Role of Pannexin 1 ATP-Permeable Channels in the Regulation of Signaling Pathways during Skeletal Muscle Unloading
Abstract
:1. Introduction
2. Results
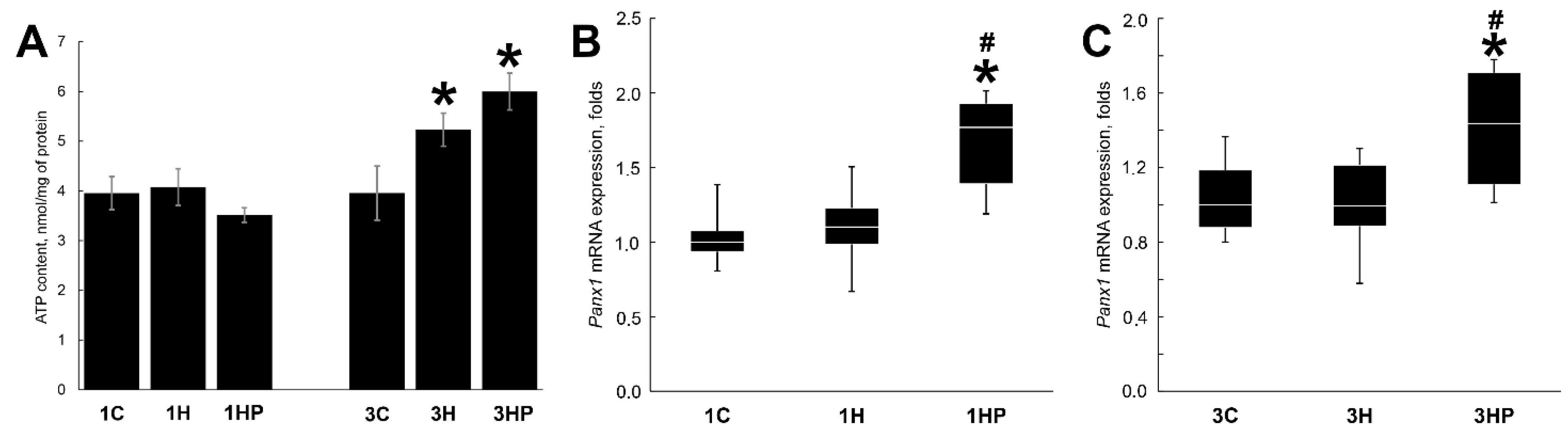
2.1. Effect of Probenecid on the Unloaded Soleus Muscle Weight, Panx1 Expression and ATP Content
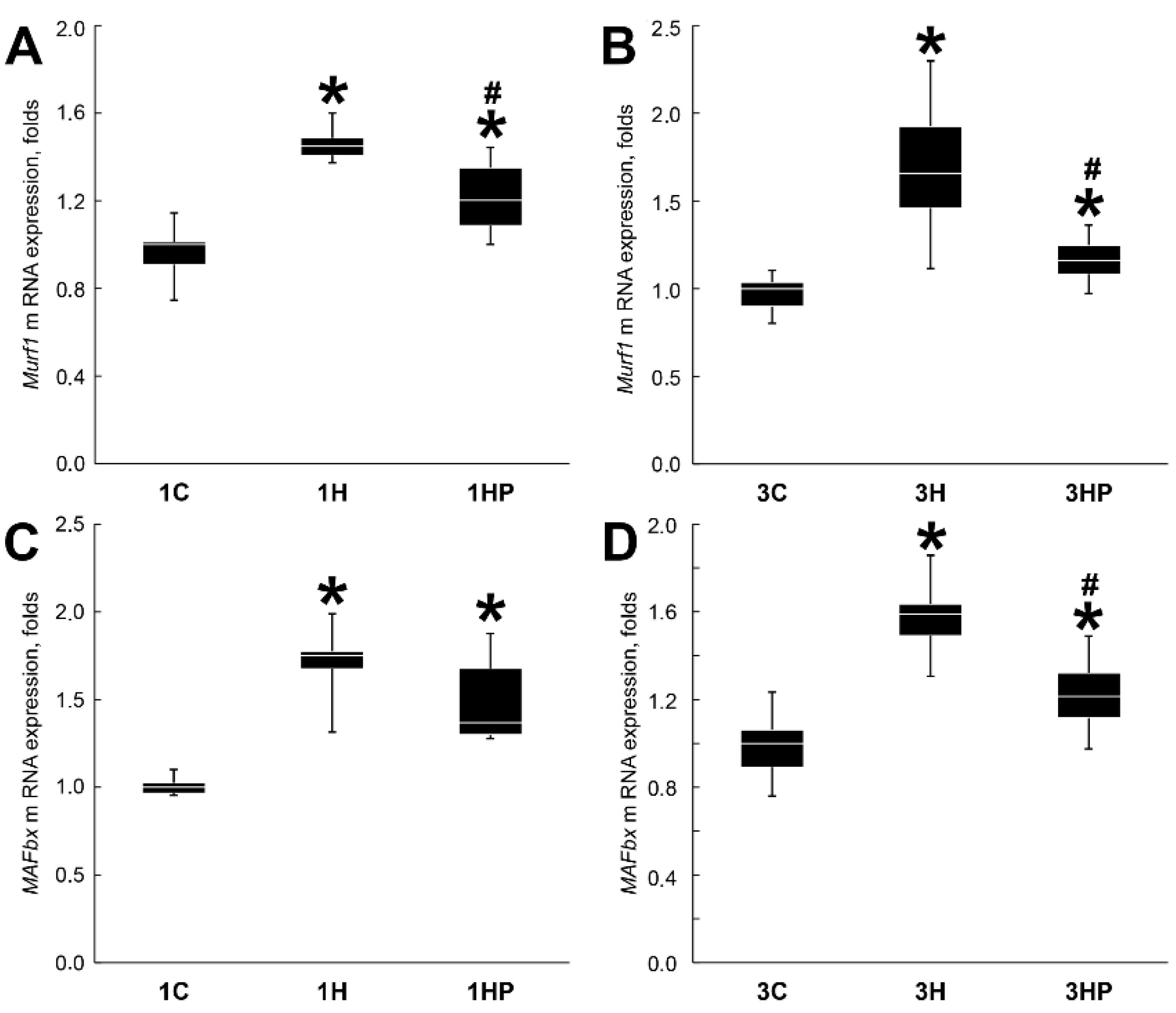
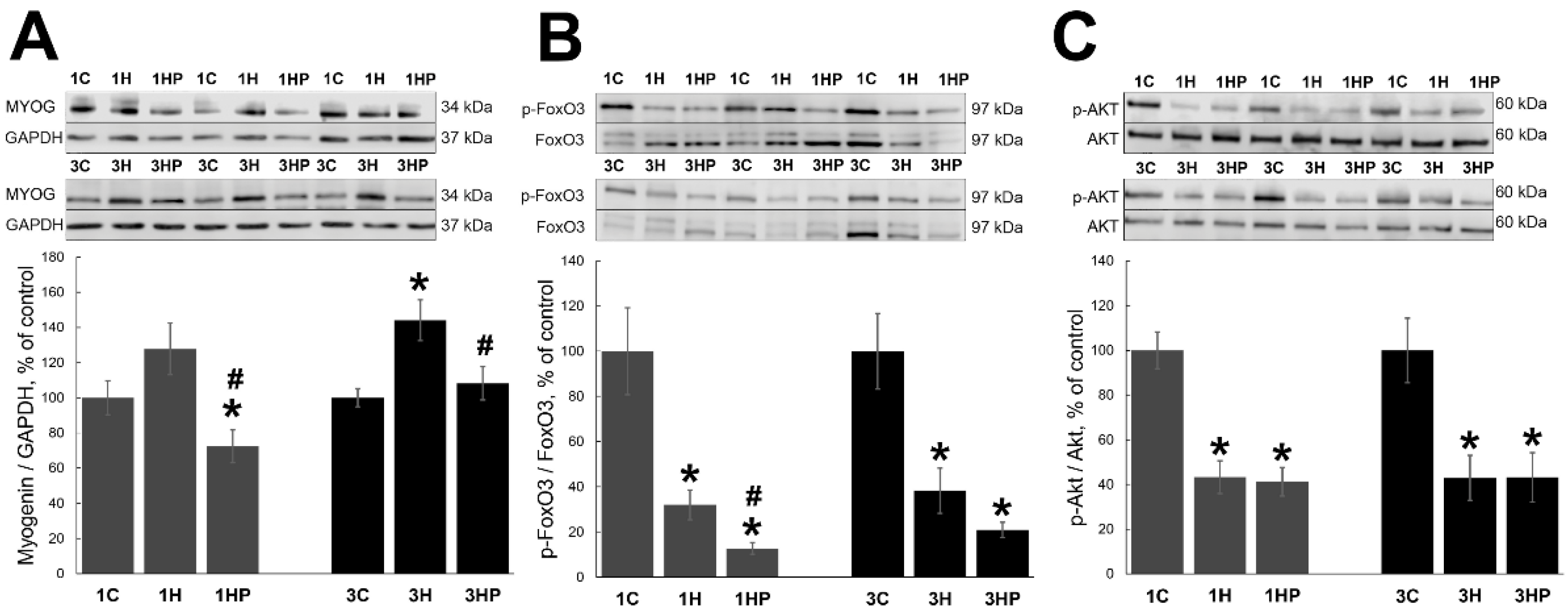
2.2. Effect of Probenecid on the Catabolic Processes in Unloaded Soleus Muscle
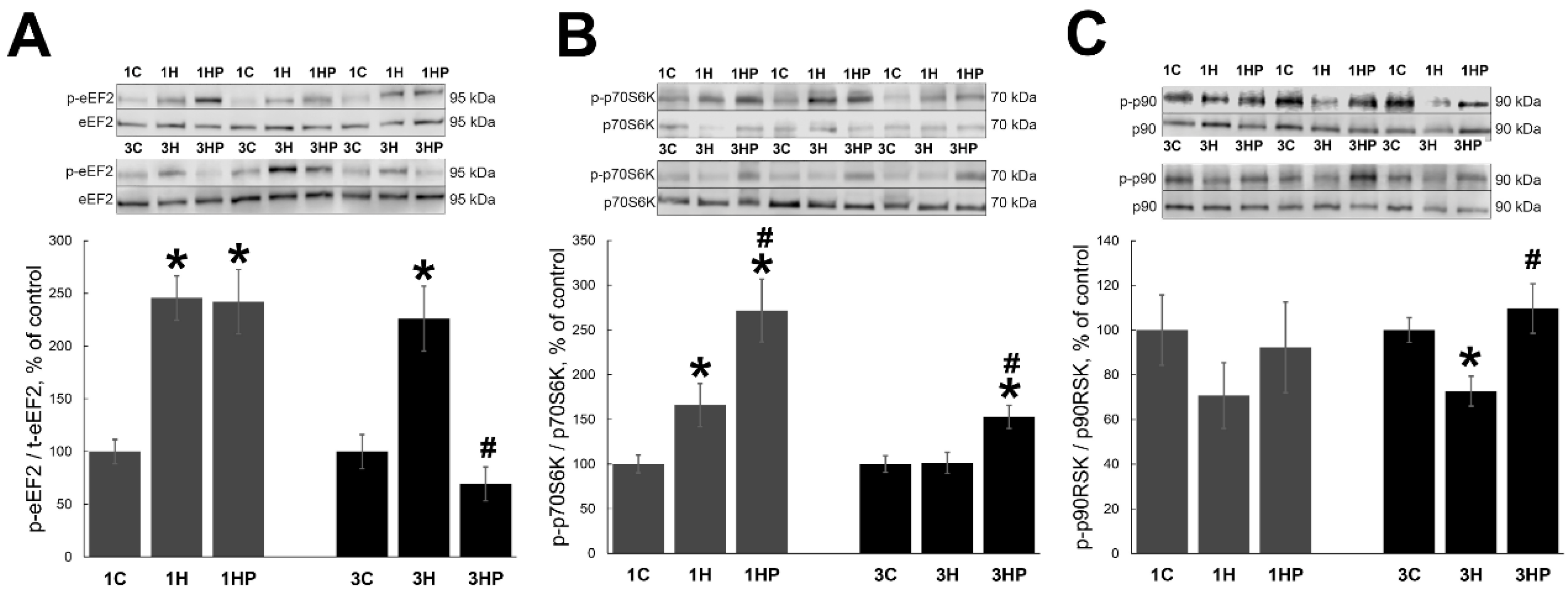
2.3. Effect of Probenecid on the Anabolic Processes in Unloaded Soleus Muscle
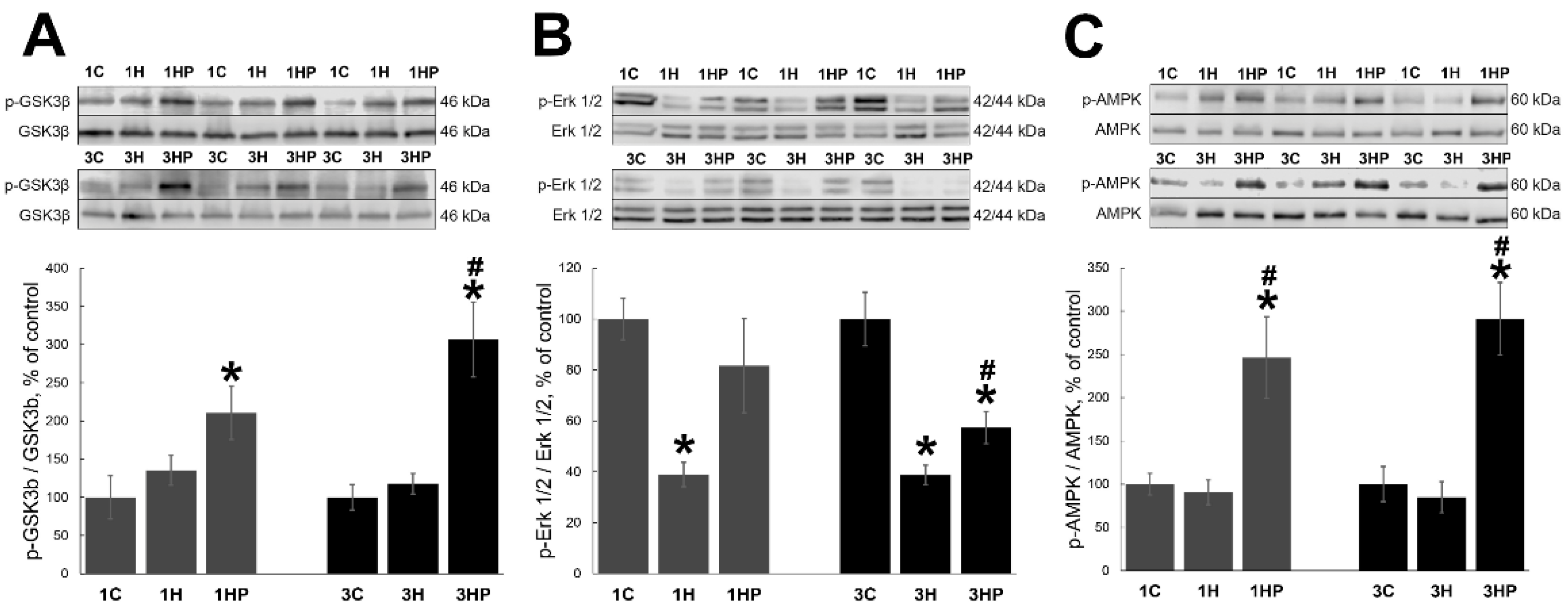
2.4. Effect of Probenecid on the Signaling Cascades Regulating Homeostasis of Unloaded Soleus Muscle
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Animal Procedures
4.3. Hindlimb Suspension
4.4. Evaluation of ATP Content
- B—amount of ATP in the sample well calculated based on the standard curve.
- V—sample volume added into the wells (50 µL).
- DDF (deproteinization dilution factor) was calculated using the following formula:
- DDF = (500 µL + volume KOH (µL))/initial sample volume PCA.
4.5. Protein Extraction and Western Blot Analysis
4.6. RNA Isolation and Reverse Transcription
4.7. Quantitative PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKT | protein kinase B alpha |
| AMPK | adenosine monophosphate-activated protein kinase |
| β-TrCP | beta-transducin repeat containing protein |
| CaMKII | Ca2+/calmodulin-dependent protein kinase II |
| CARM1 | coactivator-associated arginine methyltransferase 1 |
| cdc2 | cell division control 2 |
| DHPR | dihydropteridine receptor |
| GS | glycogen synthase |
| eEF22 | eukaryotic translation elongation factor |
| eEF2K | eukaryotic translation elongation factor 2 kinase |
| Erk1/2 | extracellular signal-regulated kinase 1/2 |
| FoxO3 | forkhead box O3 |
| GSK3β | glycogen synthase kinase-3β |
| HS | hindlimb suspension |
| IP3 | inositol trisphosphate |
| MAFbx | ubiquitin ligase muscle atrophy F-box |
| MYOG | myogenin |
| MuRF1 | muscle-specific RING finger protein 1 |
| mTORC1 | mammalian target of rapamycin complex 1 |
| PI3K | phosphatidylinositol 3-kinase |
| PCr | phosphocreatine |
| PI3K | phosphatidylinositol 3-kinase |
| PKA | protein kinase A |
| PKC | protein kinase C |
| PANX1 | pannexin channel 1 |
| PLC | phospholipase C |
| p70S6K | 70-kDa ribosomal protein S6 kinase |
| p90RSK90 | kDa ribosomal protein S6 kinase 1 |
| PRB | probenecid |
