Genome Maintenance Mechanisms at the Chromatin Level
Abstract
1. Introduction
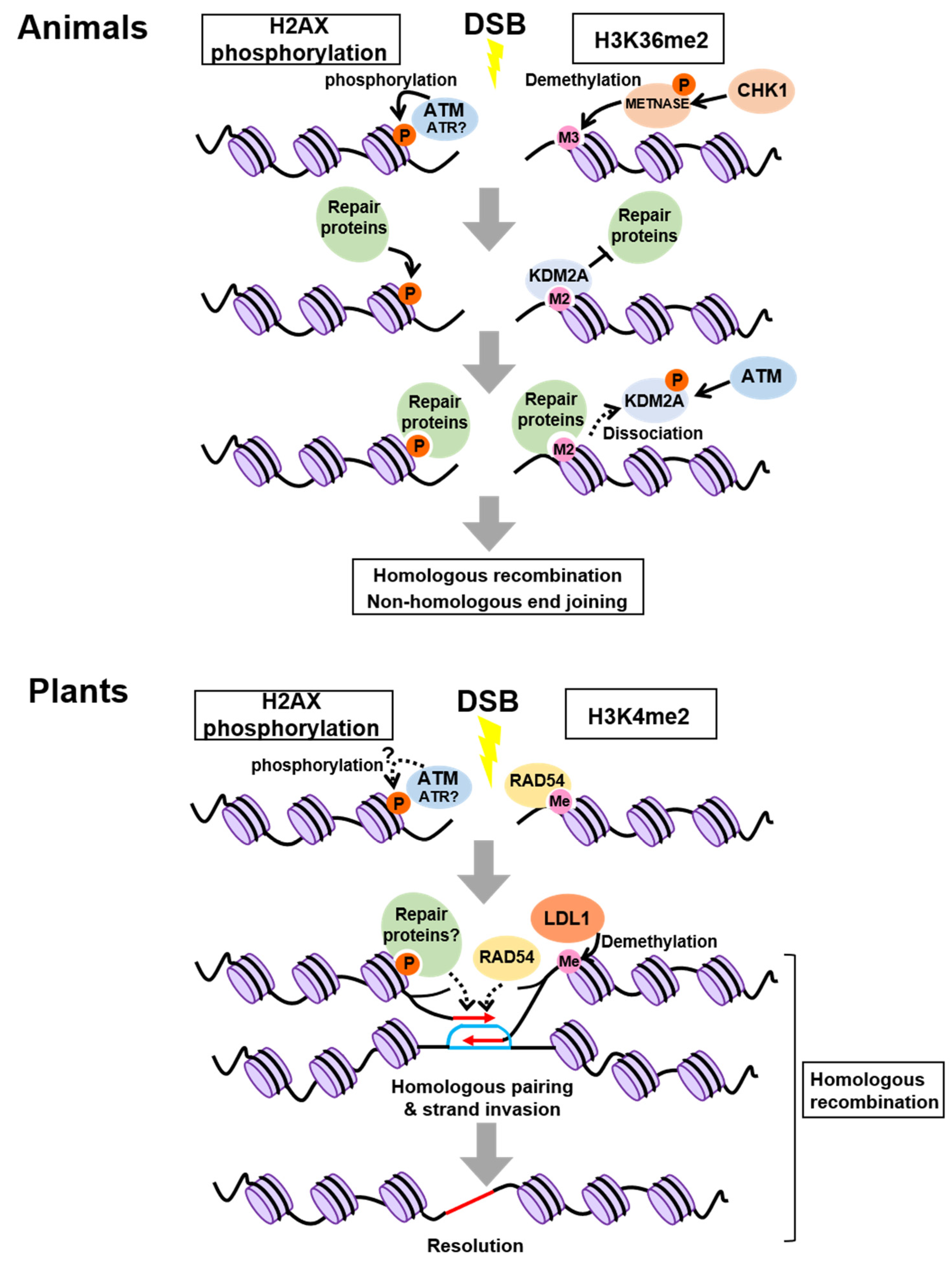
2. Control of Chromatin Modifications at Damaged Sites
2.1. In Animals
2.2. In Plants
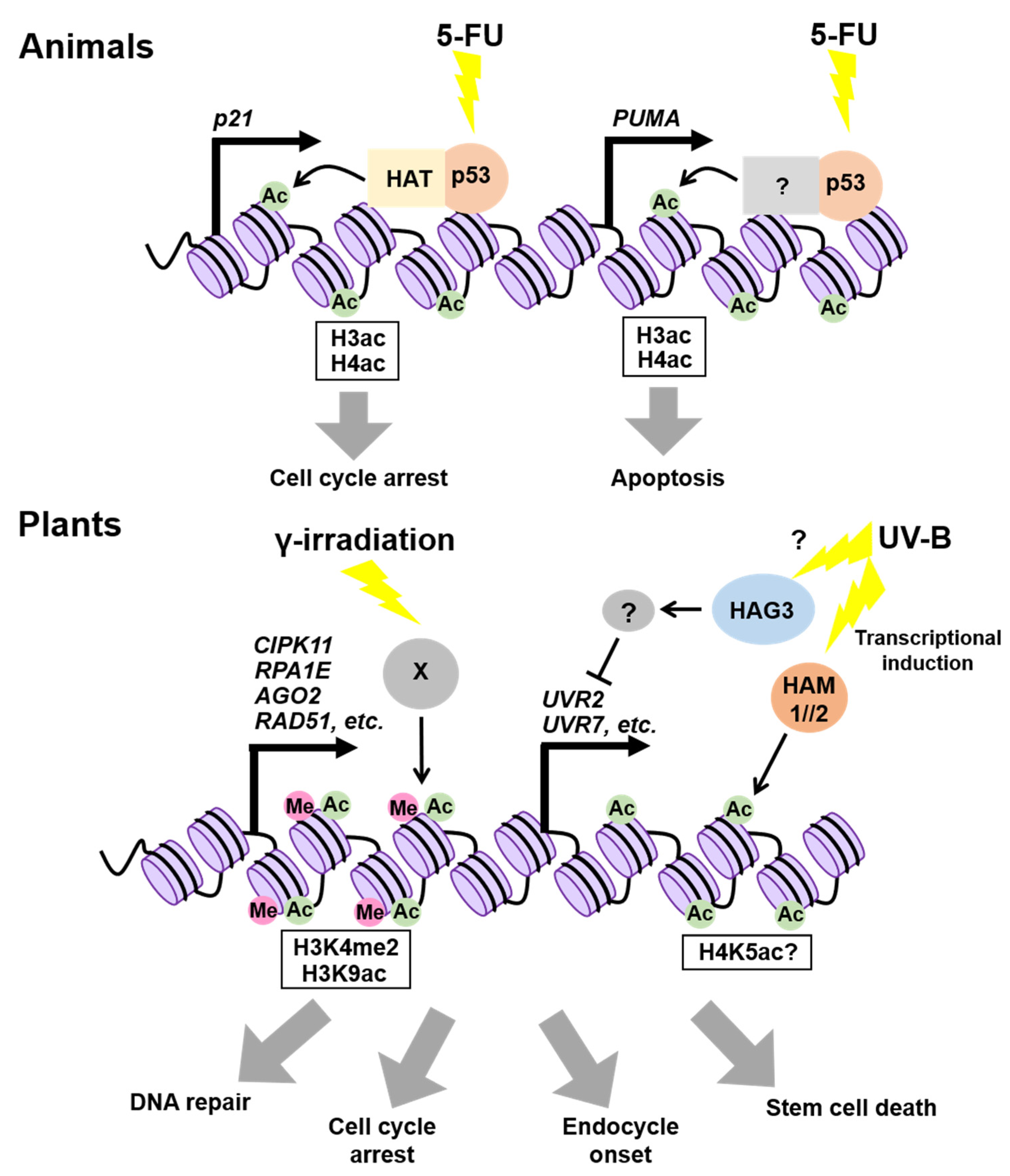
3. Epigenetic Regulation of DDR-Related Genes
3.1. In Animals
3.2. In Plants
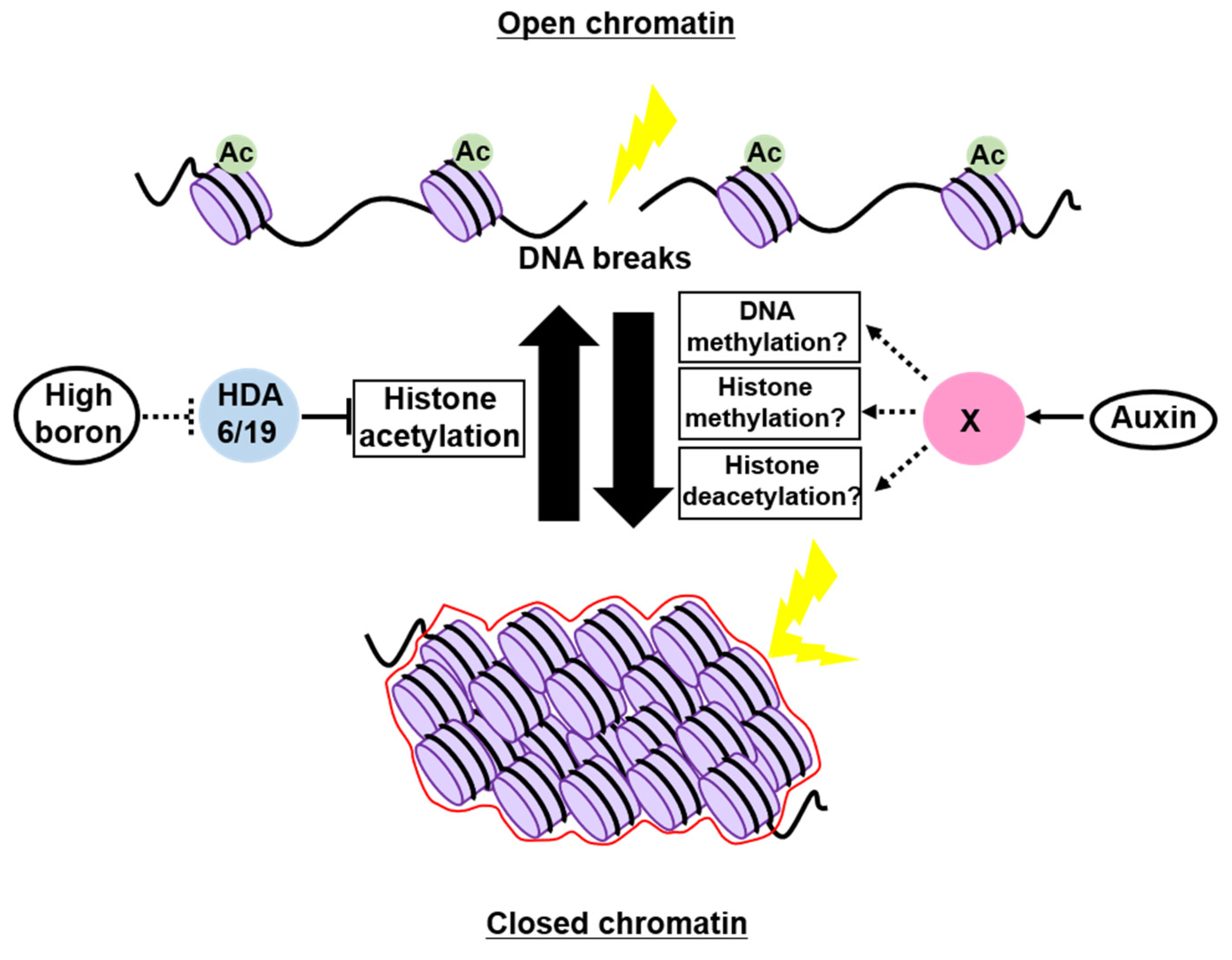
4. Possible Roles of Chromatin Structure as a Physical Barrier to Genotoxic Stress
4.1. In Animals
4.2. In Plants
5. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Britt, A.B. Repair of DNA damage induced by solar UV. Photosynth. Res. 2004, 81, 105–112. [Google Scholar] [CrossRef]
- Banaś, A.K.; Zgłobicki, P.; Kowalska, E.; Bażant, A.; Dziga, D.; Strzałka, W. All you need is light. Photorepair of uv-induced pyrimidine dimers. Genes 2020, 11, 1304. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef] [PubMed]
- Culligan, K.M.; Hays, J.B. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7 form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell 2000, 12, 991–1002. [Google Scholar] [CrossRef]
- Lario, L.D.; Ramirez-Parra, E.; Gutierrez, C.; Casati, P.; Spampinato, C.P. Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J. Exp. Bot. 2011, 62, 2925–2937. [Google Scholar] [CrossRef]
- Hohn, B.; Puchta, H. Gene therapy in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 8321–8323. [Google Scholar] [CrossRef] [PubMed]
- Waterworth, W.M.; Drury, G.E.; Bray, C.M.; West, C.E. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011, 192, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001, 15, 2177–2196. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Culligan, K.M.; Robertson, C.E.; Foreman, J.; Doerner, P.; Britt, A.B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006, 48, 947–961. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britt, A.B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Ueda, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef]
- Cools, T.; Iantcheva, A.; Weimer, A.K.; Boens, S.; Takahashi, N.; Maes, S.; van den Daele, H.; van Isterdael, G.; Schnittger, A.; de Veylder, L. The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 2011, 23, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Wang, H.; Song, J.; Cao, X.; Rogers, H.J.; Francis, D.; Jia, C.; Sun, L.; Hou, M.; Yang, Y.; et al. Cell cycle arrest mediated by Cd-induced DNA damage in Arabidopsis root tips. Ecotoxicol. Environ. Saf. 2017, 145, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Takatsuka, H.; Takahashi, N.; Kurata, R.; Fukao, Y.; Kobayashi, K.; Ito, M.; Umeda, M. