Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin
Abstract
:1. Introduction
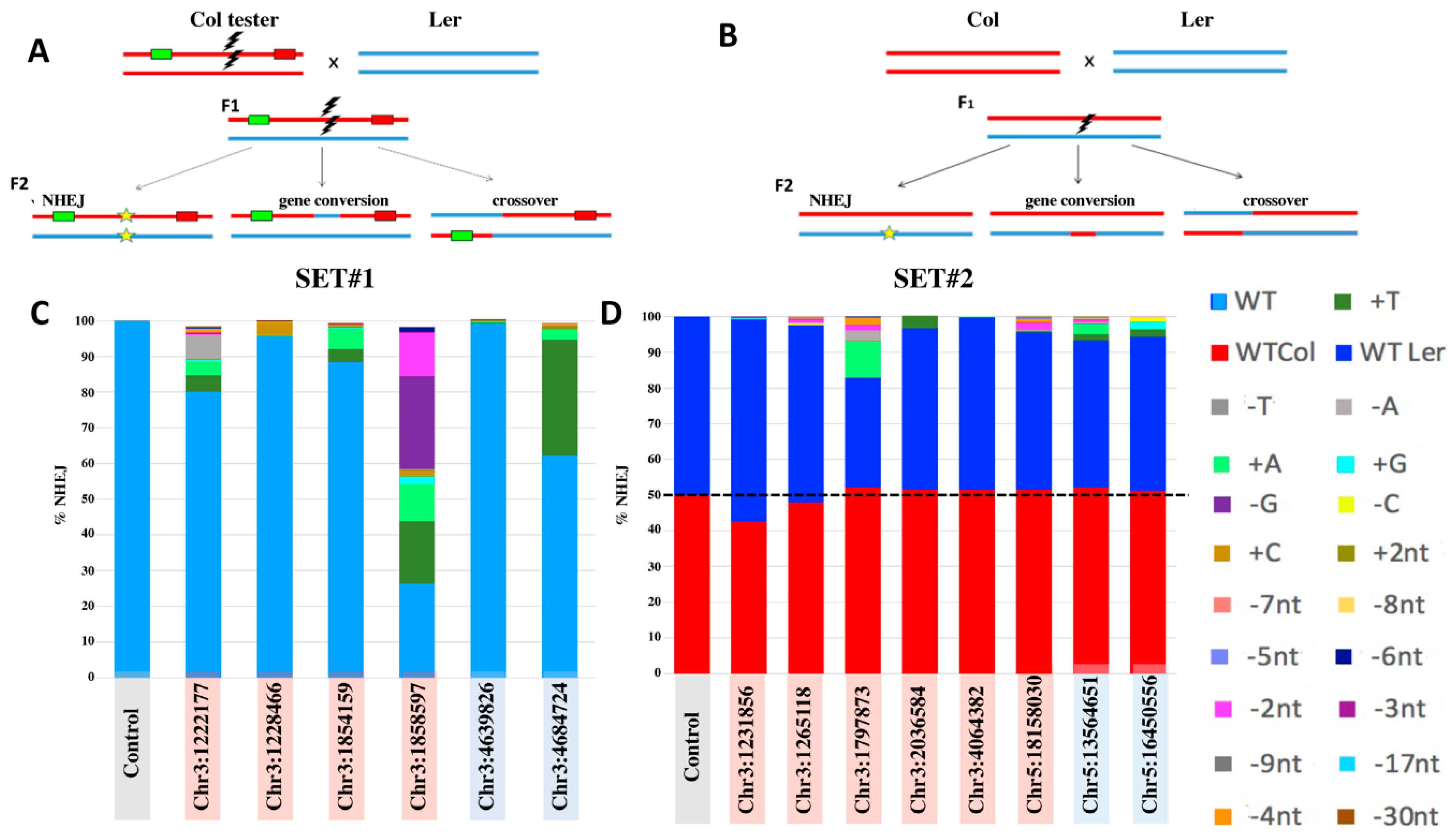
2. Results
2.1. Selection of gRNA for DSB-Induction
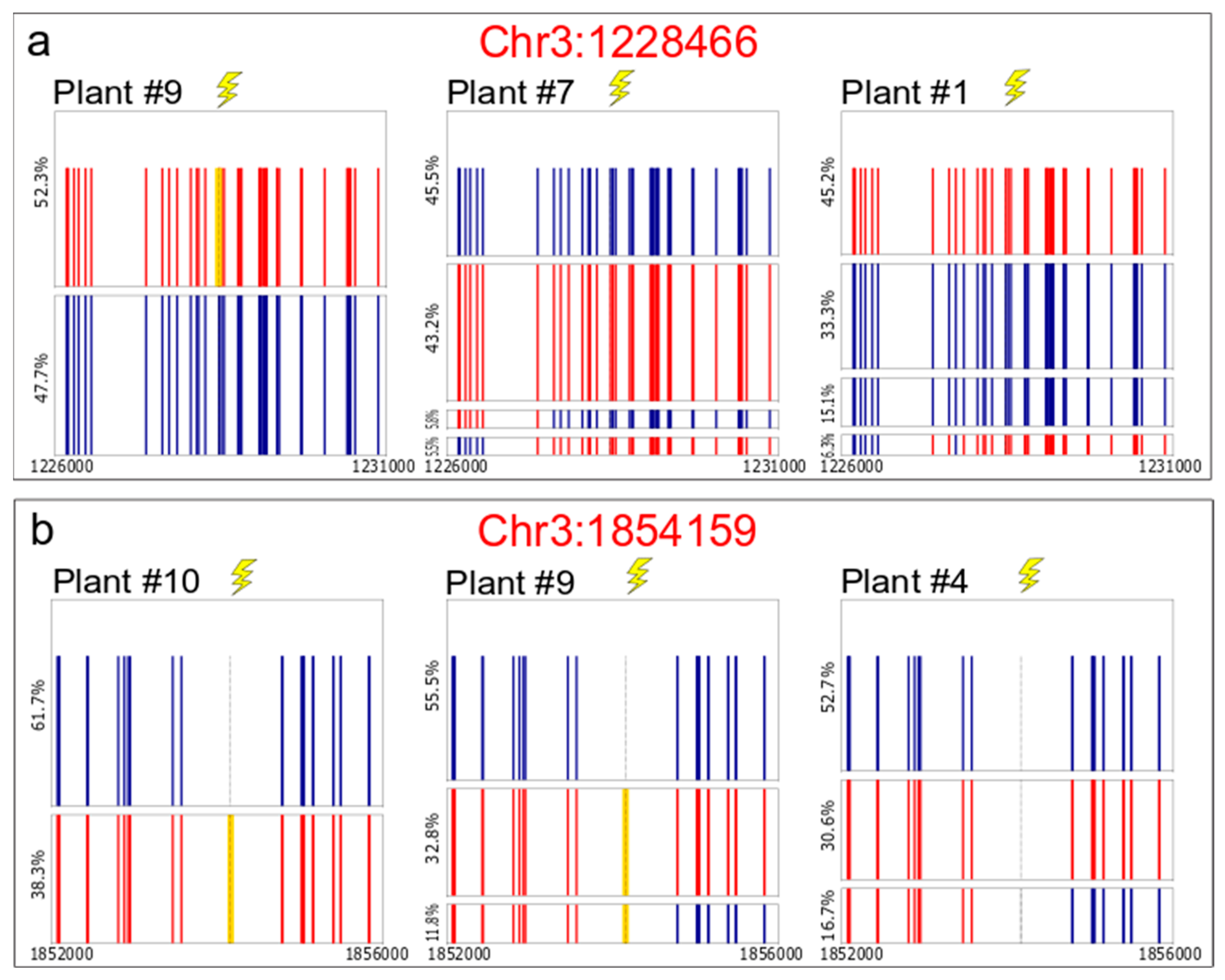
2.2. Analysis of Targeted IHR Events
2.3. PacBio® Sequencing for Somatic Recombination Analysis
3. Discussion
3.1. IHR Frequency
3.2. Conversion Tract Patterns
3.3. Somatic IHR
3.4. IHR in Euchromatin versus Heterochromatin
4. Materials and Methods
4.1. Plant Material
4.2. Plasmids and Plant Transformation
4.3. DNA Amplification and Sequencing
4.4. Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deriano, L.; Roth, D.B. Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annu. Rev. Genet 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of Homologous Recombination in Eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of Eukaryotic Homologous Recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The Molecular Biology of Meiosis in Plants. Annu. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef] [PubMed]
- Spampinato, C.P. Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.; Fauser, F.; Puchta, H. DNA recombination in somatic plant cells: Mechanisms and evolutionary consequences. Chromosome Res. 2014, 22, 191–201. [Google Scholar] [CrossRef]
- Charbonnel, C.; Allain, E.; Gallego, M.E.; White, C.I. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair 2011, 10, 611–619. [Google Scholar] [CrossRef]
- Puchta, H.; Dujon, B.; Hohn, B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 5055–5060. [Google Scholar] [CrossRef] [Green Version]
- Shalev, G.; Levy, A.A. The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 1997, 146, 1143–1151. [Google Scholar] [CrossRef]
- Marton, I.; Zuker, A.; Shklarman, E.; Zeevi, V.; Tovkach, A.; Roffe, S.; Ovadis, M.; Tzfira, T.; Vainstein, A. Nontransgenic genome modification in plant cells. Plant Physiol. 2010, 154, 1079–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Schiml, S.; Fauser, F.; Puchta, H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014, 80, 1139–1150. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef] [Green Version]
- Fauser, F.; Roth, N.; Pacher, M.; Ilg, G.; Sánchez-Fernández, R.; Biesgen, C.; Puchta, H. In planta gene targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 7535–7540. [Google Scholar] [CrossRef] [Green Version]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA Replicons for Plant Genome Engineering. Plant Cell 2014, 26, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [Green Version]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018. [Google Scholar] [CrossRef] [Green Version]
