Abstract
In recent years, brown adipose tissue (BAT), which has a high heat-producing capacity, has been confirmed to exist even in adults, and it has become a focal point for the prevention and the improvement of obesity and lifestyle-related diseases. However, the influences of obesity and physical activity (PA) on the fluid factors secreted from BAT (brown adipokines) are not well understood. In this study, therefore, we focused on brown adipokines and investigated the effects of obesity and PA. The abnormal expressions of gene fluid factors such as galectin-3 (Lgals3) and Lgals3 binding protein (Lgals3bp), whose proteins are secreted from HB2 brown adipocytes, were observed in the interscapular BAT of obese mice fed a high-fat diet for 4 months. PA attenuated the abnormalities in the expressions of these genes. Furthermore, although the gene expressions of factors related to brown adipocyte differentiation such as peroxisome proliferator-activated receptor gamma coactivator 1-α were also down-regulated in the BAT of the obese mice, PA suppressed the down-regulation of these factors. On the other hand, lipogenesis was increased more in HB2 cells overexpressing Lgals3 compared with that in control cells, and the overexpression of Lgals3bp decreased the mitochondrial mass. These results indicate that PA attenuates the obesity-induced dysregulated expression of brown adipokines and suggests that Lgals3 and Lgals3bp are involved in brown adipocyte differentiation.
1. Introduction
With the global increase in the number of lifestyle-related diseases such as type 2 diabetes, patients with obesity caused by the hypertrophy of white adipose tissue (WAT) have become a global issue [1,2]. In 2016, the World Health Organization (WHO) announced that the number of diabetes patients reached 422 million in 2014, which is almost four times higher than the case count (108 million) in 1980 [3]. Direct fatal cases of diabetes increased to 1.5 million in 2012, and higher blood glucose caused an additional 2.2 million deaths by increasing the risks of cardiovascular and other diseases [3]. Additionally, it is estimated that 43% of all deaths due to high levels of blood glucose occur before the age of 70, and the population of diabetes patients is expected to exceed 700 million by 2045 if no efficient preventative strategies can be enacted [3,4]. Hence, it is crucial to establish efficient preventative and improvement strategies for obesity and lifestyle diseases. Exercise is, in fact, widely accepted as an efficient way to prevent and improve the issues of obesity and lifestyle diseases [5,6]. For example, Helmrich et al. [7] tracked 5990 male university graduates and found that, for each 500 kcal increase in energy expenditure during exercise per week, the risk of developing type 2 diabetes was decreased by 6%. Another study of 21,271 male physicians in the United States over 5 years demonstrated that members of the group who exercised to the point of breaking into a sweat once per week had a lower risk of developing diabetes [8].
There are two types of adipose tissue. The first is WAT, which stores excess energy as triglycerides and contributes to energy supply when needed, such as during physical activity (PA) and aerobic exercise. The second is brown adipose tissue (BAT), which converts energy into heat and contributes to the maintenance of body temperature. WAT produces a wide variety of humoral factors generally called “adipokines”, such as adiponectin, tumor necrosis factor-α, and C-C motif chemokine 2 (Ccl2), which is also known as monocyte chemoattractant protein-1. A number of studies showed that insulin resistance is one of the main pathophysiological features of type 2 diabetes, and this condition is induced in skeletal muscle, the liver, and white adipocyte by WAT inflammatory changes due to the dysregulated expression of adipokines in hypertrophic WAT [9,10,11]. Thus, this condition has attracted much attention as the cause of lifestyle-related diseases, and multiple reports listed it as a target for the effect of PA and exercise [12,13].
Meanwhile, BAT is activated by noradrenaline being released during sympathetic nervous activities such as exposure to coldness [14,15]. In activated BAT, energy is converted to heat by the abundant effect of uncoupling protein 1 (UCP1) in mitochondrion [14,15]. BAT is thought to abundantly exist exclusively in small animals such as mice and in human infants, but it almost totally disappears in adults and, as such, has been assumed to carry no biological significance. However, improvements in inspection technologies recently confirmed a sufficient amount of BAT in adults, and, moreover, the degree of obesity was inversely correlated with the level of BAT activation [16,17,18]. Cold-induced, whole-body energy expenditure in humans with BAT is substantially increased compared with those without BAT, and reports documented that continuous cold stimulation reduces obesity. Therefore, BAT has become a target for the improvement of obesity [19,20,21]. Furthermore, both cold stimulation and stimulation with drugs were shown to have the same effect as noradrenaline by inducing adipocytes similar to brown adipocytes in murine subcutaneous WAT [22,23]. These cells are called beige cells, or bright cells, and are different from classic conventional brown adipocytes [22,23]. Since browning was observed in rat subcutaneous WAT even after running exercise, this phenomenon is now under active investigation [24].
In addition to WAT being reported as secreting various adipokines, humoral factors from BAT, which are called brown adipokines, or batokines, are now being reported [25,26,27]. For example, studies are showing that fibroblast growth factor 21 is secreted by BAT and acts on WAT to improve glucose and lipid metabolism, and this promotes WAT browning through increased expression of peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) [26]. Neuregulin 4 is also secreted by BAT and is reported to be involved in lipid biosynthesis in the liver [26]. The recently identified novel brown adipokine ependymin-related 1 was suggested to be important for brown fat commitment [28]. Nevertheless, little is known about the effects of obesity and PA on brown adipokines. In the present study, therefore, we identified humoral factors from HB2 brown adipocytes [29] and investigated the effects of obesity and PA on brown adipokines in mice interscapular BAT.
2. Results
2.1. Identification of Humoral Factors Secreted from HB2 Brown Adipocytes
Exhaustive analysis on humoral factors secreted from brown adipocytes was performed by various research groups in recent years [28,30,31]. We used mass spectrometry in an attempt to identify humoral factors secreted from mouse HB2 cells fully differentiated in brown adipocyte. Complement C3 and fatty acid binding protein 4 (Fabp4) protein showed high values for peptide-spectrum matches (PSMs) in the culture medium of HB2 brown adipocytes (Table 1). Furthermore, humoral factors such as transferrin, apolipoprotein E (ApoE), EGF-containing fibulin-like extracellular matrix protein 1 (Efemp1), complement factor D (Adipsin), Ccl9, galectin-1 (Lgals1), Lgals3, and Lgals3 binding protein (Lgals3bp) were confirmed in the culture medium of HB2 brown adipocytes (Table 1). Extracellular matrix proteins collagen (Col) 6a1, Col6a2, Col1a2, and fibronectin were found as well (Table 1).

Table 1.
List of identified humoral factors secreted from mature HB2 brown adipocytes.
2.2. Physical Characteristics of Animals
Physical activity is defined as any bodily movement produced by skeletal muscles that results in energy expenditure above resting levels [32]. We selected voluntary wheel running as a method of PA. Compared to forced running exercise on a treadmill, voluntary wheel running has the advantage that it can be performed under non-stress conditions and is similar to the natural running behavior of mice [33]. Voluntary wheel running has various beneficial effects in mice, such as promoting mitochondrial biogenesis in skeletal muscle, and attenuates dysregulated expression of adipokine genes in WAT due to obesity [33,34].
