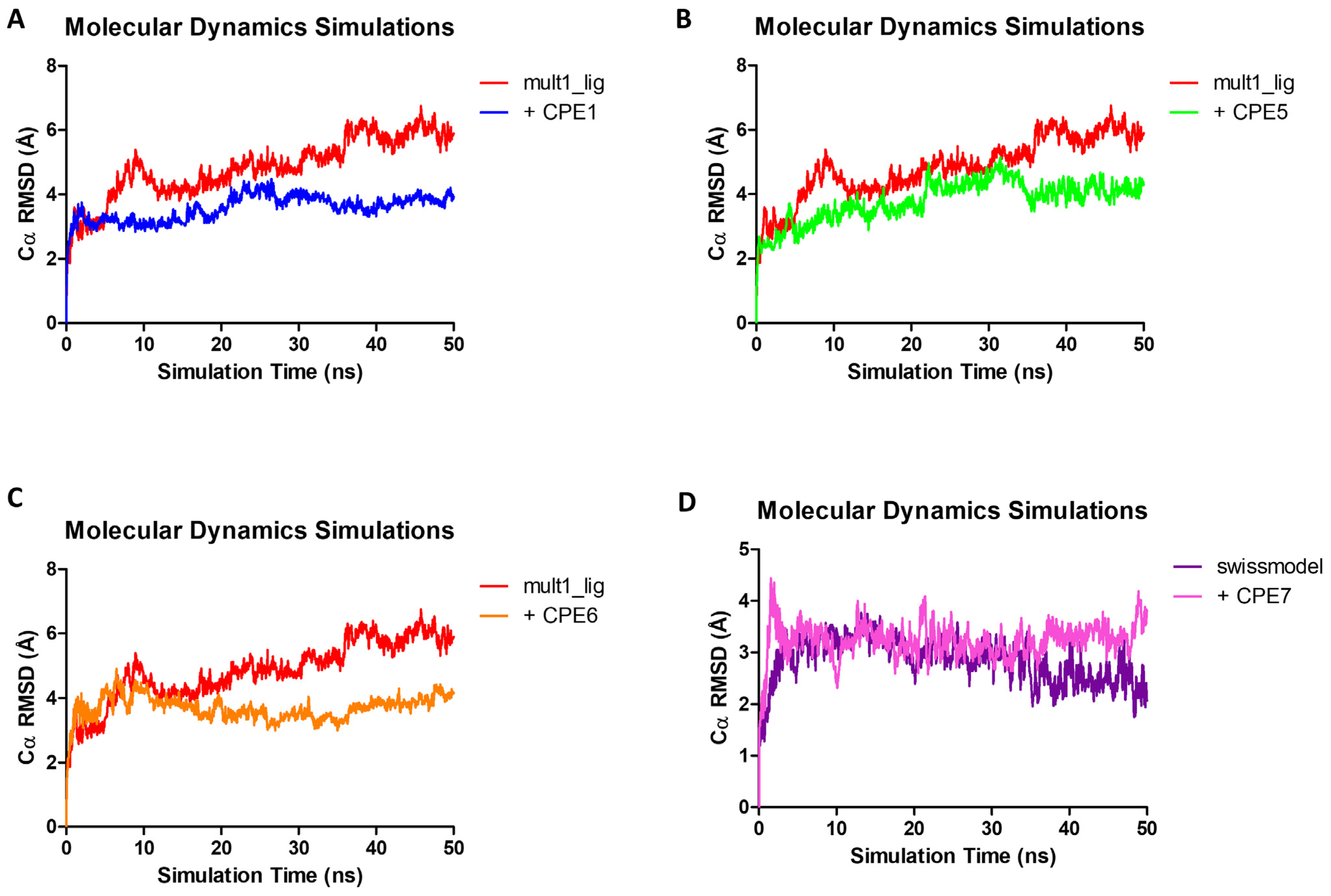
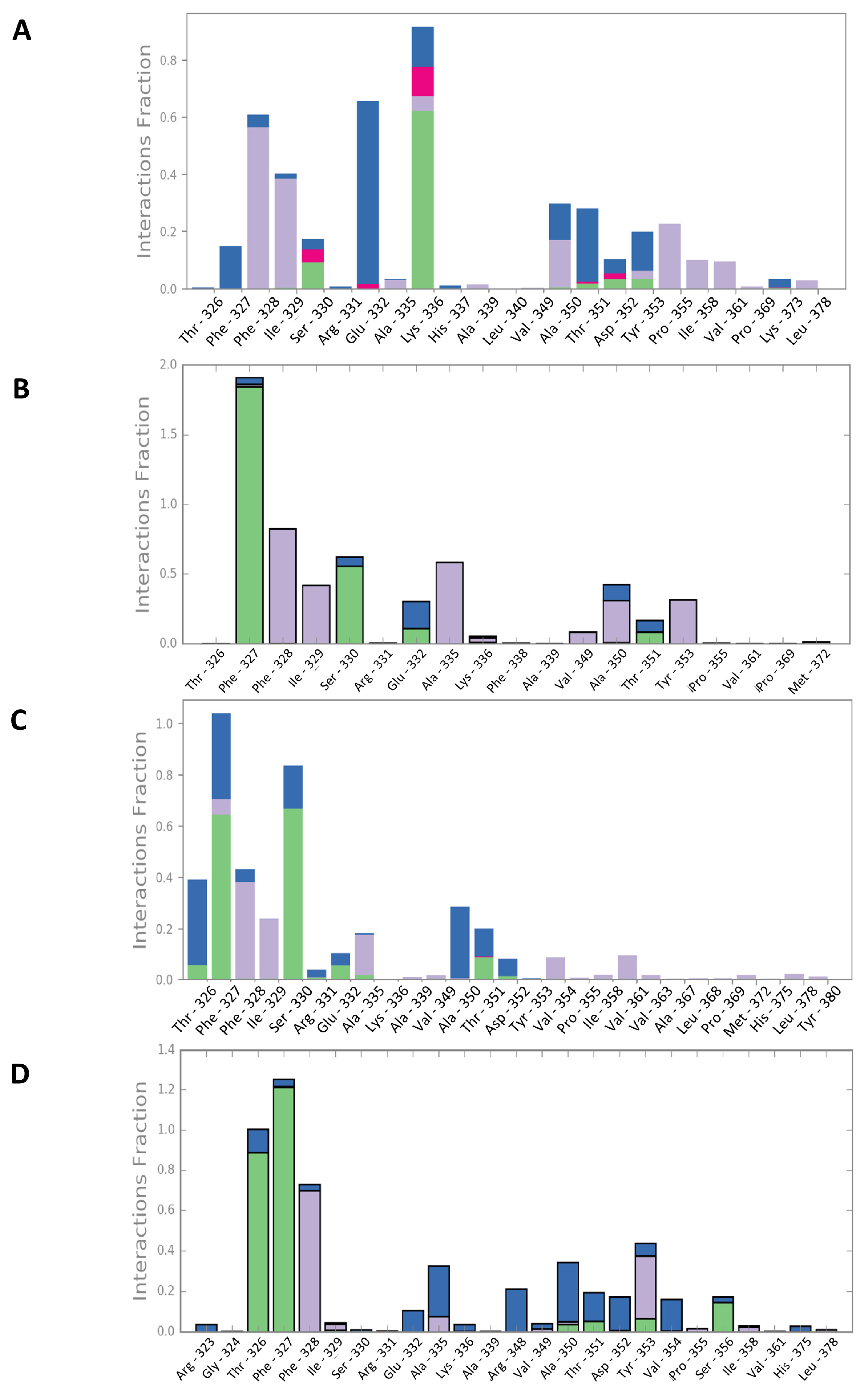
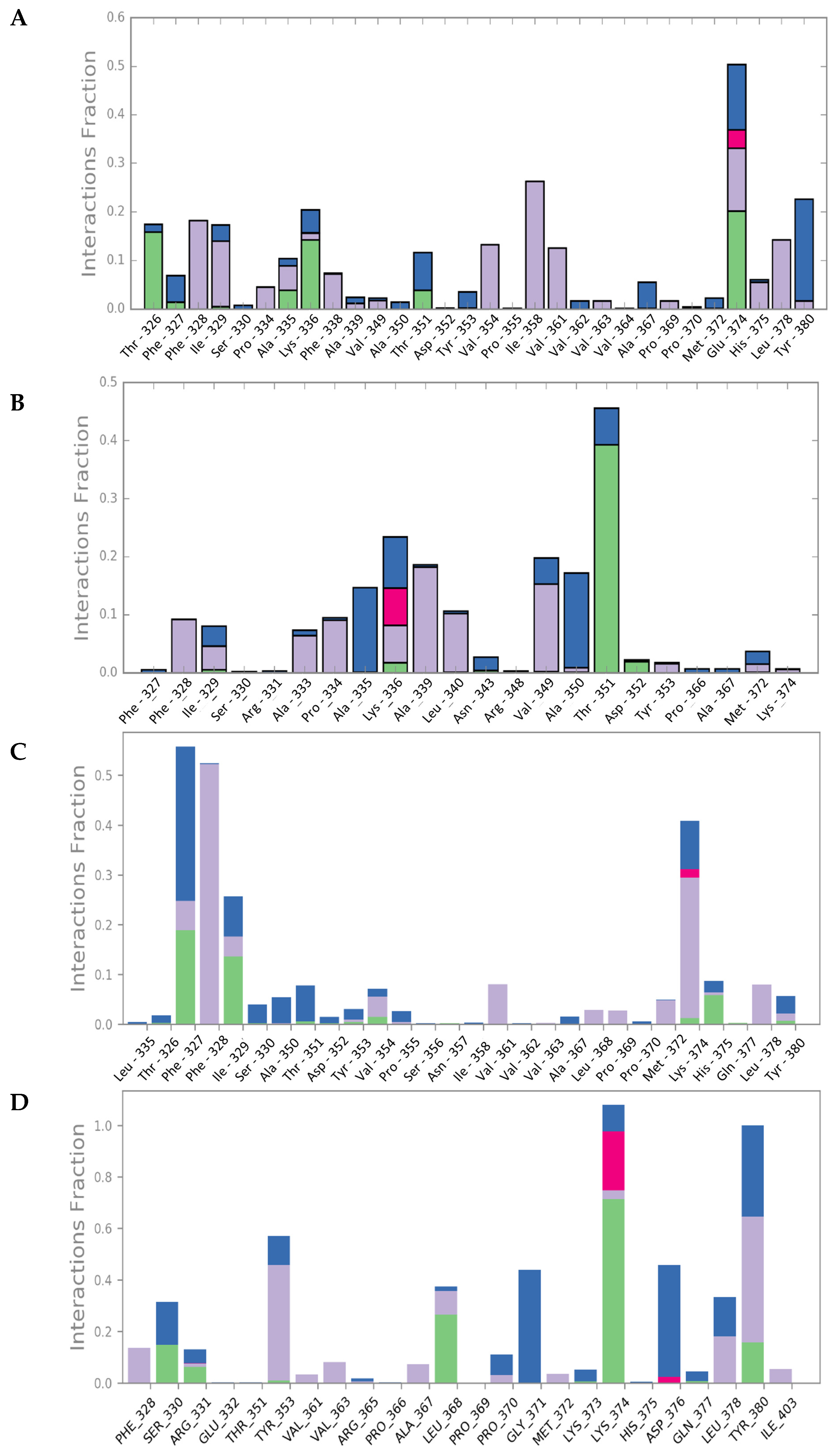
2.3. Molecular Dynamics Simulations
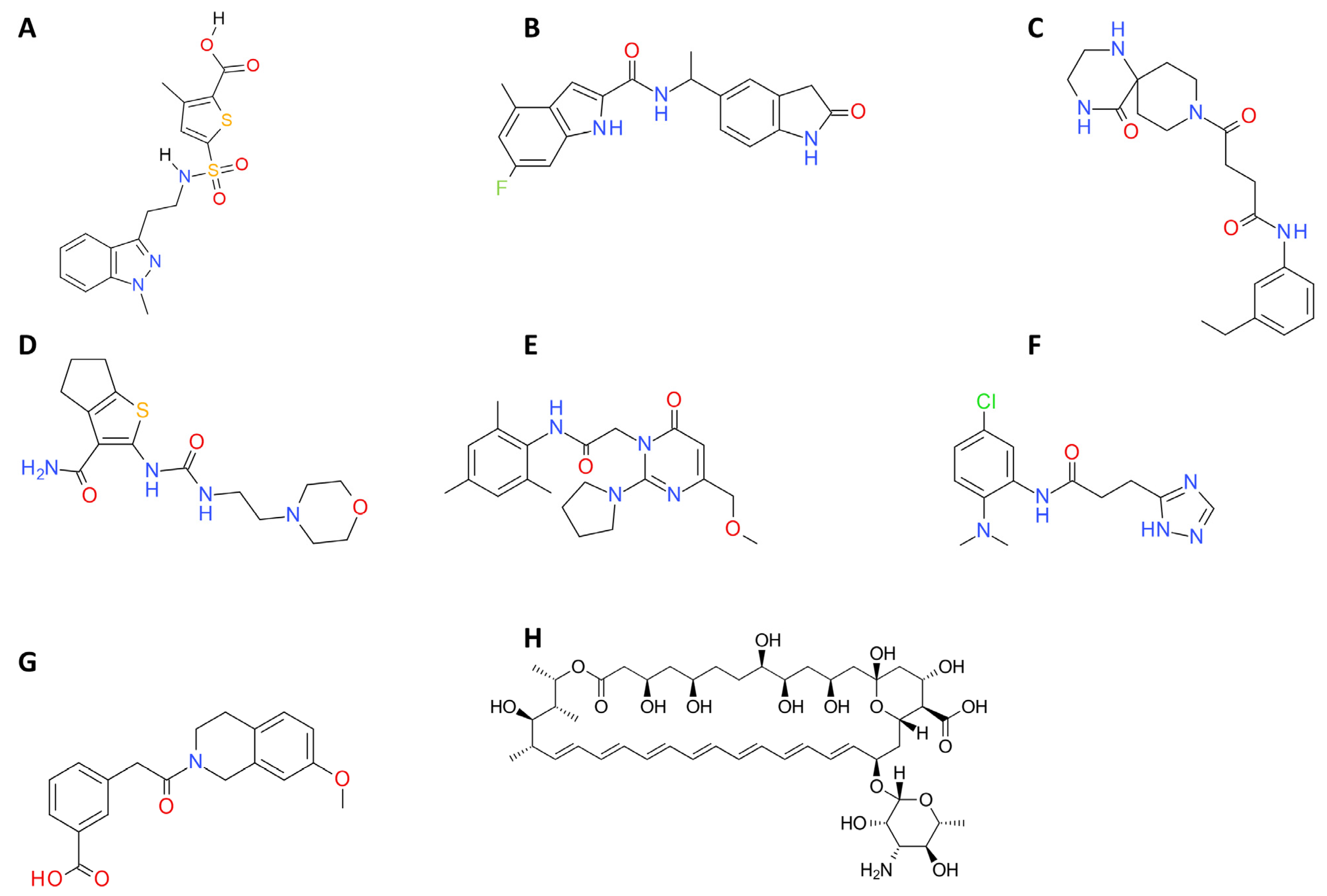
2.4. ADME and Bioavailability
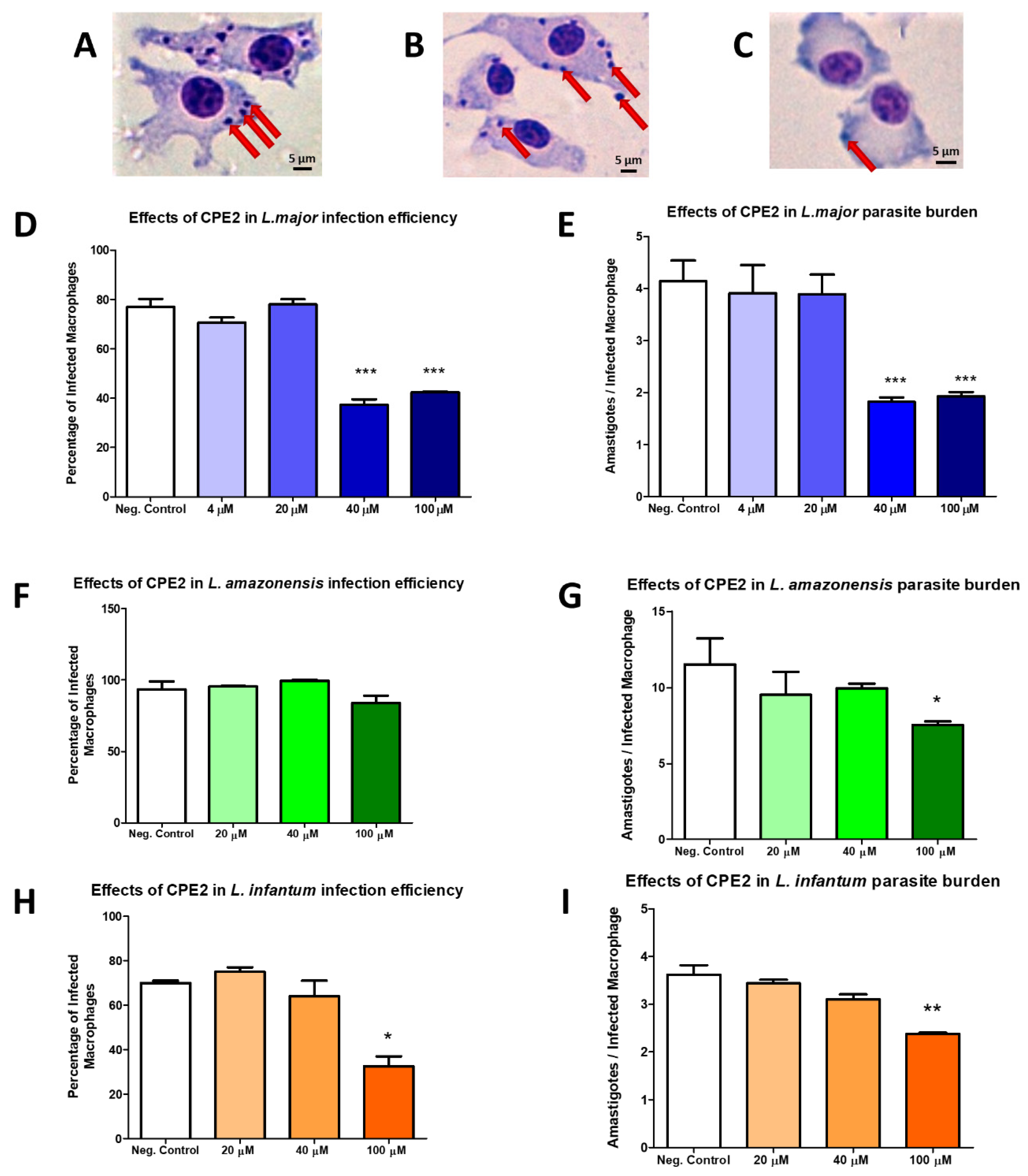
2.5. In Vitro Evaluation of Compound Activity in Leishmania major, L. amazonensis, and L. infantum
2.6. Characterization of Mult1_lr–CPE2 Interaction
2.7. CPE2 Exposure Significantly Altered the Gene Expression of Leishmania major ABC Transporter, α-Tubulin, CYCA, and Yip1
3. Discussion
4. Materials and Methods
4.1. Structure Prediction and Evaluation
4.2. Virtual Screening Analysis
4.3. Molecular Dynamics
4.4. In Vitro Biological Evaluation
4.4.1. Compounds
4.4.2. Cells and Culture Conditions
4.4.3. Leishmanicidal Activity
Activity against Promastigotes
Activity against Intracellular Amastigotes
4.4.4. In Vitro Toxicity Evaluation and Hemocompatibility
4.5. Statistical Analysis
4.6. Calculation of Selectivity Index
4.7. BRCT Sequence Identification, Orthologue Annotation, and Alignments
4.8. Gene Expression Quantification
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization WHO | Neglected Tropical Diseases. Available online: http://www.who.int/neglected_diseases/diseases/en/ (accessed on 8 October 2020).