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat. Commun. 2017, 8, 635. [Google Scholar] [CrossRef] [PubMed]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of suppressor of gamma response 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Ogita, N.; Takahashi, T.; Taniguchi, S.; Tanaka, M.; Seki, M.; Umeda, M. A regulatory module controlling stress-induced cell cycle arrest in arabidopsis. eLife 2019, 8, e43944. [Google Scholar] [CrossRef]
- Fulcher, N.; Sablowski, R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 2009, 106, 20984–20988. [Google Scholar] [CrossRef]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Kawashima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.; Schnittger, A. Endoreplication-a means to an end in cell growth and stress response. Curr. Opin. Plant Biol. 2020, 54, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.J.; Zhivotovsky, B. DNA damage-induced apoptosis. Oncogene 2004, 23, 2797–2808. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- Scheid, R.; Chen, J.; Zhong, X. Biological role and mechanism of chromatin readers in plants. Curr. Opin. Plant Biol. 2021, 61, 102008. [Google Scholar] [CrossRef]
- Hashimshony, T.; Zhang, J.; Keshet, I.; Bustin, M.; Cedar, H. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 2003, 34, 187–192. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Arfuso, F.; Arumugam, S.; Chinnathambi, A.; Jinsong, B.; Warrier, S.; Wang, L.Z.; Kumar, A.P.; Ahn, K.S.; Sethi, G.; et al. Role of novel histone modifications in cancer. Oncotarget 2018, 9, 11414–11426. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jacobsen, S.E. Epigenetic modifications in plants: An evolutionary perspective. Curr. Opin. Plant Biol. 2011, 14, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Umeda, M. Epigenetic control of cell division and cell differentiation in the root apex. Front. Plant Sci. 2015, 6, 1178. [Google Scholar] [CrossRef] [PubMed]
- Donà, M.; Mittelsten Scheid, O. DNA damage repair in the context of plant chromatin. Plant Physiol. 2015, 168, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Chromatin remodeling and epigenetic regulation in plant DNA damage repair. Int. J. Mol. Sci. 2019, 20, 4093. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef] [PubMed]