Final body masses (g, mean ± SE, n = 4 in each) were significantly higher in high-fat diet (HFD)-induced obese mice (HFD mice) (37.6 ± 1.6) and HFD mice subjected to voluntary PA (HFD + PA mice) (33.8 ± 0.8) than in control mice (26.9 ± 0.8) at the 4 month time point of HFD intake and PA (Table 2). The final percent of epididymal WAT mass per body mass (%, mean ± SE, n = 4 in each) was also significantly higher in HFD mice (5.6 ± 0.3) than in control mice (1.2 ± 0.1). The final percent of epididymal WAT mass per body mass of HFD + PA mice (4.4 ± 0.1) was also significantly higher compared with that of control mice but significantly lower than that of HFD mice (Table 2). On the other hand, no significant differences were observed in the final percent of interscapular BAT mass per body mass among control (0.24 ± 0.01), HFD (0.29 ± 0.05), and HFD + PA mice (0.23 ± 0.02) (Table 2).

Table 2.
Levels of body mass, WAT and BAT mass of control, HFD, and HFD + PA mice.
2.3. Effects of Obesity and Physical Activity on the mRNA Levels of Humoral Factors in the Interscapular BAT of Mice
In consideration of the protein in the HB2 brown adipocytes secretion shown in Table 1, we next used DNA array analysis to investigate the effects of HFD-induced obesity and PA on the expression of genes for humoral factors in the interscapular BAT of mice (Table 3). In the interscapular BAT with obesity due to the intake of an HFD, the expressions of genes for Lgals3, Lgals3bp, Ccl2, and Ccl9 were markedly upregulated compared with those of the control group. PA significantly suppressed the obesity-induced increases in Lgals3, Lgals3bp, and Ccl2 genes in BAT and tended to decrease the enhanced expression of Ccl9 genes in the BAT of HFD mice. Additionally, expression of the gene for matrix metalloproteinase 12 (MMP12) was found in brown adipose tissue. These expressions were increased with obesity, but PA significantly suppressed the obesity-induced increase. However, the secretion of MMP12 protein from HB2 cells could not be confirmed by analysis using mass spectrometry. On the other hand, HFD-induced obesity significantly reduced the expressions of genes for complement factor C3, transferrin, and resistin in the BAT compared with that of control mice, and PA had no effect on the expression of these genes. Moreover, regarding the adipsin and the ApoE in BAT, PA further augmented the inhibitory effects on gene expression caused by obesity. In addition, the expression of Efemp1 gene was low only in the BAT of the group with a combination of obesity and PA. No significant differences were observed in the expressions of genes for other humoral factors, such as Fabp4 and prosaponin, in the BAT among control, HFD, and HFD + PA mice. On the other hand, when compared with the control mice, the expressions of genes for some extracellular matrix proteins Col3a1 and Col6a1, along with fibronectin, were significantly upregulated in BAT associated with obesity, and these upregulations were attenuated by PA.

Table 3.
Expression levels of mRNA for representative humoral factors in the interscapular BAT of control, HFD, and HFD + PA mice.
2.4. Effects of Obesity and Physical Activity on the mRNA Levels of Macrophage Markers and Brown Adipocyte Differentiation-Related Factors in the Interscapular BAT of Mice
The BAT of obese mice displayed increases in the macrophage marker genes such as F4/80, CD53, and 68 expression by comparison with the control group, but there were no significant differences between the levels in the control and the HFD + PA groups (Table 4 upper). Moreover, although the significantly increased expressions of genes for macrophage-expressed gene 1 (Mpeg1) and CD11b in the BAT of HFD mice were observed, PA significantly inhibited the obesity-induced increases in the expressions of these genes (Table 4, upper).

Table 4.
Expression levels of mRNA for macrophage markers and brown adipocyte differentiation-related factors in the interscapular BAT of control, HFD, and HFD + PA mice.
Next, we noted the expression changes in the genes for brown adipocyte markers in the interscapular BAT of each group from the DNA array results (Table 4 lower). Although expression of the UCP1 gene in BAT did not differ among the three groups, the expression levels of the PGC1-α gene in HFD mice were significantly lower than those in control mice. The PGC1-αis a master transcription factor coactivator that regulates the expressions of many genes involved in energy production and heat expenditure [35]. This obesity-induced decrease in PGC1-α gene expression was not observed in HFD + PA mice. Furthermore, the expressions of genes for type II iodothyronine deiodinase (Dio2), which encodes a thyroid hormone-converting enzyme [36], and for lipoprotein lipase (LPL), which encodes a lipolytic enzyme involved in catalyzing the hydrolysis of triglycerides in chylomicrons and very low-density lipoprotein particles [37], were significantly declined in HFD and HFD + PA mice compared with that in control mice. The expression level of the very long-chain fatty acid protein 3 (Elovl3) gene, which encodes the extremely long-chain fatty acid elongase [38], was lower in HFD mice than in control mice, but there was no statistical difference between the control and the HFD + PA mice. The CCAAT/enhancer binding protein α (C/EBPα) is a critical transcription factor for brown adipocyte differentiation [39]. Expression of the gene for C/EBPα in BAT was affected by neither HFD intake nor PA. On the other hand, the PR domain containing 16 genes, which stimulates brown adipocyte differentiation from progenitor cells [40], was low in the BAT of each group, and no differences were found (data not shown).
2.5. Establishment of Brown Adipocytes Overexpressing Lgals3 and Lgals3bp
Enhanced expression of Lgals3 gene is found in the WAT and the BAT of obese mice, and the insulin signal was reported to be altered in the WAT of Lgals3 knock out (KO) mice, but the influence of Lgals3 on brown adipocytes remains unknown [41,42]. Lgals3bp was identified as a protein that binds to Lgals3 and Lgals1, but its effect on brown adipocytes is also unknown [43]. As shown in Table 3, the expressions of genes for Lgals3 and Lgals3bp are thought to be greatly influenced by HFD intake and PA. Therefore, we attempted to establish an overexpression of Flag-tagged Lgals3 and Lgals3bp in HB2 cells. Real-time PCR analysis showed a significant increase in the expressions of Lgals3 and Lgals3bp mRNAs in Flag-tagged Lgals3 (HB2-L3 cells), Lgals3bp (HB2-L3bp cells), or both (HB2-L3-L3bp cells) when compared with that in control cells (HB2-C cells) (Figure 1A). Moreover, expression of exogenous Flag-tagged proteins in cell lysate and Flag-Lgals3 or Lgals3bp protein secretion into cell culture medium were confirmed in HB2-L3, -L3bp, and -L3-L3bp cells (Figure 1B). As shown in Figure 1B,C, in the differentiation process of HB2 cells into brown adipocytes, Flag-tagged Lgals3 protein was secreted the most during the early stages of differentiation, while the secretion from mature brown adipocytes was decreased by comparison. In contrast, the secretion of the Flag-tagged Lgals3bp protein from mature brown adipocytes was the most prominent. In addition, although the Flag-tagged Lgals3 protein was secreted at each stage of brown adipocyte differentiation, most of this protein was uniformly present in the cytoplasm (Figure 1D). Furthermore, the expression of Flag-tagged Lgals3bp in the cytoplasm was heterogeneous compared with that of the Flag-tagged Lgals3 protein (Figure 1D).