- Klug, D.M.; Gelb, M.H.; Pollastri, M.P. Repurposing strategies for Tropical Disease Drug Discovery. Bioorg. Med. Chem. Lett. 2016, 26, 2569–2576. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.A.; Adlem, E.; Aert, R.; et al. The Genome of the Kinetoplastid Parasite, Leishmania major. Science 2005, 309, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Singh, B.K.; Patra, S.; Dubey, V.K. Rational Approaches for Drug Designing against Leishmaniasis. Appl. Biochem. Biotechnol. 2010, 160, 2208–2218. [Google Scholar] [CrossRef] [PubMed]
- Raj, S.; Sasidharan, S.; Balaji, S.N.; Saudagar, P. An Overview of Biochemically Characterized Drug Targets in Metabolic Pathways of Leishmania Parasite. Parasitol. Res. 2020, 119, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, F.; John, S.F.; Iniguez, E.; Montalvo, S.; Michael, K.; White, L.; Liang, D.; Olaleye, O.A.; Maldonado, R.A. In vitro and in vivo Characterization of Potent Antileishmanial Methionine Aminopeptidase 1 Inhibitors. Antimicrob. Agents Chemother. 2020, 64, e01422-19. [Google Scholar] [CrossRef]
- Das, S.; Banerjee, A.; Kamran, M.; Ejazi, S.A.; Asad, M.; Ali, N.; Chakrabarti, S. A Chemical Inhibitor of Heat Shock Protein 78 (HSP78) from Leishmania donovani Represents a Potential Antileishmanial Drug Candidate. J. Biol. Chem. 2020, 295, 9934–9947. [Google Scholar] [CrossRef]
- Algarabel-Olona, M. Discovery and Study of YinP as a New Therapeutic Target for Leishmaniasis; Universidad de Navarra: Navarra, Spain, 2019. [Google Scholar]
- Sheng, Z.Z.; Zhao, Y.Q.; Huang, J.F. Functional Evolution of BRCT Domains from Binding DNA to Protein. Evol. Bioinform. 2011, 7, 87–97. [Google Scholar] [CrossRef]
- Callebaut, I.; Mornon, J.-P. From BRCA1 to RAP1: A Widespread BRCT Module Closely Associated with DNA Repair. FEBS Lett. 1997, 400, 25–30. [Google Scholar] [CrossRef]
- Black, D.M.; Solomon, E. The Search for the Familial Breast/Ovarian Cancer Gene. Trends Genet. 1993, 9, 22–26. [Google Scholar] [CrossRef]
- Lokesh, G.L.; Muralidhara, B.K.; Negi, S.S.; Natarajan, A. Thermodynamics of Phosphopeptide Tethering to BRCT: The Structural Minima for Inhibitor Design. J. Am. Chem. Soc. 2007, 129, 10658–10659. [Google Scholar] [CrossRef]
- Yuan, Z.; Kumar, E.A.; Kizhake, S.; Natarajan, A. Structure-Activity Relationship Studies to Probe the Phosphoprotein Binding Site on the Carboxy Terminal Domains of the Breast Cancer Susceptibility Gene 1. J. Med. Chem. 2011, 54, 4264–4268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Periasamy, J.; Kurdekar, V.; Jasti, S.; Nijaguna, M.B.; Boggaram, S.; Hurakadli, M.A.; Raina, D.; Kurup, L.M.; Chintha, C.; Manjunath, K.; et al. Targeting Phosphopeptide Recognition by the Human BRCA1 Tandem BRCT Domain to Interrupt BRCA1-Dependent Signaling. Cell Chem. Biol. 2018, 25, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Na, Z.; Peng, B.; Ng, S.; Pan, S.; Lee, J.-S.; Shen, H.-M.; Yao, S.Q. A Small-Molecule Protein-Protein Interaction Inhibitor of PARP1 That Targets Its BRCT Domain. Angew. Chem. Int. Ed. 2015, 54, 2515–2519. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Ishidoh, T.; Iijima, H.; Kuriyama, I.; Shimazaki, N.; Koiwai, O.; Kuramochi, K.; Kobayashi, S.; Sugawara, F.; Sakaguchi, K.; et al. Structural Relationship of Curcumin Derivatives Binding to the BRCT Domain of Human DNA Polymerase Lambda. Genes Cells 2006, 11, 223–235. [Google Scholar] [CrossRef]
- Bork, P.; Hofmann, K.; Bucher, P.; Neuwald, A.F.; Altschul, S.F.; Koonin, E.V. A Superfamily of Conserved Domains in DNA Damage- Responsive Cell Cycle Checkpoint Proteins. FASEB J. 1997, 11, 68–76. [Google Scholar] [CrossRef]
- Zhang, X.; Moréra, S.; Bates, P.A.; Whitehead, P.C.; Coffer, A.I.; Hainbucher, K.; Nash, R.A.; Sternberg, M.J.E.; Lindahl, T.; Freemont, P.S. Structure of an XRCC1 BRCT Domain: A New Protein-Protein Interaction Module. EMBO J. 1998, 17, 6404–6411. [Google Scholar] [CrossRef]
- Krishnan, V.V.; Thornton, K.H.; Thelen, M.P.; Cosman, M. Solution structure and backbone dynamics of the human DNA ligase IIIalpha BRCT domain. Biochemistry 2001, 40, 13158–13166. [Google Scholar] [CrossRef]
- Patino, L.H.; Muskus, C.; Ramírez, J.D. Transcriptional responses of Leishmania (Leishmania) amazonensis in the presence of trivalent sodium stibogluconate. Parasites Vectors 2019, 12, 348. [Google Scholar] [CrossRef]
- Katritch, V.; Rueda, M.; Lam, P.C.H.; Yeager, M.; Abagyan, R. GPCR 3D Homology Models for Ligand Screening: Lessons Learned from Blind Predictions of Adenosine A2a Receptor Complex. Proteins Struct. Funct. Bioinform. 2010, 78, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Cavasotto, C.N.; Orry, A.J.W.; Murgolo, N.J.; Czarniecki, M.F.; Kocsi, S.A.; Hawes, B.E.; O’Neill, K.A.; Hine, H.; Burton, M.S.; Voigt, J.H.; et al. Discovery of Novel Chemotypes to a G-Protein-Coupled Receptor through Ligand-Steered Homology Modeling and Structure-Based Virtual Screening. J. Med. Chem. 2008, 51, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Evers, A.; Gohlke, H.; Klebe, G. Ligand-Supported Homology Modelling of Protein Binding-Sites Using Knowledge-Based Potentials. J. Mol. Biol. 2003, 334, 327–345. [Google Scholar] [CrossRef]