| P2Y | purinergic G protein-coupled receptor |
| Rho | Ras Homolog Family Member |
| RyR | ryanodine receptor |
| TSC2 | tuberous sclerosis complex 2 |
References
- Glass, D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nature 2003, 5, 87–90. [Google Scholar] [CrossRef]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef] [Green Version]
- Bodine, S.C.; Edward, F. Adolph distinguished lecture. Skeletal muscle atrophy: Multiple pathways leading to a common outcome. J. Appl. Physiol. 2020, 129, 272–282. [Google Scholar] [CrossRef]
- Mirzoev, T.M.; Shenkman, B.S. Regulation of protein synthesis in inactivated skeletal muscle: Signal inputs, protein kinase cascades, and ribosome biogenesis. Biochemistry 2018, 83, 1299–1317. [Google Scholar] [CrossRef]
- Shenkman, B.S. How postural muscle senses disuse? Early signs and signals. Int. J. Mol. Sci. 2020, 21, 5037. [Google Scholar] [CrossRef]
- Ohira, Y.; Yasui, W.; Kariya, F.; Wakatsuki, T.; Nakamura, K.; Asakura, T.; Edgerton, V. Metabolic adaptation of skeletal muscles to gravitational unloading. Acta Astronaut. 1994, 33, 113–117. [Google Scholar] [CrossRef]
- Ingalls, C.P.; Warren, G.L.; Armstrong, R.B. Intracellular Ca2+ transients in mouse soleus muscle after hindlimb unloading and reloading. J. Appl. Physiol. 1999, 87, 386–390. [Google Scholar] [CrossRef] [Green Version]
- Ingalls, C.P.; Wenke, J.C.; Armstrong, R.B. Time course changes in [Ca2+]i, force, and protein content in hindlimb-suspended mouse soleus muscles. Aviat. Space Environ. Med. 2001, 72, 471–476. [Google Scholar]
- Shenkman, B.S.; Nemirovskaya, T.L. Calcium-dependent signaling mechanisms and soleus fiber remodeling under gravitational unloading. J. Muscle Res. Cell Motil. 2008, 29, 221–230. [Google Scholar] [CrossRef]
- Gupta, R.C.; Misulis, K.E.; Dettbarn, W.D. Activity dependent characteristics of fast and slow muscle: Biochemical and histochemical considerations. Neurochem. Res. 1989, 14, 647–655. [Google Scholar] [CrossRef]
- Nemirovskaya, T.L.; Shenkman, B.S.; Matsievskii, D.D.; Eyu, B.; Maevskii, E.; Grishina, E. Decreased concentration of high-energy phosphates prevents a decrease in redox potential of skeletal muscles under gravitational unloading. Dokl. Biol. Sci. 2000, 370, 10–13. [Google Scholar] [PubMed]
- Vilchinskaya, N.A.; Mochalova, E.P.; Paramonova, I.I.; Belova, S.P.; Mirzoev, T.M.; Shenkman, B.S. The effect of β-GPA on the markers of anabolic and catabolic signaling pathways in rat soleus muscle at the initial stage of Hindlimb unloading. Biochem. Suppl. Ser. A Membr. Cell Biol. 2020, 14, 1–6. [Google Scholar] [CrossRef]