- Molinier, J.; Ries, G.; Bonhoeffer, S.; Hohn, B. Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 2004, 16, 342–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, G.T.H.; Cao, H.X.; Watanabe, K.; Hensel, G.; Blattner, F.R.; Kumlehn, J.; Schubert, I. Repair of Site-Specific DNA Double-Strand Breaks in Barley Occurs via Diverse Pathways Primarily Involving the Sister Chromatid. Plant Cell 2014, 26, 2156–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filler Hayut, S.; Melamed Bessudo, C.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 15605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Mudgett, M.; Rambabu, R.; Abramson, B.; Dai, X.; Michael, T.P.; Zhao, Y. Selective inheritance of target genes from only one parent of sexually reproduced F1 progeny in Arabidopsis. Nat. Commun. 2021, 12, 3854. [Google Scholar] [CrossRef]
- Shilo, S.; Melamed-Bessudo, C.; Dorone, Y.; Barkai, N.; Levy, A.A. DNA Crossover Motifs Associated with Epigenetic Modifications Delineate Open Chromatin Regions in Arabidopsis. Plant Cell 2015, 27, 2427–2436. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.; Zhao, X.; Kelly, K.A.; Venn, O.; Higgins, J.D.; Yelina, N.E.; Hardcastle, T.J.; Ziolkowski, P.A.; Copenhaver, G.P.; Chris, F.; et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 2013, 45, 1327–1336. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wang, M.; Dukowic-Schulze, S.; Zhou, A.; Tiang, C.-L.; Shilo, S.; Sidhu, G.K.; Eichten, S.; Bradbury, P.; Springer, N.M.; et al. Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc. Natl. Acad. Sci. USA 2017, 114, 12231–12236. [Google Scholar] [CrossRef] [Green Version]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Darrier, B.; Rimbert, H.; Balfourier, F.; Pingault, L.; Josselin, A.-A.; Servin, B.; Navarro, J.; Choulet, F.; Paux, E.; Sourdille, P. High-Resolution Mapping of Crossover Events in the Hexaploid Wheat Genome Suggests a Universal Recombination Mechanism. Genetics 2017, 206, 1373–1388. [Google Scholar] [CrossRef] [Green Version]