- Yu, X.; Chini, C.C.S.; He, M.; Mer, G.; Chen, J. The BRCT Domain Is a Phospho-Protein Binding Domain. Science 2003, 302, 639–642. [Google Scholar] [CrossRef]
- Pessetto, Z.Y.; Yan, Y.; Bessho, T.; Natarajan, A. Inhibition of BRCT(BRCA1)-Phosphoprotein Interaction Enhances the Cytotoxic Effect of Olaparib in Breast Cancer Cells: A Proof of Concept Study for Synthetic Lethal Therapeutic Option. Breast Cancer Res. Treat. 2012, 134, 511–517. [Google Scholar] [CrossRef] [PubMed][Green Version]
- White, E.R.; Sun, L.; Ma, Z.; Beckta, J.M.; Danzig, B.A.; Hacker, D.E.; Huie, M.; Williams, D.C.; Edwards, R.A.; Valerie, K.; et al. Peptide Library Approach to Uncover Phosphomimetic Inhibitors of the BRCA1 C-Terminal Domain. ACS Chem. Biol. 2015, 10, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Huang, Y.; Edwards, R.A.; Yang, S.; Blackford, A.N.; Niedzwiedz, W.; Glover, J.N.M. Structural insight into BLM Recognition by TopBP1. Structure 2017, 25, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Clapperton, J.A.; Manke, I.A.; Lowery, D.M.; Ho, T.; Haire, L.F.; Yaffe, M.B.; Smerdon, S.J. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004, 11, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.N.M.; Williams, R.S.; Lee, M.S. Interactions between BRCT Repeats and Phosphoproteins: Tangled up in Two. Trends Biochem. Sci. 2004, 29, 579–585. [Google Scholar] [CrossRef]
- Batista, F.A.H.; Ramos, S.L.; Tassone, G.; Leitão, A.; Montanari, C.A.; Botta, M.; Mori, M.; Borges, J.C. Discovery of Small Molecule Inhibitors of Leishmania braziliensis Hsp90 Chaperone. J. Enzym. Inhib. Med. Chem. 2020, 35, 639–649. [Google Scholar] [CrossRef]
- Istanbullu, H.; Bayraktar, G.; Akbaba, H.; Cavus, I.; Coban, G.; Debelec Butuner, B.; Kilimcioglu, A.A.; Ozbilgin, A.; Alptuzun, V.; Erciyas, E. Design, Synthesis, and in vitro Biological Evaluation of Novel Thiazolopyrimidine Derivatives as Antileishmanial Compounds. Arch. Pharm. 2020, 353, 1900325. [Google Scholar] [CrossRef]
- Stevanovic, S.; Sencanski, M.; Danel, M.; Menendez, C.; Belguedj, R.; Bouraiou, A.; Nikolic, K.; Cojean, S.; Loiseau, P.M.; Glisic, S.; et al. Synthesis, in silico, and in vitro Evaluation of Anti-Leishmanial Activity of Oxadiazoles and Indolizine Containing Compounds Flagged against Anti-Targets. Molecules 2019, 24, 1282. [Google Scholar] [CrossRef]
- Glisic, S.; Sencanski, M.; Perovic, V.; Stevanovic, S.; García-Sosa, A.T. Arginase Flavonoid Anti-Leishmanial in silico Inhibitors Flagged against Anti-Targets. Molecules 2016, 21, 589. [Google Scholar] [CrossRef]
- Meshram, R.J.; Bagul, K.T.; Aouti, S.U.; Shirsath, A.M.; Duggal, H.; Gacche, R.N. Modeling and Simulation Study to Identify Threonine Synthase as Possible Drug Target in Leishmania major. Mol. Divers. 2020, 25, 1679–1700. [Google Scholar] [CrossRef]
- Meshram, R.J.; Shirsath, A.; Aouti, S.; Bagul, K.; Gacche, R.N. Molecular Modeling and Simulation Study of Homoserine Kinase as an Effective Leishmanial Drug Target. J. Mol. Model. 2020, 26, 218. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Wang, S.M. Classification of VUS and Unclassified Variants in BRCA1 BRCT Repeats by Molecular Dynamics Simulation. Comput. Struct. Biotechnol. J. 2020, 18, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Rani, R.; Kumar, S.; Sethi, K.; Jain, S.; Batra, K.; Kumar, S.; Tripathi, B.N. Drug-Induced Reactive Oxygen Species–Mediated Inhibitory Effect on Growth of Trypanosoma evansi in Axenic Culture System. Parasitol. Res. 2020, 119, 3481–3489. [Google Scholar] [CrossRef]
- Benaim, G.; Paniz-Mondolfi, A.E.; Sordillo, E.M. Rationale for Use of Amiodarone and Its Derivatives for Treatment of Chagas’ Disease and Leishmaniasis. Curr. Pharm. Des. 2021, 27, 1825–1833. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ladias, J.A. Structural basis for the BRCA1 BRCT interaction with the proteins ATRIP and BAAT1. Biochemistry 2013, 52, 7618–7627. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.M.M.; Kang, M.; Chang, C.E.A. Mechanistic Insights into Phosphopeptide-Brct Domain Association: Preorganization, Flexibility, and Phosphate Recognition. J. Phys. Chem. B 2012, 116, 10247–10258. [Google Scholar] [CrossRef]
- Bharatham, N.; Finch, K.E.; Min, J.; Mayasundari, A.; Dyer, M.A.; Guy, R.K.; Bashford, D. Performance of a Docking/Molecular Dynamics Protocol for Virtual Screening of Nutlin-Class Inhibitors of Mdmx. J. Mol. Graph. Model. 2017, 74, 54–60. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Version 1.7.4; Schrödinger, LLC: New York, NY, USA, 2021.