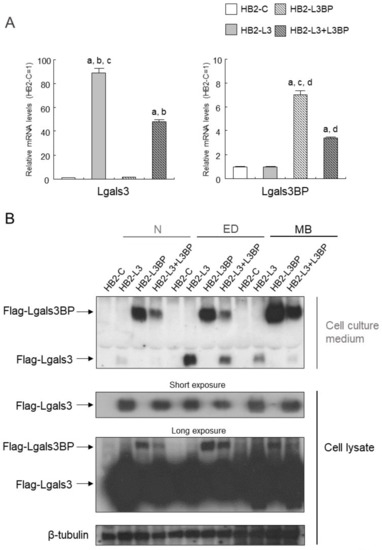
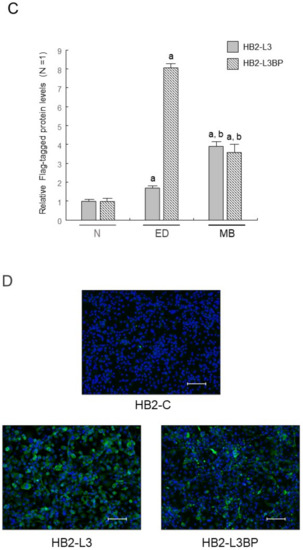
Figure 1.
Establishing HB2 cells that overexpress Lgals3 and Lgals3bp. (A) Expression levels of Lgals3 and Lgals3bp mRNA in HB2 transfectants. Total RNAs were extracted from each mature HB2 transfectant and then subjected to real time quantitative PCR. The expression levels of Lgals3 or Lgals3bp mRNA were normalized using the 18S rRNA level as the standard. The values shown in the bar graphs are related to an arbitrary unit of the HB2-C cells (set to 1), and the means ± SE values (n = 3) are given: a p < 0.05 vs. HB2-C cells; b p < 0.05 vs. HB2-L3bp cells; c p < 0.05 vs. HB2-L3-L3bp cells; and d p < 0.05 vs. HB2-L3 cells. (B) Expression levels of Flag-Lgals3 and Flag-Lgals3bp proteins in the cell lysate and the culture medium of HB2 transfectants. Cellular protein extracts and cell culture media of each HB2 transfectant at different differentiation stages (N: not differentiated; ED: early phases of brown adipocyte differentiation; MB: mature brown adipocyte) were collected and then subjected to Western blot analysis. Flag-tag proteins were detected by anti-DYKDDDDK (Flag) antibody. β-tubulin probe was used as a loading control. (C) Relative expression levels of Flag-Lgals3 or -Lgals3bp protein in culture medium of HB2-L3 or -L3bp cells. The values shown in the bar graphs are related to an arbitrary unit of the N stage (set to 1), and the means ± SE values (n = 3) are given: a p < 0.05 vs. N stage; b p < 0.05 vs. ED stage. (D) Flag-tag protein in mature HB2-C, HB2-L3, or HB2-L3bp cells was stained using an anti-Flag antibody. Representative photomicrographs of each cell are shown. Each scale bar indicates 50 μm.
2.6. Effects of Lgals3 and Lgals3bp on Brown Adipocyte Differentiation
Intracellular lipid droplet formation after mature brown adipocyte differentiation of HB2-L3 and HB2-L3-L3bp cells was significantly enhanced compared with that of HB2-C and HB2-L3bp cells (Figure 2). On the other hand, the amounts of intracellular mitochondria in HB2-L3bp cells were significantly decreased compared with those of HB2-C and HB2-L3 cells in the early differentiation stage and also by comparison with all other cell groups after mature brown adipocyte differentiation (Figure 3A,B). Furthermore, the copy number of mitochondrial DNA (mtDNA) in each mature HB2 transfectant was examined using primers that were reported to specifically amplify mouse mtDNA [44]. The results showed that the copy number of mtDNA in HB2-L3bp cells was significantly lower than that in HB2-C cells (Figure 3C). In addition, when we examined mitochondrial function by measuring the oxygen consumption rate (OCR), we found that the OCR of HB2-L3bp was significantly suppressed when stimulated by isoproterenol compared with that of either HB2-C or HB2-L3-L3bp cells, which was also the case when stimulated with the potent uncoupler carbonyl cyanide-p-trifluoro-methoxyphenylhydrazone (FCCP) compared with that of HB2-C cells (Figure 3D).
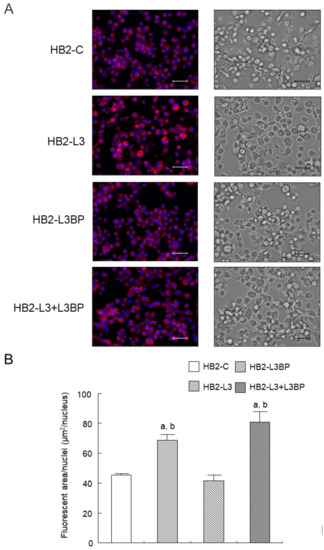
Figure 2.
Effects of Lgals3 and Lgals3bp on lipogenesis in HB2 cells. (A) Lipid droplets in HB2 transfectants after mature differentiation were stained red using a Nile red solution. Representative photomicrographs (left) and corresponding bright field images (right) are shown. Each scale bar indicates 50 μm. (B) Measured areas of staining with Nile red per the number of Hoechst positive nuclei are calculated in three different fields of view. The mean ± SE values (n = 3) are given: a p < 0.05 vs. HB2-C cells; and b p < 0.05 vs. HB2-L3bp cells.
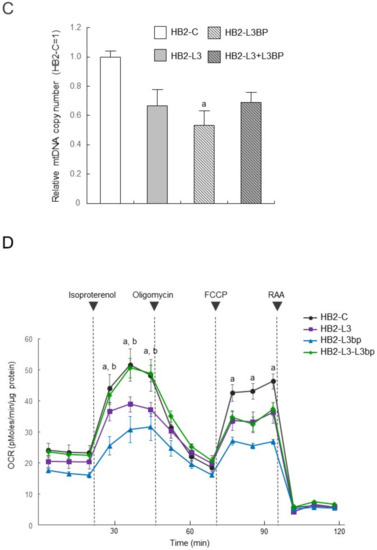
Figure 3.
Effects of Lgals3 and Lgals3bp on the mitochondrial content in HB2 cells. (A) Intracellular mitochondria in HB2 transfectants during or after differentiation were stained red using a MitoTracker solution. Representative photomicrographs are shown. Each scale bar indicates 50 μm. ED: early phases of brown adipocyte differentiation; MB: mature brown adipocyte. (B) Measured areas of staining using a MitoTracker per the number of Hoechst positive nuclei are calculated in three different fields of view. The mean ± SE values (n = 3) are given: a p < 0.05 vs. HB2-C cells; b p < 0.05 vs. HB2-L3-L3bp cells; and c p < 0.05 vs. HB2-L3 cells. (C) The mtDNA copy number in mature HB2 transfectants. The mtDNA was amplified using the PCR protocol with primers that specifically amplify mouse mtDNA. The values shown in the bar graphs are related to an arbitrary unit of the HB2-C cells (set to 1), and the means ± SE values (n = 3) are given: a p < 0.05 vs. HB2-C cells. (D) OCR in mature HB2 transfectants. OCRs were measured with the sequential injection of isoproterenol (1 µm), oligomycin (5 µm), FCCP (10 µM), rotenone (R; 0.5 µm), and antimycin A (AA: 0.5 µm) at the indicated times. The means ± SE values (n = 5) are given: a p < 0.05, HB2-L3bp cells vs. HB2-C cells; b p < 0.05, HB2-L3bp cells vs. HB2-L3-L3bp cells.