- Yelina, N.E.; Lambing, C.; Hardcastle, T.J.; Zhao, X.; Santos, B.; Henderson, I.R. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 2015, 29, 2183–2202. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.; Zhao, X.; Tock, A.J.; Lambing, C.; Underwood, C.J.; Hardcastle, T.J.; Serra, H.; Kim, J.; Cho, H.S.; Kim, J.; et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018, 28, 532–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Keeney, S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn. Stab. 2008, 2, 81–123. [Google Scholar] [PubMed] [Green Version]
- Lange, J.; Yamada, S.; Tischfield, S.E.; Pan, J.; Kim, S.; Zhu, X.; Socci, N.D.; Jasin, M.; Keeney, S. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell 2016, 167, 695–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Roudier, F.; Ahmed, I.; Bérard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Lim, K.; Kim, J.-S.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef] [Green Version]
- Melamed-Bessudo, C.; Yehuda, E.; Stuitje, A.R.; Levy, A.A. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 2005, 43, 458–466. [Google Scholar] [CrossRef]
- Leenay, R.T.; Beisel, C.L. Deciphering, Communicating, and Engineering the CRISPR PAM. J. Mol. Biol. 2017, 429, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Puchta, H.; Swoboda, P.; Hohn, B. Induction of intrachromosomal homologous recombination in whole plants. Plant J. 1995, 7, 203–210. [Google Scholar] [CrossRef]
- Orel, N.; Kyryk, A.; Puchta, H. Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 2003, 35, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Gisler, B.; Salomon, S.; Puchta, H. The role of double-strand break-induced allelic homologous recombination in somatic plant cells. Plant J. 2002, 32, 277–284. [Google Scholar] [CrossRef] [Green Version]
- Ben Shlush, I.; Samach, A.; Melamed-Bessudo, C.; Ben-Tov, D.; Dahan-Meir, T.; Filler-Hayut, S.; Levy, A.A. CRISPR/Cas9 Induced Somatic Recombination at the CRTISOLocus in Tomato. Genes 2021, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Mancera, E.; Bourgon, R.; Brozzi, A.; Huber, W.; Steinmetz, L.M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 2008, 454, 479–485. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Dominska, M.; Yim, E.; Petes, T.D. High-resolution mapping of heteroduplex DNA formed during UV-induced and spontaneous mitotic recombination events in yeast. eLife 2017, 6, e28069. [Google Scholar] [CrossRef]