- Fernández-Rubio, C.; Larrea, E.; Guerrero, J.P.; Herrero, E.S.; Gamboa, I.; Berrio, C.; Plano, D.; Amin, S.; Sharma, A.K.; Nguewa, P.A. Leishmanicidal Activity of Isoselenocyanate Derivatives. Antimicrob. Agents Chemother. 2019, 63, e00904-18. [Google Scholar] [CrossRef]
- Calvo, A.; Moreno, E.; Larrea, E.; Sanmartín, C.; Irache, J.M.; Espuelas, S. Berberine-Loaded Liposomes for the Treatment of Leishmania infantum-Infected BALB/c Mice. Pharmaceutics 2020, 12, 858. [Google Scholar] [CrossRef]
- Hnik, P.; Wasan, E.K.; Wasana, K.M. Safety, Tolerability, and Pharmacokinetics of a Novel Oral Amphotericin B Formulation (ICo-019) Following Single-Dose Administration to Healthy Human Subjects: An Alternative Approach to Parenteral Amphotericin B Administration. Antimicrob. Agents Chemother. 2020, 64, e01450-20. [Google Scholar] [CrossRef]
- Murray, H.W. Clinical and Experimental Advances in Treatment of Visceral Leishmaniasis. Antimicrob. Agents Chemother. 2001, 45, 2185–2197. [Google Scholar] [CrossRef]
- Pathak, M.K.; Yi, T. Sodium Stibogluconate Is a Potent Inhibitor of Protein Tyrosine Phosphatases and Augments Cytokine Responses in Hemopoietic Cell Lines. J. Immunol. 2001, 167, 3391–3397. [Google Scholar] [CrossRef]
- Muniz-Junqueira, M.I.; de Paula-Coelho, V.N. Meglumine Antimonate Directly Increases Phagocytosis, Superoxide Anion and TNF-α Production, but Only via TNF-α It Indirectly Increases Nitric Oxide Production by Phagocytes of Healthy Individuals, in vitro. Int. Immunopharmacol. 2008, 8, 1633–1638. [Google Scholar] [CrossRef]
- Hilgard, P.; Klenner, T.; Stekar, J.; Nössner, G.; Kutscher, B.; Engel, J. D-21266, a New Heterocyclic Alkylphospholipid with Antitumour Activity. Eur. J. Cancer Part A 1997, 33, 442–446. [Google Scholar] [CrossRef]
- Zeisig, R.; Rudolf, M.; Eue, I.; Arndt, D. Influence of Hexadecylphosphocholine on the Release of Tumor Necrosis Factor and Nitroxide from Peritoneal Macrophages in vitro. J. Cancer Res. Clin. Oncol. 1995, 121, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wadhone, P.; Maiti, M.; Agarwal, R.; Kamat, V.; Martin, S.; Saha, B. Miltefosine Promotes IFN-γ-Dominated Anti-Leishmanial Immune Response. J. Immunol. 2009, 182, 7146–7154. [Google Scholar] [CrossRef] [PubMed]
- Hochhuth, C.H.; Vehmeyer, K.; Eibl, H.; Unger, C. Hexadecylphosphocholine Induces Interferon-γ Secretion and Expression of GM-CSF MRNA in Human Mononuclear Cells. Cell. Immunol. 1992, 141, 161–168. [Google Scholar] [CrossRef]
- Eue, I. Hexadecylphosphocholine Selectively Upregulates Expression of Intracellular Adhesion Molecule-1 and Class I Major Histocompatibility Complex Antigen in Human Monocytes. J. Exp. Ther. Oncol. 2002, 2, 333–336. [Google Scholar] [CrossRef]
- Goodwin, L.G.; Page, J.E. A Study of the Excretion of Organic Antimonials Using a Polarographic Procedure. Biochem. J. 1943, 37, 198–209. [Google Scholar] [CrossRef]
- Burguera, J.L.; Burguera, M.; DE PENA, Y.; Lugo, A.; Anez, N. Selective Determination of Antimony (III) and Antimony (V) in Serum and Urine and of Total Antimony in Skin Biopsies of Patients with Cutaneous Leishmaniasis Treated with Meglumine Antimonate. Trace Elem. Med. 1993, 10, 66–70. [Google Scholar]
- Shaked-Mishant, P.; Ulrich, N.; Ephros, M.; Zilberstein, D. Novel Intracellular Sbv Reducing Activity Correlates with Antimony Susceptibility in Leishmania donovani. J. Biol. Chem. 2001, 276, 3971–3976. [Google Scholar] [CrossRef]
- Dos Santos Ferreira, C.; Silveira Martins, P.; Demicheli, C.; Brochu, C.; Ouellette, M.; Frézard, F. Thiol-Induced Reduction of Antimony(V) into Antimony(III): A Comparative Study with Trypanothione, Cysteinyl-Glycine, Cysteine and Glutathione. BioMetals 2003, 16, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Frézard, F.; Demicheli, C.; Ferreira, C.S.; Costa, M.A.P. Glutathione-Induced Conversion of Pentavalent Antimony to Trivalent Antimony in Meglumine Antimoniate. Antimicrob. Agents Chemother. 2001, 45, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Li, F.; Ding, K.; Sun, H. Reduction of Pentavalent Antimony by Trypanothione and Formation of a Binary and Ternary Complex of Antimony(III) and Trypanothione. J. Biol. Inorg. Chem. 2003, 8, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Alcântara, L.M.; Ferreira, T.C.S.; Gadelha, F.R.; Miguel, D.C. Challenges in Drug Discovery Targeting TriTryp Diseases with an Emphasis on Leishmaniasis. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 430–439. [Google Scholar] [CrossRef]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.C.; Barrett, M.P.; López-Vélez, R.; García-Hernández, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug Resistance and Treatment Failure in Leishmaniasis: A 21st Century Challenge. PLoS Negl. Trop. Dis. 2017, 11, e0006052. [Google Scholar] [CrossRef]
- MacLean, R.C.; Hall, A.R.; Perron, G.G.; Buckling, A. The Population Genetics of Antibiotic Resistance: Integrating Molecular Mechanisms and Treatment Contexts. Nat. Rev. Genet. 2010, 11, 405–414. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Andrejšali, A.A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Do, R.K.G.; Ali, A.S. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Vernon, R.; Thompson, J.; Tyka, M.; Sadreyev, R.; Pei, J.; Kim, D.; Kellogg, E.; Dimaio, F.; Lange, O.; et al. Structure Prediction for CASP8 with All-Atom Refinement Using Rosetta. Proteins Struct. Funct. Bioinform. 2009, 77, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Dimaio, F.; Wang, R.Y.R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Pieper, U.; Eswar, N.; Webb, B.M.; Eramian, D.; Kelly, L.; Barkan, D.T.; Carter, H.; Mankoo, P.; Karchin, R.; Marti-Renom, M.A.; et al. MODBASE, a Database of Annotated Comparative Protein Structure Models and Associated Resources. Nucleic Acids Res. 2009, 37, D347–D354. [Google Scholar] [CrossRef] [PubMed]
- ModWeb. Available online: https://modbase.compbio.ucsf.edu/modweb/ (accessed on 25 February 2021).