- Ward, I.M.; Chen, J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001, 276, 47759–47762. [Google Scholar] [CrossRef] [PubMed]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. γh2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Fnu, S.; Williamson, E.A.; De Haro, L.P.; Brenneman, M.; Wray, J.; Shaheen, M.; Radhakrishnan, K.; Lee, S.H.; Nickoloff, J.A.; Hromas, R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2011, 108, 540–545. [Google Scholar] [CrossRef]
- Hromas, R.; Williamson, E.A.; Fnu, S.; Lee, Y.J.; Park, S.J.; Beck, B.D.; You, J.S.; Laitao, A.; Nickoloff, J.A.; Lee, S.H. Chk1 phosphorylation of Metnase enhances DNA repair but inhibits replication fork restart. Oncogene 2012, 31, 4245–4254. [Google Scholar] [CrossRef]
- Cao, L.L.; Wei, F.; Du, Y.; Song, B.; Wang, D.; Shen, C.; Lu, X.; Cao, Z.; Yang, Q.; Gao, Y.; et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 2016, 35, 301–313. [Google Scholar] [CrossRef]
- Wei, S.; Li, C.; Yin, Z.; Wen, J.; Meng, H.; Xue, L.; Wang, J. Histone methylation in DNA repair and clinical practice: New findings during the past 5-years. J. Cancer 2018, 9, 2072–2081. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Noon, A.T.; Deckbar, D.; Ziv, Y.; Shiloh, Y.; Löbrich, M.; Jeggo, P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 2008, 31, 167–177. [Google Scholar] [CrossRef]
- Noon, A.T.; Shibata, A.; Rief, N.; Löbrich, M.; Stewart, G.S.; Jeggo, P.A.; Goodarzi, A.A. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 2010, 12, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. Roles for the DNA-PK complex and 53BP1 in protecting ends from resection during DNA double-strand break repair. J. Radiat. Res. 2020, 61, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef]
- Falk, M.; Hausmann, M. A paradigm revolution or just better resolution—Will newly emerging superresolution techniques identify chromatin architecture as a key factor in radiation-induced dna damage and repair regulation? Cancers 2021, 13, 18. [Google Scholar] [CrossRef]
- Janssen, A.; Colmenares, S.U.; Lee, T.; Karpen, G.H. Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4A. Genes Dev. 2019, 33, 103–115. [Google Scholar] [CrossRef]
- Charbonnel, C.; Allain, E.; Gallego, M.E.; White, C.I. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 2011, 10, 611–619. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Wilson, M.; Wang, D.; Nuhse, T.; Warward, S.; Selley, J.; West, C.E. Phosphoproteomic analysis reveals plant DNA damage signalling pathways with a functional role for histone H2AX phosphorylation in plant growth under genotoxic stress. Plant J. 2019, 100, 1007–1021. [Google Scholar] [CrossRef]