As for the expression levels of mRNA for brown adipocyte differentiation-related factors in HB2 transfectants (Figure 4A), in the early stages of brown adipocyte differentiation, expression of the PGC1-α gene was significantly upregulated in HB2-L3bp cells compared with that of HB2-C cells. Moreover, Elovl3 gene expression was significantly increased in the Lgals3bp-transfected cells. After differentiation of the mature brown adipocytes, the expression of the Dio2 gene was significantly decreased by the introduction of Lgals3bp into HB2 cells. No significant effect of Lgals3 or Lgals3bp overexpression was observed in the expression of other genes such as UCP1 (Figure 4A). On the other hand, the protein expression of PGC-1α and UCP1 in the early phases of brown adipocyte differentiation and after maturation was not significantly different among the HB2 transfectants (Figure 4B).
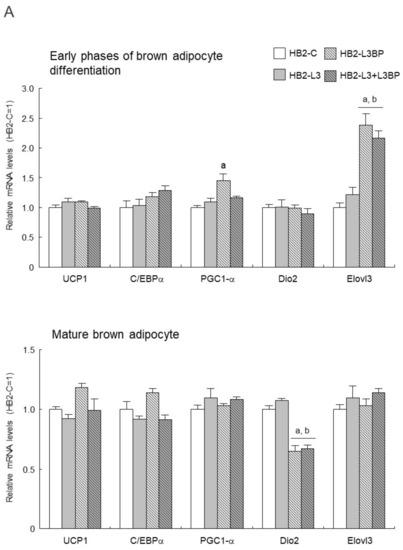
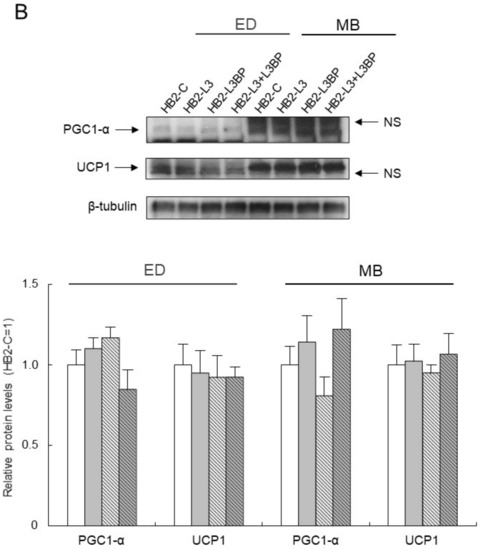
Figure 4.
Effect of Lgals3 and Lgals3bp on the expression of brown adipocyte differentiation-related factors. (A) Expression levels of brown adipocyte differentiation-related mRNAs in HB2 transfectants. Total RNAs were extracted during or after differentiation from each HB2 transfectant and then subjected to real time quantitative PCR. The expression level of each mRNA was normalized using the 18S rRNA level as the standard. The values shown in the bar graphs are related to an arbitrary unit of the HB2-C cells (set to 1), and the means ± SE values (n = 3) are given: a p < 0.05 vs. HB2-C cells; and b p < 0.05 vs. HB2-L3 cells. (B) Expression levels of PGC1-αand UCP1 protein in HB2 transfectants. Representative data from western blot analyses are shown in the superior region. Expression levels of PGC1-α or UCP1 proteins were normalized to those of the expression levels of β-tubulin. The data are expressed as ratios with the value of the HB2-C cells = 1. The mean ± SE (n = 3) is given. ED: early phases of brown adipocyte differentiation; MB: mature brown adipocyte; NS: nonspecific band.
3. Discussion
In the present study, we identified humoral factors from HB2 brown adipocytes similar to those reported by Villarroya et al. [30] using murine brown preadipocytes in BAT. Among these humoral factors, Ccl9, Lgals3, and Lgals3bp were found to be brown adipokines with gene expressions that were largely influenced by obesity and PA. Ccl9, which is also known as macrophage inflammatory protein 1-γ, is a chemokine belonging to the CC chemokine family [45] and is known to play an important role in anti-leukemia and bone resorption procedures. Although there are very few reports of the effect of Ccl9 on adipocytes, it is known to inhibit the differentiation of white adipocytes [46].
The Lgals3 protein (galectin-3) is involved in biological processes such as cell adhesion, inflammation, and apoptosis. Lgals3 is upregulated in the WAT and the BAT of obese mice and can attenuate insulin signaling in white adipocytes [41]. Furthermore, obese and diabetic individuals were shown to have higher blood levels of the Lgals3 protein, which parallels the deterioration of glucose homeostasis and suggests that Lgals3 may be involved in the development of obesity and type 2 diabetes [41]. In addition, studies using Lgals3 KO mice reported decreases in body and fat masses in HFD-fed Lgals3 KO mice by comparison with control mice [47]. However, another study using Lgals3 KO mice demonstrated that Lgals3 deficiency accelerates adiposity, levels of adipose tissue, and systemic inflammation associated with altered glucose homeostasis [48,49]. Additionally, Lgals3 is known to stimulate the differentiation of preadipocytes into mature white adipocytes in vitro [47]. On the other hand, human Lgals3bp has long been regarded as an important clinical tumor biomarker associated with disease diagnosis, negative prognosis, and poor response to therapy [50]. Lgals3bp is also secreted by human visceral WAT and inhibits white adipocyte differentiation [51]. Lgals3 and Lgals3bp proteins are secreted by macrophages and are involved in immune responses. Inflammatory stimuli to macrophages promote the secretion of Lgals3 protein, which is known to be involved in infection and inflammation as an immune regulator with functions such as regulating leukocyte function as well as inducing angiogenesis in cancer cells and regulating apoptosis in tumor cells [52]. Lgals3 protein is also related to the onset of tissue fibrosis [53]. On the other hand, Lgals3bp is abundantly expressed in human M1 macrophages and is suggested to be a macrophage inflammatory marker in carotid artery disease in women with human immunodeficiency virus or hepatitis C virus infections [54]. Results of the present study suggest that inflammatory changes due to macrophage infiltration may be occurring in the BAT of obese mice. In fact, Krautbauer et al. [42] found that the expression of pro-inflammatory cytokine and macrophage marker genes was upregulated in the BAT of HFD-induced obese mice and genetically ob/ob mice. Therefore, the gene expressions of Lgals3 and Lgals3bp in macrophages infiltrating BAT may be partially responsible for the elevated expressions of those genes in the BAT of obese mice. In addition, the enhanced expressions of fibronectin and collagen genes, which play important roles in tissue fibrosis, were also observed in the BAT of obese mice. These observations suggest the possibility that Lgals3 brings about increases in fibrosis-related factors in the BAT of obese subjects.