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-New Features and Functionality. Nucleic Acids Res. 2016, 45, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated Comparative Protein Structure Modeling with SWISS-MODEL and Swiss-PdbViewer: A Historical Perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Huang, N.; Shoichet, B.K.; Irwin, J.J. Benchmarking sets for molecular docking. J. Med. Chem. 2006, 49, 6789–6801. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Modification of the TRI Reagent(TM) Procedure for Isolation of RNA from Polysaccharide- and Proteoglycan-Rich Sources. Biotechniques 1995, 19, 942–945. [Google Scholar]
- García-Sosa, A.T. Designing Ligands for Leishmania, Plasmodium, and Aspergillus N-Myristoyl Transferase with Specificity and Anti-Target-Safe Virtual Libraries. Curr. Comput. Aided Drug Des. 2018, 14, 131–141. [Google Scholar] [CrossRef]
- Viira, B.; Selyutina, A.; García-Sosa, A.T.; Karonen, M.; Sinkkonen, J.; Merits, A.; Maran, U. Design, Discovery, Modelling, Synthesis, and Biological Evaluation of Novel and Small, Low Toxicity s-Triazine Derivatives as HIV 1 Nonnucleoside Reverse Transcriptase Inhibitors. Bioorg. Med. Chem. 2016, 24, 2519–2529. [Google Scholar] [CrossRef]
- García-Sosa, A.T.; Sild, S.; Maran, U. Docking and Virtual Screening Using Distributed Grid Technology. QSAR Comb. Sci. 2009, 28, 815–821. [Google Scholar] [CrossRef]
- Anjana, R.; Vaishnavi, M.K.; Sherlin, D.; Kumar, S.P.; Naveen, K.; Kanth, P.S.; Sekar, K. Aromatic-Aromatic Interactions in Structures of Proteins and Protein-DNA Complexes: A Study Based on Orientation and Distance. Bioinformation 2012, 8, 1220–1224. [Google Scholar] [CrossRef]
- D. E. Shaw Research. Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, USA, 2020. [Google Scholar]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC ’06), Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Grant, B.J.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R Package for the Comparative Analysis of Protein Structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, D.S.; Rabinovitch, M. Nitric Oxide Partially Controls Coxiella Burnetii Phase II Infection in Mouse Primary Macrophages. Infect. Immun. 2003, 71, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Rubio, C.; Campbell, D.; Vacas, A.; Ibañez, E.; Moreno, E.; Espuelas, S.; Calvo, A.; Palop, J.A.; Plano, D.; Sanmartin, C.; et al. Leishmanicidal Activities of Novel Methylseleno-Imidocarbamates. Antimicrob. Agents Chemother. 2015, 59, 5705–5713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sacks, D.L.; Perkins, P.V. Identification of an Infective Stage of Leishmania Promastigotes. Science 1984, 223, 1417–1419. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. Selective Lysis of Bacteria but Not Mammalian Cells by Diastereomers of Melittin: Structure−Function Study. Biochemistry 1997, 36, 1826–1835. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Abio Madeira, F.; Mi Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Toni, L.S.; Garcia, A.M.; Jeffrey, D.A.; Jiang, X.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Optimization of Phenol-Chloroform RNA Extraction. MethodsX 2018, 5, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Vacas, A.; Fernández-Rubio, C.; Larrea, E.; Peña-Guerrero, J.; Nguewa, P.A. LmjF.22.0810 from Leishmania major Modulates the TH2-Type Immune Response and Is Involved in Leishmaniasis Outcome. Biomedicines 2020, 8, 452. [Google Scholar] [CrossRef]



| Maximum RMSD (Å) | Average RMSD (Å) | Average Interacting Residues RMSD (Å) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Model | Model | |||||||
| Ligand | Mult1_lr | Mult1_lig | SwissModel_2 | Mult1_lr | Mult1_lig | SwissModel_2 | Mult1_lr | Mult1_lig | SwissModel_2 |
| (None) | 3.84 | 6.75 | 3.77 | 3.02 | 4.83 | 2.84 | |||
| CPE1 | 5.26 | 4.48 | 3.87 | 3.57 | 1.46 | 1.37 | |||
| CPE2 | 4.65 | 3.51 | 1.00 | ||||||
| CPE3 | 5.12 | 4.10 | 2.00 | ||||||
| CPE4 | 4.04 | 3.26 | 1.96 | ||||||
| CPE5 | 5.14 | 3.80 | 1.71 | ||||||
| CPE6 | 4.89 | 3.69 | 1.73 | ||||||
| CPE7 | 4.43 | 3.23 | 1.73 | ||||||
| Molecule | GI Absorption | BBB Permeant | P-gp Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|---|---|
| CPE1 | Low | No | No | No | No | Yes | No | No |
| CPE2 | High | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CPE3 | High | No | Yes | No | No | No | No | No |
| CPE4 | High | No | Yes | No | No | No | No | No |