- Lazarowski, E.R.; Boucher, R.C.; Harden, T.K. Mechanisms of release of nucleotides and integration of their action as P2X- and P2Y-receptor activating molecules. Mol. Pharmacol. 2003, 64, 785–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casas, M.; Buvinic, S.; Jaimovich, E. ATP signaling in skeletal muscle: From fiber plasticity to regulation of metabolism. Exerc. Sport Sci. Rev. 2014, 42, 110–116. [Google Scholar] [CrossRef]
- Panchina, Y.; Kelmanson, I.; Matz, M.; Lukyanov, K.; Usman, N.; Lukyanov, S. A ubiquitous family of putative gap junction molecules. Curr. Biol. 2000, 10, R473–R474. [Google Scholar] [CrossRef] [Green Version]
- Penuela, S.; Gehi, R.; Laird, D.W. The biochemistry and function of pannexin channels. Biochim. Biophys. Acta Biomembr. 2013, 1828, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Shestopalov, V.I.; Panchin, Y. Pannexins and gap junction protein diversity. Experientia 2007, 65, 376–394. [Google Scholar] [CrossRef]
- MacVicar, B.; Thompson, R. Non-junction functions of pannexin-1 channels. Trends Neurosci. 2010, 33, 93–102. [Google Scholar] [CrossRef]
- Dahl, G. ATP release through pannexon channels. Philos. Trans. R. Soc. Lond. Biol. Sci. 2015, 370, 1672. [Google Scholar] [CrossRef]
- May, C.; Weigl, L.; Karel, A.; Hohenegger, M. Extracellular ATP activates ERK1/ERK2 via a metabotropic P2Y1 receptor in a Ca2+ independent manner in differentiated human skeletal muscle cells. Biochem. Pharmacol. 2006, 71, 1497–1509. [Google Scholar] [CrossRef]
- Jaimovich, E.; Carrasco, M.A. IP3 dependent Ca2+ signals in muscle cells are involved in regulation of gene expression. Biol. Res. 2002, 35, 195–202. [Google Scholar] [CrossRef]
- Jaimovich, E.; Reyes, R.; Liberona, J.L.; Powell, J.A. IP3 receptors, IP3 transients, and nucleus-associated Ca2+ signals in cultured skeletal muscle. Am. J. Physiol. Physiol. 2000, 278, C998–C1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valladares, D.; Utreras-Mendoza, Y.; Campos, C.; Morales, C.; Vegas, A.D.; Contreras-Ferrat, A.; Westermeier, F.; Jaimovich, E.; Marchi, S.; Pinton, P.; et al. IP3 receptor blockade restores autophagy and mitochondrial function in skeletal muscle fibers of dystrophic mice. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3685–3695. [Google Scholar] [CrossRef] [PubMed]