Overexpression of Lgals3 in HB2 cells resulted in enhanced intracellular lipogenesis, suggesting that Lgals3 may play an important role in the differentiation process of brown adipocytes, particularly in lipogenesis. On the other hand, the mitochondrial content of HB2-L3bp cells was reduced compared with control cells in the early phases of brown adipocyte differentiation and after maturation. Furthermore, after the differentiation of mature brown adipocytes, the copy numbers of mtDNA and OCR were significantly lower than those of HB2-C cells when stimulated with isoproterenol or the potent uncoupler FCCP. Therefore, Lgals3bp is suggested to suppress mitochondrial biogenesis. On the other hand, the expression levels of Elovl3 genes were significantly increased in HB2-L3bp cells in the early stages of differentiation compared with control cells, and the expression levels of PGC-1α proteins in HB2-L3 cells during brown adipocyte differentiation did not differ significantly from the other three HB2 transfectants. Nonetheless, mitochondrial content was decreased. The reason for this is unknown, but the expressions of PGC-1α and Elovl3 did not seem to affect the mitochondrial biogenesis of HB2-L3bp cells. The Dio2 gene encodes type II deiodinase, which converts thyroxine (T4) to triiodothyronine (T3). In particular, in skeletal muscles and BAT, the converted T3 stimulates protein synthesis relating to mitochondrial biogenesis, which results in higher energy expenditure [36]. Moreover, when Christoffolete et al. [55] generated Dio2 KO mice and examined their BAT, they showed a cold-induced overexpression of the UCP1 gene in their BAT, but they did not exhibit increases in BAT lipogenesis or adaptive thermogenesis. In the present study, we found that Lgals3bp decreased the expression of the Dio2 gene in mature brown adipocytes. Therefore, it is possible that the reduced expression of Dio2 gene is involved in the reduced mitochondrial content that was observed in the HB2-L3bp cells. Nevertheless, although Dio2 gene expression was decreased in mature HB2-L3-L3bp cells, the mitochondrial content was not significantly different from that of control cells. Thus, the repressive effect of mitochondrial biogenesis with Lgals3bp was not simply due to the decreased expression of Dio2 gene but also to other causes. In this study, we mainly observed the mRNA expression levels of brown adipocyte differentiation markers. In order to elucidate the molecular mechanism of the effect of Lgals3bp on mitochondrial biogenesis, it will be necessary to further investigate not only mRNA expression levels but also changes in related protein modifications.
As described above, Lgals3 was expected to enhance lipogenesis in brown adipocytes, while Lgals3bp was expected to inhibit mitochondrial biogenesis by stimulating differentiation. However, in the present study, although Lgals3bp proteins were identified as a factor in binding to Lgals3 protein and enhancing Lgals3 protein activity [37], Lgals3 and Lgals3bp did not appear to work in concert. This observation could be attributed to the fact that simultaneous infection of HB2 cells by Lgals3 and Lgals3bp via lentiviruses reduces gene transfer to about half that of a single infection. However, the Flag-Lgals3 protein is mainly found in the cytoplasm, and its secretion appears to be lower than that of the Flag-Lgals3bp protein. It is likely that each protein functions independently due to its different localization. Nevertheless, since it cannot be denied that Lgals3 and Lgals3bp bind and exert cooperative effects on brown adipocytes, further investigation is needed.
It is well known that PA and exercise are recognized as effective means of preventing and improving obesity and lifestyle-related diseases. The purpose of our investigation was to examine the effect of PA on obesity-induced remodeling in BAT and to find novel effects of PA on the prevention and the improvement of obesity and lifestyle-related diseases. Therefore, we did not establish a group that exclusively engaged in PA.
A wide variety of adipokines secreted from WAT have been identified, and it has become clear that dysregulated expression of adipokines due to obesity is closely related to the onset of lifestyle-related diseases [9,10,11]. The effects of PA and exercise on adipokine expression in WAT were reported in many studies [12,13]. The influence of PA and exercise on adipokine expression in WAT and in blood levels has not always been consistent among reports due to differences in experimental subjects, exercise intensity, and duration [12,13]. However, PA and exercise is believed to have a positive effect. With respect to BAT, the present study revealed that obesity causes an abnormal expression of several fluid factors, presumably brown adipokines, and a decrease in the expression of genes for brown adipocyte differentiation markers such as PGC1-α. Therefore, it is inferred that obesity causes dysfunction in BAT as well as WAT obesity. Physical activity seems to partially suppress the dysfunction of BAT caused by obesity. Nevertheless, there is disagreement regarding the effects of exercise on the functions of BAT. For example, in an 8 week study of running exercise in rats using treadmills, decreases in BAT mass and UCP1 expression were observed [56]. By contrast, swimming exercise at a water temperature that does not cause heat production by shivering enhances the function of BAT [57]. Furthermore, several studies demonstrated increased thermogenic and metabolic activities of BAT and UCP1 expression after exercise in rodents [58]. Thus, further studies including the examination of brown adipokines are needed to clarify the effects of PA and exercise on BAT.
In the present study, we observed the dysregulated expressions of many genes, which included brown adipokines and differentiation-related factors, in the BAT of obese mice subjected to a HFD intake. PA attenuated those abnormal gene expressions, albeit only partially. Specifically, the expressions of genes for putative brown adipokines such as Ccl9, Lgals3, and Lgals3bp were substantially upregulated by obesity, and PA reduced their expressions. In brown adipocytes, Lgals3 caused increased lipogenesis, and Lgals3bp induced decreases in the mitochondrial content. However, overexpression of Lgals3 or Lgals3bp in HB2 cells did not correspond to BAT in mice subjected to a HFD intake and PA. These results suggest that Lgals3 or Lgals3bp may exert their functions in brown adipocytes through functions such as protein modification instead of the regulation of gene expression. Further investigations into the effects of Lgals3 and Lgals3bp on BAT are necessary, and these include elucidation of the molecular mechanisms as well as in vivo studies.
4. Methods
4.1. Cell Culture
The brown preadipocytes of HB2 mice were kindly provided by Professor M. Saito (Tenshi University, Sapporo, Japan) [29] and were maintained in Dulbecco’s modified eagle medium (DMEM) (Sigma Chemical, St. Louis, MO, USA) supplemented with 10% fetal calf serum (FCS). Differentiation to brown adipocytes was induced by treatment with 1 μM dexamethasone (Sigma Chemical) and 0.5 mM 3-isobutyl-1-methylxanthine (Wako, Osaka, Japan) for 48 h (early differentiation phase). The treated cells were maintained in DMEM containing 10 μg/mL of insulin (Wako) and T3 (Wako) for 96 h to accumulate triglycerides (inducing mature brown adipocytes). Fully differentiated HB2 cells were incubated with phenol red and FCS-free DMEM supplemented with Insulin-Transferrin-Selenium (Thermo Fisher Scientific, Waltham, MA, USA) for 24 h. Incubated medium was concentrated using Amicon Ultra (Merck Millipore, Darmstadt, Germany), and protein identification in the concentrated medium was performed via mass spectrometry.
4.2. Proteomic Analysis for the Identification of Fluid Factors from Brown Adipocytes
Proteomic analyses were performed as described previously [59]. All of the fractionated peptides described above were injected into a trap column (C18, 0.3 × 5 mm; L-column, Chemicals Evaluation and Research Institute, Tokyo, Japan) and an analytical column (C18, 0.075 × 120 mm; Nikkyo Technos, Tokyo, Japan), and both were attached to a nano liquid chromatography-tandem mass spectrometry (nanoLCMS/MS) system. The nanoLC-MS/MS analysis was conducted using an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoLC interface (KYA, Tokyo, Japan) and a nano high-performance liquid chromatography (nanoHPLC) system (DiNa; KYA). Purified peptides from the nanoLC were introduced into the LTQ Orbitrap Velos, a hybrid ion-trap Fourier transform mass spectrometer. Full MS and MS/MS scans were followed by higher energy collisionally activated dissociation. The database search engines Proteome Discoverer 1.4 (Thermo Scientific) and MASCOT 2.6 (Matrix Science, Tokyo, Japan) were used to identify and quantify proteins from MS, MS/MS, and reporter ion spectra of peptides. Peptide mass data were matched by searching the murine protein database (UniprotKB/Swiss-prot). The false discovery rate (FDR) [60] was calculated via peptide sequence analysis using a Percolator [61]. High-confidence peptide identifications were obtained by setting a target FDR threshold of ≤1.0% at the peptide level. The numbers of identified peptide spectra matches (PSM) appear in Table 1.