| CPE5 | High | No | Yes | No | No | No | Yes | Yes |
| CPE6 | High | Yes | No | Yes | No | No | No | No |
| CPE7 | High | Yes | No | No | Yes | No | Yes | No |
| Amphotericin B | Low | No | Yes | No | No | No | No | No |
| Molecule | Bioavailability Score | PAINS #Alerts | Lead-likeness #Violations | Synthetic Accessibility |
|---|---|---|---|---|
| CPE1 | 0.56 | 0 | 1 | 3.27 |
| CPE2 | 0.55 | 0 | 1 | 3.09 |
| CPE3 | 0.55 | 0 | 1 | 3.51 |
| CPE4 | 0.55 | 0 | 0 | 3.47 |
| CPE5 | 0.55 | 0 | 1 | 3.54 |
| CPE6 | 0.55 | 0 | 0 | 2.41 |
| CPE7 | 0.85 | 0 | 0 | 2.36 |
| Amphotericin B | 0.17 | 0 | 1 | 10 |
| Molecule | Canonical SMILES | MW | #H-Bond Acceptors | #H-Bond Donors | Consensus logP | Lipinski #Violations | TPSA (Ų) |
|---|---|---|---|---|---|---|---|
| CPE1 | OC(=O)c1sc(cc1C)S(=O)(=O)NCCc1nn(c2c1cccc2)C | 379.45 | 6 | 2 | 2.31 | 0 | 137.91 |
| CPE2 | O=C1Nc2c(C1)cc(cc2)C(NC(=O)c1cc2c([nH]1)cc(cc2C)F)C | 351.37 | 3 | 3 | 3.11 | 0 | 73.99 |
| CPE3 | CCc1cccc(c1)NC(=O)CCC(=O)N1CCC2(CC1)NCCNC2=O | 372.46 | 4 | 3 | 1.18 | 0 | 90.54 |
| CPE4 | O=C(Nc1sc2c(c1C(=O)N)CCC2)NCCN1CCOCC1 | 338.43 | 4 | 3 | 1.19 | 0 | 124.93 |
| CPE5 | COCc1cc(=O)n(c(n1)N1CCCC1)CC(=O)Nc1c(C)cc(cc1C)C | 384.47 | 4 | 1 | 2.44 | 0 | 76.46 |
| CPE6 | O=C(Nc1cc(Cl)ccc1N(C)C)CCc1ncn[nH]1 | 293.75 | 3 | 2 | 1.69 | 0 | 73.91 |
| CPE7 | COc1ccc2c(c1)CN(CC2)C(=O)Cc1cccc(c1)C(=O)O | 325.36 | 4 | 1 | 2.45 | 0 | 66.84 |
| Amphotericin B | OC1CCC(O)C(O)CC(O)CC2(O)CC(O)C(C(O2)CC(C=CC=CC=CC=CC=CC=CC=CC(C(C(C(OC(=O)CC(C1)O)C)C)O)C)OC1OC(C)C(C(C1O)N)O)C(=O)O | 924.08 | 18 | 12 | −0.39 | 3 | 319.61 |
| Mφ | L. major | L. amazonensis | L. infantum | ||||
|---|---|---|---|---|---|---|---|
| Compound | EC50 (μM) | EC50 (μM) | SI | EC50 (μM) | SI | EC50 (μM) | SI |
| CPE-2 | >200 | 58.13 ± 1.72 | >3.4 | 62.44 ± 2.12 | >3.2 | 101.85 ± 0.35 | >2 |
| Amphotericin B | 4.1 ± 0.7 | 0.1 ± 0.03 | 41 | 0.2 ± 0.06 | 20.5 | 1.4 ± 0.3 | 2.9 |
| Code | Structure/Sequence | Canonical SMILES | MW (g/mol) | Name (Literature) | References |
|---|---|---|---|---|---|
| Gossypol | 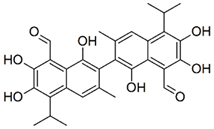 | CC1=CC2=C(C(=C(C(=C2C(C)C)O)O)C=O)C(=C1C3=C(C4=C(C=C3C)C(=C(C(=C4C=O)O)O)C(C)C)O)O | 518.6 | Gossypol | PubChem CID: 3503 [15] |
| Peptide 1 | pSPTF (pS = phosphoserine) | 530.47 | Peptide 1 | [80] | |
| Peptide M | DFDEYRFRKT (F = 4-fluoro-L-phenylalanine) | 1412.47 | Peptide 8.6 | [26] |
| Model Name | Used Method | Used Template (PDB Code) | QMEAN4 | Verify3d | ERRAT | EF1 | % of Aa in Ramachandran Favored Regions |
|---|---|---|---|---|---|---|---|
| SwissModel_2 | SwissModel web server, YASARA force field | 3PC6 | 0.71 | 84.95% | 89.28 | na | 92.31% |
| Mult1_lr | MODELLER, using multiple alignment of protein sequences, de novo modeling of loops | 2EP8 3U3Z 4BU0 2COU 4ID3 3OLC | 0.64 | 68.24% | 38.89 | 13.55 | 92.31% |
| Mult1_lig | MODELLER, multiple alignment of protein sequences, modeling with ligands in the binding site | Same as Mult1_lr +5U6K | 0.33 | 68.75% | 16.67 | 3.39 | 75.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña-Guerrero, J.; Fernández-Rubio, C.; Burguete-Mikeo, A.; El-Dirany, R.; García-Sosa, A.T.; Nguewa, P. Discovery and Validation of Lmj_04_BRCT Domain, a Novel Therapeutic Target: Identification of Candidate Drugs for Leishmaniasis. Int. J. Mol. Sci. 2021, 22, 10493. https://doi.org/10.3390/ijms221910493
Peña-Guerrero J, Fernández-Rubio C, Burguete-Mikeo A, El-Dirany R, García-Sosa AT, Nguewa P. Discovery and Validation of Lmj_04_BRCT Domain, a Novel Therapeutic Target: Identification of Candidate Drugs for Leishmaniasis. International Journal of Molecular Sciences. 2021; 22(19):10493. https://doi.org/10.3390/ijms221910493
Chicago/Turabian StylePeña-Guerrero, José, Celia Fernández-Rubio, Aroia Burguete-Mikeo, Rima El-Dirany, Alfonso T. García-Sosa, and Paul Nguewa. 2021. "Discovery and Validation of Lmj_04_BRCT Domain, a Novel Therapeutic Target: Identification of Candidate Drugs for Leishmaniasis" International Journal of Molecular Sciences 22, no. 19: 10493. https://doi.org/10.3390/ijms221910493
APA StylePeña-Guerrero, J., Fernández-Rubio, C., Burguete-Mikeo, A., El-Dirany, R., García-Sosa, A. T., & Nguewa, P. (2021). Discovery and Validation of Lmj_04_BRCT Domain, a Novel Therapeutic Target: Identification of Candidate Drugs for Leishmaniasis. International Journal of Molecular Sciences, 22(19), 10493. https://doi.org/10.3390/ijms221910493