- Friesner, J.D.; Liu, B.; Culligan, K.; Britt, A.B. Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 2005, 16, 2566–2576. [Google Scholar] [CrossRef]
- Hirakawa, T.; Kuwata, K.; Gallego, M.E.; White, C.I.; Nomoto, M.; Tada, Y.; Matsunaga, S. LSD1-LIKE1-mediated H3K4me2 demethylation is required for homologous recombination repair. Plant Physiol. 2019, 181, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Mazin, A.V.; Alexeev, A.A.; Kowalczykowski, S.C. A novel function of Rad54 protein: Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003, 278, 14029–14036. [Google Scholar] [CrossRef]
- Jacob, Y.; Feng, S.; LeBlanc, C.A.; Bernatavichute, Y.V.; Stroud, H.; Cokus, S.; Johnson, L.M.; Pellegrini, M.; Jacobsen, S.E.; Michaels, S.D. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 763–768. [Google Scholar] [CrossRef]
- Jacob, Y.; Stroud, H.; Leblanc, C.; Feng, S.; Zhuo, L.; Caro, E.; Hassel, C.; Gutierrez, C.; Michaels, S.D.; Jacobsen, S.E. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 2010, 466, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.E.; Michaels, S.D. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar] [CrossRef]
- Costa, R.M.A.; Lima, W.C.; Vogel, C.I.G.; Berra, C.M.; Luche, D.D.; Medina-Silva, R.; Galhardo, R.S.; Menck, C.F.M.; Oliveira, V.R. DNA repair-related genes in sugarcane expressed sequence tags (ESTs). Genet. Mol. Biol. 2001, 24, 131–140. [Google Scholar] [CrossRef]
- Singh, S.K.; Roy, S.; Choudhury, S.R.; Sengupta, D.N. DNA repair and recombination in higher plants: Insights from comparative genomics of arabidopsis and rice. BMC Genom. 2010, 11, 443. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Zhang, T.; Wang, Y.; Cui, Z.; He, Y. Zmrad51c is essential for double-strand break repair and homologous recombination in maize meiosis. Int. J. Mol. Sci. 2019, 20, 5513. [Google Scholar] [CrossRef]
- Horn, H.F.; Vousden, K.H. Coping with stress: Multiple ways to activate p53. Oncogene 2007, 26, 1306–1316. [Google Scholar] [CrossRef]
- Soussi, T. The history of p53. A perfect example of the drawbacks of scientific paradigms. EMBO Rep. 2010, 11, 822–826. [Google Scholar] [CrossRef]
- Barnoud, T.; Indeglia, A.; Murphy, M.E. Shifting the paradigms for tumor suppression: Lessons from the p53 field. Oncogene 2021, 40, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Hernández Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta -Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Barlev, N.A.; Liu, L.; Chehab, N.H.; Mansfield, K.; Harris, K.G.; Halazonetis, T.D.; Berger, S.L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 2001, 8, 1243–1254. [Google Scholar] [CrossRef]