- Moresi, V.; Williams, A.H.; Meadows, E.; Flynn, J.M.; Potthoff, M.; McAnally, J.; Shelton, J.M.; Backs, J.; Klein, W.H.; Richardson, J.A.; et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 2010, 143, 35–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bricceno, K.V.; Sampognaro, P.J.; Van Meerbeke, J.P.; Sumner, C.J.; Fischbeck, K.H.; Burnett, B.G. Histone deacetylase inhibition suppresses myogenin-dependent atrogene activation in spinal muscular atrophy mice. Hum. Mol. Genet. 2012, 21, 4448–4459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furlow, J.D.; Watson, M.L.; Waddell, D.; Neff, E.S.; Baehr, L.M.; Ross, A.P.; Bodine, S. Altered gene expression patterns in muscle ring finger 1 null mice during denervation- and dexamethasone-induced muscle atrophy. Physiol. Genom. 2013, 45, 1168–1185. [Google Scholar] [CrossRef] [Green Version]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo Transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Clavel, S.; Siffroi-Fernandez, S.; Coldefy, A.S.; Boulukos, K.; Pisani, D.; Dérijard, B. Regulation of the intracellular localization of Foxo3a by stress-activated protein kinase signaling pathways in skeletal muscle cells. Mol. Cell. Biol. 2010, 30, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.K.; Webb, A.E. Regulation of FOXO factors in mammalian cells. Curr. Top. Dev. Biol. 2017, 127, 165–192. [Google Scholar] [CrossRef]
- Kodani, N.; Nakae, J. Tissue-specific metabolic regulation of FOXO-binding protein: FOXO does not act alone. Cells 2020, 9, 702. [Google Scholar] [CrossRef] [Green Version]
- Knight, J.R.P.; Garland, G.; Poyry, T.; Mead, E.; Vlahov, N.; Sfakianos, A.; Grosso, S.; De-Lima-Hedayioglu, F.; Mallucci, G.R.; von der Haar, T.; et al. Control of translation elongation in health and disease. Dis. Model Mech. 2020, 13, dmm043208. [Google Scholar] [CrossRef] [Green Version]
- Arias-Calderón, M.; Almarza, G.; Díaz-Vegas, A.; Contreras-Ferrat, A.; Valladares, D.; Casas, M.; Toledo, H.; Jaimovich, E.; Buvinic, S. Characterization of a multiprotein complex involved in excitation-transcription coupling of skeletal muscle. Skelet. Muscle 2016, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Silverman, W.; Locovei, S.; Dahl, G. Probenecid, a gout remedy, inhibits pannexin 1 channels. Am. J. Physiol.-Cell Physiol. 2008, 295, C761–C767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.-M.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Ralevic, V.; Knight, G.; Burnstock, G. Effects of hibernation and arousal from hibernation on mesenteric arterial responses of the golden hamster. J. Pharmacol. Exp. Ther. 1998, 287. [Google Scholar]