4.3. Animal Care
Six-week-old male C57BL/6J mice (Sankyo Labo Service Corporation, Tokyo, Japan) were housed in cages in a temperature-controlled environment at 23 °C under a 12:12 h light–dark cycle. The mice were divided randomly into three groups (n = 4 each): (i) C mice—control mice fed normal chow for four months; (ii) HFD mice—mice given an HFD for four months; and, (iii) HFD + PA mice—PA mice given an HFD for four months. The HFD consisted of 60.7% (calorie ratio) fat, 17.9% protein, and 21.4% nitrogen-free extract (HFD-60; Oriental Yeast Co., ltd., Tokyo, Japan). HFD + PA mice were subjected to wheel running. The wheels were equipped with a permanent magnetic switch that activated a digital counter of wheel revolutions (Melquest Ltd., Toyama, Japan). Total wheel revolutions were recorded daily, and the total distance run per day was determined by multiplying the number of wheel rotations by wheel circumference. All experiments conducted in the present study were approved by the Experimental Animal Ethics Committee of the Kyorin University School of Medicine, Mitaka.
4.4. RNA Isolation and DNA Array Analysis
Total RNAs in the interscapular BAT of mice or HB2 brown adipocytes were prepared by using Isogen (Nippon Gene, Toyama, Japan). The concentration and the purity of extracted total RNA of BAT were measured using the NanoDrop system (Thermo Fisher Scientific, Waltham, MA, USA). In addition, the RNA integrity number (RIN) was measured using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA) to confirm the quality of extracted RNA. cDNA synthesis via purified total RNA, fragmentation, and labeling was performed using a GeneChip WT PLUS Reagent Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Subsequently, the gene expression profiles of each group were determined using Clarion S arrays for mice (Thermo Fisher Scientific).
4.5. Real Time Quantitative PCR
First-strand cDNA of HB2 brown adipocytes was obtained by incubating total RNA samples (2 μg) with reverse transcriptase (PrimeScrip II 1st strand cDNA Synthesis Kit, Takara Bio, Kusatsu, Japan) in a reaction mixture (20 μL). The RT product was amplified using a KAPA SYBR Fast qPCR Kit (Kapa Biosystems, Wilmington, MA, USA) in triplicate in a 7500 Real Time PCR System (Applied Biosystems). PCR conditions were 50 °C for 2 min and 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. An 18s rRNA gene was used as an internal control, and the primer sequences were referenced from previous studies. Quantitative real-time PCR was performed for brown adipocyte marker genes such as 18S rRNA, Lgals3, Lgals3bp, UCP1, PGC-1α, Dio2, Elovl3, and C/EBPα using their specific primers. Each primer sequence was referenced in previous studies [28,43,62,63]. The 18S rRNA was used as an internal control.
The mtDNA in fully differentiated HB2 transfectants was extracted using an mtDNA Extractor CT kit (Wako) according to the manufacturer’s instructions. The extracted mtDNA was amplified using the PCR protocol described above with primer sets that were reported to specifically amplify mouse mitochondrial DNA [44]. Each value was corrected for the corresponding nDNA (β2- macroglobulin DNA) value.
4.6. Lentiviral Vector Construction and Lentivirus Production
Full-length mouse Lgals3 and Lgals3bp cDNA with a FLAG sequence at the 3-terminus was subcloned into the pLVSIN-EF1α vector (Takara Bio) at HindIII and XhoI sites. The lentiviral expression vectors of Lgals3, Lgals3bp, and a mock version were produced using the Lentiviral High Titer Packaging Mix (Takara Bio) and Lenti-X 293T cells (Takara Bio) according to the manufacturer’s recommendations. A cell culture medium containing each lentivirus was concentrated using a Lenti-X™ Concentrator (Takara Bio), and the lentiviral titer was measured using Lenti-X™ GoStix™ Plus (Takara Bio). Almost 1 × 106 of IFU/mL lentivirus was infected with HB2 cells. Infected cells were selected in medium containing 1mg/mL of G418 (Geneticin; Wako).
4.7. Western Blot Analysis
Cell culture medium and lysate from lentivirus-infected HB2 cells was subjected to 10% TGX FastCast gel (Bio-Rad Laboratories, Hercules, CA, USA) electrophoresis and transferred onto a nylon membrane. The membrane was blocked using TBS T (20 mmol/L of Tris HCl, pH 7.5, 137 mM of NaCl, and 0.1% Tween 20) containing 5% nonfat dry milk and was then probed with an anti DYKDDDDK (Flag) tag (WAKO), PGC1-α (Santa Cruz Biotechnology, Santa Cruz, CA, USA), UCP1 (Abclonal, Tokyo, Japan), or β-tubulin (WAKO) antibody. After washing with TBS T, the bound antibody was detected using an ECL system (Amersham, Buckinghamshire, UK).
4.8. Cell Staining
After the induction of differentiation into brown adipocytes, HB2 cells infected with lentivirus were washed with phosphate buffer saline (PBS) and then fixed with 3.7% paraformaldehyde in PBS for 15 min. After fixation, these cells were permeabilized with 0.1% Triton X-100 in PBS containing 5% normal goat serum (NGS-PBSTX). The cells were then incubated with 5 µg/mL of the anti- DYKDDDDK (Flag)-tag antibody in NGS-PBSTX for 1.5 h at room temperature. After this incubation, the cells were washed and incubated for another 1 h with the Alexa Flour 488-conjugated anti-mouse IgG antibody (Molecular Probes, Thermo Fisher Scientific). The stained cells were observed with a fluorescent microscope (model BZ-X700, Keyence corporation, Osaka, Japan).
The intracellular lipid accumulations and mitochondria in HB2 cells were analyzed by Nile red (Wako) and MitoTracker (Thermo Fisher Scientific) staining, respectively. HB2 cells were washed twice with PBS and then were stained either with Nile red (500 nM) for 15 min at room temperature or with MitoTracker (200 nM) for 60 min at 37 °C and washed twice with PBS. After Nile red or MitoTracker staining, the nuclei in these cells were stained with Hoechst 33342 (Thermo Fisher Scientific). The stained cells were observed under a fluorescent microscope, and the light-emitting area was then measured using a BZ-X Analyzer (Keyence corporation). Moreover, we chose three fields of view for each of the groups stained with either Nail red or MitoTracker and calculated the light-emitting area per number of nuclei.
4.9. Measurement of OCR
Fully differentiated cells HB2-C, -L3, L3bp, and -L3-L3bp were seeded into collagen I coated (KOKEN, Tokyo, Japan) XFe24 Microplates (Seahorse Bioscience, North Billerica, MA, USA) at a density of 50,000 cells/well and were incubated in a CO2 incubator for 24 h, and then cells were washed three times and incubated in Seahorse XF DMEM Medium, pH 7.4 (Seahorse Bioscience) supplemented with 25 mM glucose and 1 mM sodium pyruvate for 1 h. The OCR was measured using a Seahorse XFe24 flux analyzer (Seahorse Bioscience). The compounds used to determine OCR included 1 μM isoproterenol (Sigma), 5 μM oligomycin (Sigma), 10 μM FCCP, 0.5 μM antimycin A (Sigma), and 0.5 μM rotenone (Sigma). OCRs were normalized to the total protein content.
4.10. Statistical Analysis
Values are represented as the means ± standard error (SE). The significance of multiple comparisons was determined using one-way analysis of variance (ANOVA) followed by a Bonferroni’s post hoc test. p < 0.05 was considered statistically significant.