- Kaeser, M.D.; Iggo, R.D. Promoter-specific p53-dependent histone acetylation following DNA damage. Oncogene 2004, 23, 4007–4013. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaneshiro, K.; Tsutsumi, S.; Tsuji, S.; Shirahige, K.; Aburatani, H. An integrated map of p53-binding sites and histone modification in the human ENCODE regions. Genomics 2007, 89, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.O. SOG1: A master regulator of the DNA damage responsein plants. Genes Genet. Syst. 2016, 90, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.O.; Kaminoyama, K.; Sakamoto, T.; Kimura, S. Increased phosphorylation of Ser-Gln sites on SUPPRESSOR OF GAMMA RESPONSE1 strengthens the DNA damage response in Arabidopsis thaliana. Plant Cell 2017, 29, 3255–3268. [Google Scholar] [CrossRef]
- Furukawa, T.; Curtis, M.J.; Tominey, C.M.; Duong, Y.H.; Wilcox, B.W.L.; Aggoune, D.; Hays, J.B.; Britt, A.B. A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair 2010, 9, 940–948. [Google Scholar] [CrossRef]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The plant-specific CDKB -CYCB 1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef] [PubMed]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef]
- Mondal, S.; Go, Y.S.; Lee, S.S.; Chung, B.Y.; Kim, J.H. Characterization of histone modifications associated with DNA damage repair genes upon exposure to gamma rays in Arabidopsis seedlings. J. Radiat. Res. 2016, 57, 646–654. [Google Scholar] [CrossRef]
- Servet, C.; Conde, E. Silva, N.; Zhou, D.X. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed]
- Britt, A.B. DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Campi, M.; D’Andrea, L.; Emiliani, J.; Casati, P. Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol. 2012, 158, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhang, H.; Xing, L.; Xu, S.; Liu, H.; Chong, K.; Xu, Y. Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J. Plant Physiol. 2013, 170, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Fina, J.P.; Casati, P. HAG3, a histone acetyltransferase, affects UV-B responses by negatively regulating the expression of DNA repair enzymes and sunscreen content in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 1388–1400. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, Y.; Mori, T.; Magome, N.; Hibino, K.; Yoshikawa, K. DNA compaction plays a key role in radioprotection against double-strand breaks as revealed by single-molecule observation. Chem. Phys. Lett. 2008, 456, 80–83. [Google Scholar] [CrossRef]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin compaction protects genomic DNA from radiation damage. PLoS ONE 2013, 8, e75622. [Google Scholar] [CrossRef]