- Yoshihara, T.; Machida, S.; Kurosaka, Y.; Kakigi, R.; Sugiura, T.; Naito, H. Immobilization induces nuclear accumulation of HDAC4 in rat skeletal muscle. J. Physiol. Sci. 2016, 66, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Mochalova, E.P.; Belova, S.P.; Kostrominova, T.Y.; Shenkman, B.S.; Nemirovskaya, T.L. Differences in the role of HDACs 4 and 5 in the modulation of processes regulating MAFbx and MuRF1 expression during muscle unloading. Int. J. Mol. Sci. 2020, 21, 4815. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Li, Y.-F. Distinct signal transductions in fast- and slow- twitch muscles upon denervation. Physiol. Rep. 2018, 6, e13606. [Google Scholar] [CrossRef] [PubMed]
- Shenkman, B.S.; Belova, S.P.; Lomonosova, Y.N.; Kostrominova, T.Y.; Nemirovskaya, T.L. Calpain-dependent regulation of the skeletal muscle atrophy following unloading. Arch. Biochem. Biophys. 2015, 584, 36–41. [Google Scholar] [CrossRef]
- Lomonosova, Y.N.; Belova, S.P.; Mirzoev, T.M.; Kozlovskaya, I.B.; Shenkman, B.S. Eukaryotic elongation factor 2 kinase activation in M. soleus under 14-day hindlimb unloading of rats. Dokl. Biochem. Biophys. 2017, 474, 165–167. [Google Scholar] [CrossRef]
- Tyganov, S.; Mochalova, E.P.; Belova, S.P.; Sharlo, K.A.; Rozhkov, S.V.; Vilchinskaya, N.A.; Paramonova, I.I.; Mirzoev, T.; Shenkman, B.S. Effects of plantar mechanical stimulation on anabolic and catabolic signaling in rat postural muscle under short-term simulated gravitational unloading. Front. Physiol. 2019, 10, 1252. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Williams, M.R.; Terada, N.; Alessi, D.; Proud, C.G. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 2001, 20, 4370–4379. [Google Scholar] [CrossRef]
- Ahmad, F.; Woodgett, J.R. Emerging roles of GSK-3α in pathophysiology: Emphasis on cardio-metabolic disorders. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118616. [Google Scholar] [CrossRef]
- Robertson, H.; Hayes, J.D.; Sutherland, C. A partnership with the proteasome; the destructive nature of GSK3. Biochem. Pharmacol. 2017, 147, 77–92. [Google Scholar] [CrossRef]