The intensities of bands from the Western blot analyses were quantified using the National Institute of Health Image software.
Author Contributions
T.S. performed most of the experiments with assistance from T.F., S.Y., E.N.; T.S. wrote the manuscript; T.K. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by Grants-in-Aid for Scientific Research of Japan (18K10824). We are also grateful for financial support from the Kyorin University School of Medicine, Tokyo, Japan.
Institutional Review Board Statement
All animals were carefully handled during the experiment under the auspices of the Experimental Animal Ethics Committee of Kyorin University School of Medicine (Approval number: 76), approved on 1 April 2018.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Adipsin | complement factor D |
| ApoE | apolipoprotein E |
| BAT | brown adipose tissue |
| C/EBPα | CCAAT/enhancer binding protein α |
| Ccl9 | C-C motif chemokine 9 |
| Dio2 | type II iodothyronine deiodinase |
| Efemp1 | EGF-containing fibulin-like extracellular matrix protein 1 |
| Elovl3 | elongation of very long chain fatty acid protein 3 |
| Fabp4 | fatty acid binding protein 4 |
| FCCP | carbonyl cyanide-p-trifluoro-methoxyphenylhydrazone |
| HFD | high-fat diet |
| mtDNA | mitochondrial DNA |
| Lgals3 | lectin, galactose-binding, soluble 3 |
| Lgals3bp | Lgals3 binding protein |
| OCR | oxygen consumption rate |
| PA | Physical activity |
| PGC-1α | peroxisome proliferator-activated receptor γ coactivator-1α |
| PSM’s | peptide-spectrum matches |
| RAA | rotenone-antimycin A |
| UCP1 | uncoupling protein 1 |
| WAT | white adipose tissue |
References
- Prentice, A.M. The emerging epidemic of obesity in developing countries. Int. J. Epidemiol. 2006, 35, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Rössner, S. Obesity: The disease of the twenty-first century. Int. J. Obes. Relat. Metab. Disord. 2002, 26, S2–S4. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Report on Diabetes; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; IDF: Brussels, Belgium, 2019. [Google Scholar]
- Bassuk, S.S.; Manson, J. Epidemiological evidence for the role of physical activity in reducing risk of type 2 diabetes and cardiovascular disease. J. Appl. Physiol. 2005, 99, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, M.J.; Blair, S.N.; Church, T.S. Physical activity and diabetes prevention. J. Appl. Physiol. 2005, 99, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, S.P.; Ragland, D.R.; Leung, R.W.; Paffenbarger, R.S., Jr. Physical activity and reduced occurrence of non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 1991, 325, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Manson, J.E.; Nathan, D.M.; Krolewski, A.S.; Stampfer, M.J.; Willett, W.C.; Hennekens, C.H. A prospective study of exercise and incidence of diabetes among US male physicians. JAMA 1992, 268, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Moschen, A.R. Adipocytokines: Mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 2006, 6, 772–783. [Google Scholar] [CrossRef]
- Rabe, K.; Lehrke, M.; Parhofer, K.G.; Broedl, U.C. Adipokines and insulin resistance. Mol. Med. 2008, 14, 741–751. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Saltiel, A.R. Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 2010, 121, 2111–2117. [Google Scholar] [CrossRef]
- Sakurai, T.; Ogasawara, J.; Shirato, K.; Izawa, T.; Oh-ishi, S.; Ishibashi, Y.; Radák, Z.; Ohno, H.; Kizaki, T. Exercise training attenuates the dysregulated expression of adipokines and oxidative stress in white adipose tissue. Oxid. Med. Cell. Longev. 2017, 2017, 9410954. [Google Scholar] [CrossRef]
- Sakurai, T.; Ogasawara, J.; Kizaki, T.; Sato, S.; Ishibashi, Y.; Takahashi, M.; Kobayashi, O.; Oh-Ishi, S.; Nagasawa, J.; Takahashi, K.; et al. The effects of exercise training on obesity- induced dysregulated expression of adipokines in white adipose tissue. Int. J. Endocrinol. 2013, 2013, 801743. [Google Scholar] [CrossRef]
- Bahler, L.; Molenaars, R.J.; Verberne, H.J.; Holleman, F. Role of the autonomic nervous system in activation of human brown adipose tissue: A review of the literature. Diabetes Metab. 2015, 41, 437–445. [Google Scholar] [CrossRef]
- Bargut, T.C.L.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Brown adipose tissue: Updates in cellular and molecular biology. Tissue Cell 2016, 48, 452–460. [Google Scholar] [CrossRef]
- Van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerbäck, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Kameya, T.; Nakada, K.; Kawai, Y.; Saito, M. Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity 2011, 19, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Kayahara, T.; Kameya, T.; Kawai, Y.; Iwanaga, T.; Saito, M. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 2013, 123, 3404–3408. [Google Scholar]
- Mulya, A.; Kirwan, J.P. Brown and beige adipose tissue: Therapy for obesity and its comorbidities? Endocrinol. Metab. Clin. N. Am. 2016, 45, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, J.; Seale, P. Medicine. Beige can be slimming. Science 2010, 328, 1113–1114. [Google Scholar] [CrossRef]
- Dewal, R.S.; Stanford, K.I. Effects of exercise on brown and beige adipocytes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, F.; Cereijo, R.; Villarroya, J.; Giralt, M. Brown adipose tissue as a secretory organ. Nat. Rev. Endocrinol. 2017, 13, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.X.; Zhao, X.Y.; Lin, J.D. The brown fat secretome: Metabolic functions beyond thermogenesis. Trends Endocrinol. Metab. 2015, 26, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, J.; Cereijo, R.; Gavaldà-Navarro, A.; Peyrou, M.; Giralt, M.; Villarroya, F. New insights into the secretory functions of brown adipose tissue. J. Endocrinol. 2019, 243, R19–R27. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.S.; Peijs, L.; Beaudry, J.L.; Jespersen, N.Z.; Nielsen, C.H.; Ma, T.; Brunner, A.D.; Larsen, T.J.; Bayarri-Olmos, R.; Prabhakar, B.S.; et al. Proteomics-based comparative mapping of the secretomes of human brown and white adipocytes reveals EPDR1 as a novel batokine. Cell Metab. 2019, 30, 963–975. [Google Scholar] [CrossRef]
- Irie, Y.; Asano, A.; Cañas, X.; Nikami, H.; Aizawa, S.; Saito, M. Immortal brown adipocytes from p53-knockout mice: Differentiation and expression of uncoupling proteins. Biochem. Biophys. Res. Commun. 1999, 255, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, J.; Cereijo, R.; Giralt, M.; Villarroya, F. Secretory proteome of brown adipocytes in response to cAMP-mediated thermogenic activation. Front. Physiol. 2019, 10, 67. [Google Scholar] [CrossRef]