- Brambilla, F.; Garcia-Manteiga, J.M.; Monteleone, E.; Hoelzen, L.; Zocchi, A.; Agresti, A.; Bianchi, M.E. Nucleosomes effectively shield DNA from radiation damage in living cells. Nucleic Acids Res. 2020, 48, 8993–9006. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Yan, W.X.; Mirzazadeh, R.; Garnerone, S.; Scott, D.; Schneider, M.W.; Kallas, T.; Custodio, J.; Wernersson, E.; Li, Y.; Gao, L.; et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017, 8, 15058. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Schuster, B.S.; Reed, E.H.; Parthasarathy, R.; Jahnke, C.N.; Caldwell, R.M.; Bermudez, J.G.; Ramage, H.; Good, M.C.; Hammer, D.A. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 2018, 9, 2985. [Google Scholar] [CrossRef]
- Shireen, F.; Nawaz, M.A.; Chen, C.; Zhang, Q.; Zheng, Z.; Sohail, H.; Sun, J.; Cao, H.; Huang, Y.; Bie, Z. Boron: Functions and approaches to enhance its availability in plants for sustainable agriculture. Int. J. Mol. Sci. 2018, 19, 1856. [Google Scholar] [CrossRef]
- Sakamoto, T.; Inui, Y.T.; Uraguchi, S.; Yoshizumi, T.; Matsunaga, S.; Mastui, M.; Umeda, M.; Fukui, K.; Fujiwara, T. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in arabidopsis. Plant Cell 2011, 23, 3533–3546. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Tsujimoto-Inui, Y.; Sotta, N.; Hirakawa, T.; Matsunaga, T.M.; Fukao, Y.; Matsunaga, S.; Fujiwara, T. Proteasomal degradation of BRAHMA promotes Boron tolerance in Arabidopsis. Nat. Commun. 2018, 9, 5285. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yu, C.W.; Chen, F.F.; Zhao, L.; Tian, G.; Liu, X.; Cui, Y.; Yang, J.Y.; Wu, K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 2012, 8, e1003114. [Google Scholar] [CrossRef]
- Chen, W.Q.; Drapek, C.; Li, D.X.; Xu, Z.H.; Benfey, P.N.; Bai, S.N. Histone deacetylase HDA19 affects root cortical cell fate by interacting with SCARECROW. Plant Physiol. 2019, 180, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, J.; Sakamoto, T.; Fujimoto, S.; Yamashita, T.; Suzuki, T.; Matsunaga, S. Auxin decreases chromatin accessibility through the TIR1/AFBs auxin signaling pathway in proliferative cells. Sci. Rep. 2018, 8, 7773. [Google Scholar] [CrossRef]