- Aweida, D.; Rudesky, I.; Volodin, A.; Shimko, E.; Cohen, S. GSK3-beta promotes calpain-1-mediated desmin filament depolymerization and myofibril loss in atrophy. J. Cell Biol. 2018, 217, 3698–3714. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S. Role of calpains in promoting desmin filaments depolymerization and muscle atrophy. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118788. [Google Scholar] [CrossRef]
- Jefferson, L.S.; Fabian, J.R.; Kimball, S.R. Glycogen synthase kinase-3 is the predominant insulin-regulated eukaryotic initiation factor 2B kinase in skeletal muscle. Int. J. Biochem. Cell Biol. 1999, 31, 191–200. [Google Scholar] [CrossRef]
- Léger, B.; Cartoni, R.; Praz, M.; Lamon, S.; Dériaz, O.; Crettenand, A.; Gobelet, C.; Rohmer, P.; Konzelmann, M.; Luthi, F.; et al. Akt signalling through GSK-3β, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J. Physiol. 2006, 576, 923–933. [Google Scholar] [CrossRef]
- Fang, X.; Yu, S.X.; Lu, Y.; Bast, R.; Woodgett, J.; Mills, G.B. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. USA 2000, 97, 11960–11965. [Google Scholar] [CrossRef] [Green Version]
- Song, B.; Lai, B.; Zheng, Z.; Zhang, Y.; Luo, J.; Wang, C.; Chen, Y.; Woodgett, J.; Li, M. Inhibitory phosphorylation of GSK-3 by CaMKII couples depolarization to neuronal survival. J. Biol. Chem. 2010, 285, 41122–41134. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Gao, L.; Han, Q.; Li, A.; Yu, H.; Liu, D.; Pang, Q. GSK-3beta at the crossroads in regulating protein synthesis and lipid deposition in zebrafish. Cells 2019, 8, 205. [Google Scholar] [CrossRef] [Green Version]
- Mirzoev, T.; Tyganov, S.; Vilchinskaya, N.; Lomonosova, Y.; Shenkman, B. Key markers of mTORC1-dependent and mTORC1-independent signaling pathways regulating protein synthesis in rat soleus muscle during early stages of hindlimb unloading. Cell. Physiol. Biochem. 2016, 39, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.; Ballif, B.A.; Anjum, R.; Gygi, S.P.; Blenis, J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 13489–13494. [Google Scholar] [CrossRef] [Green Version]
- Carrière, A.; Cargnello, M.; Julien, L.-A.; Gao, H.; Bonneil, E.; Thibault, P.; Roux, P. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 2008, 18, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Aronson, D.; Violan, M.; Dufresne, S.D.; Zangen, D.; Fielding, R.; Goodyear, L.J. Exercise stimulates the mitogen-activated protein kinase pathway in human skeletal muscle. J. Clin. Investig. 1997, 99, 1251–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Blomstrand, E.; Chibalin, A.V.; Krook, A.; Zierath, J.R. Marathon running increases ERK1/2 and p38 MAP kinase signalling to downstream targets in human skeletal muscle. J. Physiol. 2001, 536, 273–282. [Google Scholar] [CrossRef]