- Ali Khan, A.; Hansson, J.; Weber, P.; Foehr, S.; Krijgsveld, J.; Herzig, S.; Scheideler, M. Comparative secretome analyses of primary murine white and brown adipocytes reveal novel adipokines. Mol. Cell Proteom. 2018, 17, 2358–2370. [Google Scholar] [CrossRef]
- Caspersen, C.J.; Powell, K.E.; Christenson, G.M. Physical activity, exercise, and physical fitness: Definitions and distinctions for health-related research. Publ. Health Rep. 1985, 100, 126–131. [Google Scholar]
- Manzanares, G.; Brito-da-Silva, G.; Gandra, P.G. Voluntary wheel running: Patterns and physiological effects in mice. Braz. J. Med. Biol. Res. 2018, 52, e7830. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.L.; Jeon, J.Y.; Liu, F.F.; Maratos-Flier, E. Voluntary exercise improves insulin sensitivity and adipose tissue inflammation in diet-induced obese mice. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E586–E594. [Google Scholar] [CrossRef] [PubMed]
- Uldry, M.; Yang, W.; St-Pierre, J.; Lin, J.; Seale, P.; Spiegelman, B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, B.; Li, M.; Speakman, J.R. Switching on the furnace: Regulation of heat production in brown adipose tissue. Mol. Aspects Med. 2019, 68, 60–73. [Google Scholar] [CrossRef]
- Kobayashi, J. Pre-heparin lipoprotein lipase mass. J Atheroscler. Thromb. 2004, 11, 1–5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Carmona, C.M.; Iglesias, R.; Obregón, M.J.; Darlington, D.J.; Villarroya, F.; Giralt, M. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein α. J. Biol. Chem. 2002, 277, 21489–21498. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef]
- Menini, S.; Iacobini, C.; Blasetti Fantauzzi, C.; Pesce, C.M.; Pugliese, G. Role of galectin-3 in obesity and impaired glucose homeostasis. Oxid. Med. Cell Longev. 2016, 2016, 9618092. [Google Scholar] [CrossRef]
- Krautbauer, S.; Eisinger, K.; Hader, Y.; Buechler, C. Free fatty acids and IL-6 induce adipocyte galectin-3 which is increased in white and brown adipose tissues of obese mice. Cytokine 2014, 69, 263–271. [Google Scholar] [CrossRef]
- Sato, S.; Hughes, R.C. Regulation of secretion and surface expression of Mac-2, galactoside-binding protein of macrophages. J. Biol. Chem. 1994, 269, 4424–4430. [Google Scholar] [CrossRef]
- Malik, A.N.; Czajka, A.; Cunningham, P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 2016, 29, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; von Stebut, M. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 2004, 36, 1882–1886. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.S.; Kawada, T.; Yoo, H.; Kwon, B.S.; Yu, R. Macrophage inflammatory protein-related protein-2, a novel CC chemokine, can regulate preadipocyte migration and adipocyte differentiation. FEBS Lett. 2003, 548, 125–130. [Google Scholar] [CrossRef]
- Baek, J.H.; Kim, S.J.; Kang, H.G.; Lee, H.W.; Kim, J.H.; Hwang, K.A.; Song, J.; Chun, K.H. Galectin-3 activates PPARγ and supports white adipose tissue formation and high-fat diet-induced obesity. Endocrinology 2015, 156, 147–156. [Google Scholar] [CrossRef]
- Pejnovic, N.N.; Pantic, J.M.; Jovanovic, I.P.; Radosavljevic, G.D.; Milovanovic, M.Z.; Nikolic, I.G.; Zdravkovic, N.S.; Djukic, A.L.; Arsenijevic, N.N.; Lukic, M.L. Galectin-3 deficiency accelerates high-fat diet-induced obesity and amplifies inflammation in adipose tissue and pancreatic islets. Diabetes 2013, 62, 1932–1944. [Google Scholar] [CrossRef]
- Pang, J.; Rhodes, D.H.; Pini, M.; Akasheh, R.T.; Castellanos, K.J.; Cabay, R.J.; Cooper, D.; Perretti, M.; Fantuzzi, G. Increased adiposity, dysregulated glucose metabolism and systemic inflammation in galectin-3 KO mice. PLoS ONE 2013, 8, e57915. [Google Scholar] [CrossRef]
- Grassadonia, A.; Tinari, N.; Iurisci, I.; Piccolo, E.; Cumashi, A.; Innominato, P.; D’Egidio, M.; Natoli, C.; Piantelli, M.; Iacobelli, S. 90K (Mac-2 BP) and galectins in tumor progression and metastasis. Glycoconj. J. 2002, 19, 551–556. [Google Scholar] [CrossRef]
- Challa, T.D.; Straub, L.G.; Balaz, M.; Kiehlmann, E.; Donze, O.; Rudofsky, G.; Ukropec, J.; Ukropcova, B.; Wolfrum, C. Regulation of De Novo Adipocyte Differentiation Through Cross Talk Between Adipocytes and Preadipocytes. Diabetes 2015, 64, 4075–4087. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, M.; Hu, Q.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy. Int. J. Mol. Med. 2018, 41, 599–614. [Google Scholar] [CrossRef]
- Li, L.H.; Li, J.; Gao, J. Functions of galectin-3 and its role in fibrotic diseases. J. Pharmacol. Exp. Ther. 2014, 351, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Shaked, I.; Hanna, D.B.; Gleißner, C.; Marsh, B.; Plants, J.; Tracy, D.; Anastos, K.; Cohen, M.; Golub, E.T.; Karim, R.; et al. Macrophage inflammatory markers are associated with subclinical carotid artery disease in women with human immunodeficiency virus or hepatitis C virus infection. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Christoffolete, M.A.; Linardi, C.C.; de Jesus, L.; Ebina, K.N.; Carvalho, S.D.; Ribeiro, M.O.; Rabelo, R.; Curcio, C.; Martins, L.; Kimura, E.T.; et al. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes 2004, 53, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.V.; Bikopoulos, G.; Hung, S.; Ceddia, R.B. Thermogenic capacity is antagonistically regulated in classical brown and white subcutaneous fat depots by high fat diet and endurance training in rats. J. Biol. Chem. 2014, 289, 34129–34140. [Google Scholar] [CrossRef]
- Oh-Ishi, S.; Kizaki, T.; Toshinai, K.; Haga, S.; Fukuda, K.; Nagata, N.; Ohno, H. Swimming training improves brown-adipose-tissue activity in young and old mice. Mech. Ageing Dev. 1996, 89, 67–78. [Google Scholar] [CrossRef]
- Lehnig, A.C.; Stanford, K.I. Exercise-induced adaptations to white and brown adipose tissue. J. Exp. Biol. 2018, 221 (Suppl. S1), jeb161570. [Google Scholar] [CrossRef]
- Yoshida, S.; Fukutomi, T.; Kimura, T.; Sakurai, H.; Hatano, R.; Yamamoto, H.; Mukaisho, K.; Hattori, T.; Sugihara, H.; Asano, S. Comprehensive proteome analysis of brush border membrane fraction of ileum of ezrin knockdown mice. Biomed. Res. 2016, 37, 127–139. [Google Scholar] [CrossRef]
- Wang, G.; Wu, W.W.; Zhang, Z.; Masilamani, S.; Shen, R.F. Decoy methods for assessing false positives and false discovery rates in shotgun proteomics. Anal Chem. 2009, 81, 146–159. [Google Scholar] [CrossRef]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Van Dam, A.D.; Nahon, K.J.; Kooijman, S.; van den Berg, S.M.; Kanhai, A.A.; Kikuchi, T.; Heemskerk, M.M.; van Harmelen, V.; Lombès, M.; van den Hoek, A.M.; et al. Salsalate activates brown adipose tissue in mice. Diabetes 2015, 64, 1544–1554. [Google Scholar] [CrossRef]
- Xu, G.; Xia, Z.; Deng, F.; Liu, L.; Wang, Q.; Yu, Y.; Wang, F.; Zhu, C.; Liu, W.; Ceng, Z.; et al. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLoS Pathog. 2019, 15, e1008002. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).