- Armengot, L.; Moreno-Romero, J. Micrococcal Nuclease (MNase) assay of Arabidopsis thaliana nuclei. Bio-Protocol 2013, 3, 3–8. [Google Scholar] [CrossRef]
- Orzechowska, M.; Stępień, K.; Kamińska, T.; Siwińska, D. Chromosome variations in regenerants of Arabidopsis thaliana derived from 2- and 6-week-old callus detected using flow cytometry and FISH analyses. Plant Cell. Tissue Organ Cult. 2013, 112, 263–273. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018, 23, 235–247. [Google Scholar] [CrossRef]
- Vanneste, S.; Maes, L.; De Smet, I.; Himanen, K.; Naudts, M.; Inzé, D.; Beeckman, T. Auxin regulation of cell cycle and its role during lateral root initiation. Physiol. Plant. 2005, 123, 139–146. [Google Scholar] [CrossRef]
- Ishida, T.; Adachi, S.; Yoshimura, M.; Shimizu, K.; Umeda, M.; Sugimoto, K. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 2010, 137, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Desvoyes, B.; Fernández-Marcos, M.; Sequeira-Mendes, J.; Otero, S.; Vergara, Z.; Gutierrez, C. Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 2014, 5, 369. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Inagaki, S.; Nishimura, K.; Sakakibara, H. Alterations in hormonal signals spatially coordinate distinct responses to DNA double-strand breaks in Arabidopsis roots. Sci. Adv. 2021, 7, eabg0993. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Kannivadi Ramakanth, K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell 2017, 170, 102–113. [Google Scholar] [CrossRef]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318. [Google Scholar] [CrossRef]
- Yadav, R.K.; Girke, T.; Pasala, S.; Xie, M.; Reddy, G.V. Gene expression map of the Arabidopsis shoot apical meri stem stem cell niche. Proc. Natl. Acad. Sci. USA 2009, 106, 4941–4946. [Google Scholar] [CrossRef] [PubMed]
- Mihandoost, E.; Shirazi, A.; Mahdavi, S.R.; Aliasgharzadeh, A. Consequences of lethal-whole-body gamma radiation and possible ameliorative role of melatonin. Sci. World J. 2014, 2014, 621570. [Google Scholar] [CrossRef] [PubMed]
- Einset, J.; Collins, A.R. Genome size and sensitivity to DNA damage by X-rays-Plant comets tell the story. Mutagenesis 2018, 33, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Tu, X.; Chu, P.Y.; Lü, P.; Zhu, N.; Grierson, D.; Du, B.; Li, P.; Zhong, S. 3D Chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 2017, 10, 1497–1509. [Google Scholar] [CrossRef]