- Vilchinskaya, N.A.; Mochalova, E.P.; Nemirovskaya, T.L.; Mirzoev, T.M.; Turtikova, O.V.; Shenkman, B.S. Rapid decline in MyHC I(beta) mRNA expression in rat soleus during hindlimb unloading is associated with AMPK dephosphorylation. J. Physiol. 2017, 595, 7123–7134. [Google Scholar] [CrossRef] [Green Version]
- Stouth, D.W.; Manta, A.; Ljubicic, V. Protein arginine methyltransferase expression, localization, and activity during disuse-induced skeletal muscle plasticity. Am. J. Physiol. Physiol. 2018, 314, C177–C190. [Google Scholar] [CrossRef] [Green Version]
- Stouth, D.W.; VanLieshout, T.L.; Ng, S.Y.; Webb, E.K.; Manta, A.; Moll, Z.; Ljubicic, V. CARM1 regulates AMPK signaling in skeletal muscle. iScience 2020, 23, 101755. [Google Scholar] [CrossRef]
- Mathew, T.; Ferris, R.; Downs, R.; Kinsey, S.; Baumgarner, B. Caffeine promotes autophagy in skeletal muscle cells by increasing the calcium-dependent activation of AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 2014, 453, 411–418. [Google Scholar] [CrossRef]
- Grundy, D. Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology. Exp. Physiol. 2015, 100, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Burma, N.E.; Bonin, R.P.; Leduc-Pessah, H.; Baimel, C.; Cairncross, Z.F.; Mousseau, M.; Shankara, J.V.; Stemkowski, P.L.; Baimoukhametova, D.; Bains, J.S.; et al. Blocking microglial pannexin-1 channels alleviates morphine withdrawal in rodents. Nat. Med. 2017, 23, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrillo-Mora, P.; Méndez-Cuesta, L.A.; La Cruz, V.P.-D.; Der Goes, T.I.F.-V.; Santamaría, A. Protective effect of systemic l-kynurenine and probenecid administration on behavioural and morphological alterations induced by toxic soluble amyloid beta (25–35) in rat hippocampus. Behav. Brain Res. 2010, 210, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Navis, K.E.; Fan, C.Y.; Trang, T.; Thompson, R.J.; Derksen, D.J. Pannexin 1 channels as a therapeutic target: Structure, inhibition, and outlook. ACS Chem. Neurosci. 