- Rukść, A.; Bell-Rogers, P.L.; Smith, J.D.L.; Baker, M.D. Analysis of Spontaneous Gene Conversion Tracts within and between Mammalian Chromosomes. J. Mol. Biol. 2008, 377, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Bitoun, E.; Altemose, N.; Davies, R.W.; Davies, B.; Myers, S.R. A high-resolution map of non-crossover events reveals impacts of genetic diversity on mammalian meiotic recombination. Nat. Commun. 2019, 10, 3900. [Google Scholar] [CrossRef] [Green Version]
- Lesecque, Y.; Mouchiroud, D.; Duret, L. GC-biased gene conversion in yeast is specifically associated with crossovers: Molecular mechanisms and evolutionary significance. Mol. Biol. Evol. 2013, 30, 1409–1419. [Google Scholar] [CrossRef] [Green Version]
- Brown, T.C.; Jiricny, J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell 1988, 54, 705–711. [Google Scholar] [CrossRef]
- Nasmyth, K.; Haering, C.H. Cohesin: Its Roles and Mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hum, Y.F.; Jinks-Robertson, S. Mismatch recognition and subsequent processing have distinct effects on mitotic recombination intermediates and outcomes in yeast. Nucleic Acids Res. 2019, 47, 4554–4568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piazza, A.; Wright, W.D.; Heyer, W.-D. Multi-invasions Are Recombination Byproducts that Induce Chromosomal Rearrangements. Cell 2017, 170, 760–773. [Google Scholar] [CrossRef]
- Gorbunova, V.; Levy, A.A. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997, 25, 4650–4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, E.; Levy, A.A. Abortive gap repair: Underlying mechanism for Ds element formation. Mol. Cell. Biol. 1997, 17, 6294–6302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Knapp, S. Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 1991, 96, 985–989. [Google Scholar] [CrossRef] [Green Version]
- Underwood, C.J.; Choi, K.; Lambing, C.; Zhao, X.; Serra, H.; Borges, F.; Simorowski, J.; Ernst, E.; Jacob, Y.; Henderson, I.R.; et al. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 2018, 28, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, E.J.; Griffin, C.H.; Henderson, I.R. Modification of meiotic recombination by natural variation in plants. J. Exp. Bot. 2017, 68, 5471–5483. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Giraut, L.; Falque, M.; Drouaud, J.; Pereira, L.; Martin, O.C.; Mézard, C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011, 7, e1002354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallimasioti-Pazi, E.M.; Chathoth, K.T.; Taylor, G.C.; Meynert, A.; Ballinger, T.; Kelder, M.J.E.; Lalevée, S.; Sanli, I.; Feil, R.; Wood, A.J. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 2018, 16, e2005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.; Breuer, G.A.; Brinkman, E.K.; van der Meulen, A.I.; Borden, S.V.; van Steensel, B.; Bindra, R.S.; LaRocque, J.R.; Karpen, G.H. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and Euchromatin. Genes Dev. 2016, 30, 1645–1657. [Google Scholar] [CrossRef] [Green Version]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPRCas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, D.; Yong, K.; Zhang, D. Varied Transcriptional Efficiencies of Multiple Arabidopsis U6 Small Nuclear RNA. Genes 2007, 49, 222–229. [Google Scholar] [CrossRef]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, S.I.; Juárez, P.; Fernández-del-Carmen, A.; Granell, A.; Orzaez, D. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLoS ONE 2011, 6, e21622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Blecher-Gonen, R.; Barnett-Itzhaki, Z.; Jaitin, D.; Amann-Zalcenstein, D.; Lara-Astiaso, D.; Amit, I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013, 8, 539–554. [Google Scholar] [CrossRef]
- Fulton, T.M.; Chunzoongse, J.; Tanksley, S.D. Microprep Protocol for Extraction of DNA from Tomato and other Herbaceous Plants. Plant Mol. Biol. Rep. 1195, 13, 207–209. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filler-Hayut, S.; Kniazev, K.; Melamed-Bessudo, C.; Levy, A.A. Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin. Int. J. Mol. Sci. 2021, 22, 12096. https://doi.org/10.3390/ijms222212096
Filler-Hayut S, Kniazev K, Melamed-Bessudo C, Levy AA. Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin. International Journal of Molecular Sciences. 2021; 22(22):12096. https://doi.org/10.3390/ijms222212096
Chicago/Turabian StyleFiller-Hayut, Shdema, Kiril Kniazev, Cathy Melamed-Bessudo, and Avraham A. Levy. 2021. "Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin" International Journal of Molecular Sciences 22, no. 22: 12096. https://doi.org/10.3390/ijms222212096
APA StyleFiller-Hayut, S., Kniazev, K., Melamed-Bessudo, C., & Levy, A. A. (2021). Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin. International Journal of Molecular Sciences, 22(22), 12096. https://doi.org/10.3390/ijms222212096