- Liu, C.; Cheng, Y.J.; Wang, J.W.; Weigel, D. Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 2017, 3, 742–748. [Google Scholar] [CrossRef]
- Feng, S.; Cokus, S.J.; Schubert, V.; Zhai, J.; Pellegrini, M.; Jacobsen, S.E. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 2014, 55, 694–707. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Roukos, V.; Misteli, T. The biogenesis of chromosome translocations. Nat. Cell Biol. 2014, 16, 293–300. [Google Scholar] [CrossRef]
- Avramova, Z.V. Heterochromatin in animals and plants. Similarities and differences. Plant Physiol. 2002, 129, 40–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dong, P.; Tu, X.; Liang, Z.; Kang, B.H.; Zhong, S. Plant and animal chromatin three-dimensional organization: Similar structures but different functions. J. Exp. Bot. 2020, 71, 5119–5128. [Google Scholar] [CrossRef] [PubMed]
- Kochanova, N.Y.; Schauer, T.; Mathias, G.P.; Lukacs, A.; Schmidt, A.; Flatley, A.; Schepers, A.; Thomae, A.W.; Imhof, A. A multi-layered structure of the interphase chromocenter revealed by proximity-based biotinylation. Nucleic Acids Res. 2021, 48, 4161–4178. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; Scheid, O.M. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Kurita, K.; Sakamoto, T.; Yagi, N.; Sakamoto, Y.; Ito, A.; Nishino, N.; Sako, K.; Yoshida, M.; Kimura, H.; Seki, M.; et al. Live imaging of H3K9 acetylation in plant cells. Sci. Rep. 2017, 7, 45894. [Google Scholar] [CrossRef]
- Yadav, V.K.; Santos-González, J.; Köhler, C. INT-Hi-C reveals distinct chromatin architecture in endosperm and leaf tissues of Arabidopsis. Nucleic Acids Res. 2021, 49, 4371–4385. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takatsuka, H.; Shibata, A.; Umeda, M. Genome Maintenance Mechanisms at the Chromatin Level. Int. J. Mol. Sci. 2021, 22, 10384. https://doi.org/10.3390/ijms221910384
Takatsuka H, Shibata A, Umeda M. Genome Maintenance Mechanisms at the Chromatin Level. International Journal of Molecular Sciences. 2021; 22(19):10384. https://doi.org/10.3390/ijms221910384
Chicago/Turabian StyleTakatsuka, Hirotomo, Atsushi Shibata, and Masaaki Umeda. 2021. "Genome Maintenance Mechanisms at the Chromatin Level" International Journal of Molecular Sciences 22, no. 19: 10384. https://doi.org/10.3390/ijms221910384
APA StyleTakatsuka, H., Shibata, A., & Umeda, M. (2021). Genome Maintenance Mechanisms at the Chromatin Level. International Journal of Molecular Sciences, 22(19), 10384. https://doi.org/10.3390/ijms221910384