2020, 11, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Liu, D.; Long, Y.; Wang, X.; Yao, B. The pannexin-1 channel inhibitor probenecid attenuates skeletal muscle cellular energy crisis and histopathological injury in a rabbit endotoxemia model. Inflammation 2018, 41, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Morey-Holton, E.; Globus, R.K.; Kaplansky, A.; Durnova, G. The Hindlimb unloading rat model: Literature overview, technique update and comparison with space flight data. Adv. Space Biol. Med. 2005, 10, 7–40. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
| Group | Soleus Muscle Weight |
|---|---|
| 1C | 89.0 ± 3.8 mg |
| 1H | 84.2 ± 1.5 mg |
| 1HP | 84.4 ± 2.6 mg |
| 3C | 90.6 ± 3.0 mg |
| 3H | 73.7 ± 1.7 * mg |
| 3HP | 73.6 ± 2.9 * mg |
| Gene | Accession Number | Forward Primer | Reverse Primer |
|---|---|---|---|
| b-actin | NM_031144.3 | 5′-TCATGAAGTGTGACGTTGACATCC-3′ | 5′-GTAAAACGCAGCTCAGTAACAGTC-3′ |
| Gapdh | NM_017008.4 | 5′-ACGGCAAGTTCAACGGCACAGTCAA-3′ | 5′-GCTTTCCAGAGGGGCCATCCACA-3′ |
| MAFbx | NM_133521.1 | 5′-CTACGATGTTGCAGCCAAGA-3′ | 5′-GGCAGTCGAGAAGTCCAGTC-3′ |
| MuRF1 | NM_080903.2 | 5′-GCCAATTTGGTGCTTTTTGT-3′ | 5′-AAATTCAGTCCTCTCCCCGT-3′ |
| Panx1 | NM_199397.2 | 5′-CGATGCTGGAGCAGTACTTGAAGA-3′ | 5′-AGGAGAGGCTGAAGTAGTAGCT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaripova, K.A.; Kalashnikova, E.P.; Belova, S.P.; Kostrominova, T.Y.; Shenkman, B.S.; Nemirovskaya, T.L. Role of Pannexin 1 ATP-Permeable Channels in the Regulation of Signaling Pathways during Skeletal Muscle Unloading. Int. J. Mol. Sci. 2021, 22, 10444. https://doi.org/10.3390/ijms221910444
Zaripova KA, Kalashnikova EP, Belova SP, Kostrominova TY, Shenkman BS, Nemirovskaya TL. Role of Pannexin 1 ATP-Permeable Channels in the Regulation of Signaling Pathways during Skeletal Muscle Unloading. International Journal of Molecular Sciences. 2021; 22(19):10444. https://doi.org/10.3390/ijms221910444
Chicago/Turabian StyleZaripova, Ksenia A., Ekaterina P. Kalashnikova, Svetlana P. Belova, Tatiana Y. Kostrominova, Boris S. Shenkman, and Tatiana L. Nemirovskaya. 2021. "Role of Pannexin 1 ATP-Permeable Channels in the Regulation of Signaling Pathways during Skeletal Muscle Unloading" International Journal of Molecular Sciences 22, no. 19: 10444. https://doi.org/10.3390/ijms221910444
APA StyleZaripova, K. A., Kalashnikova, E. P., Belova, S. P., Kostrominova, T. Y., Shenkman, B. S., & Nemirovskaya, T. L. (2021). Role of Pannexin 1 ATP-Permeable Channels in the Regulation of Signaling Pathways during Skeletal Muscle Unloading. International Journal of Molecular Sciences, 22(19), 10444. https://doi.org/10.3390/ijms221910444








