Differential Association of 4E-BP2-Interacting Proteins Is Related to Selective Delayed Neuronal Death after Ischemia
Abstract
:1. Introduction
2. Results
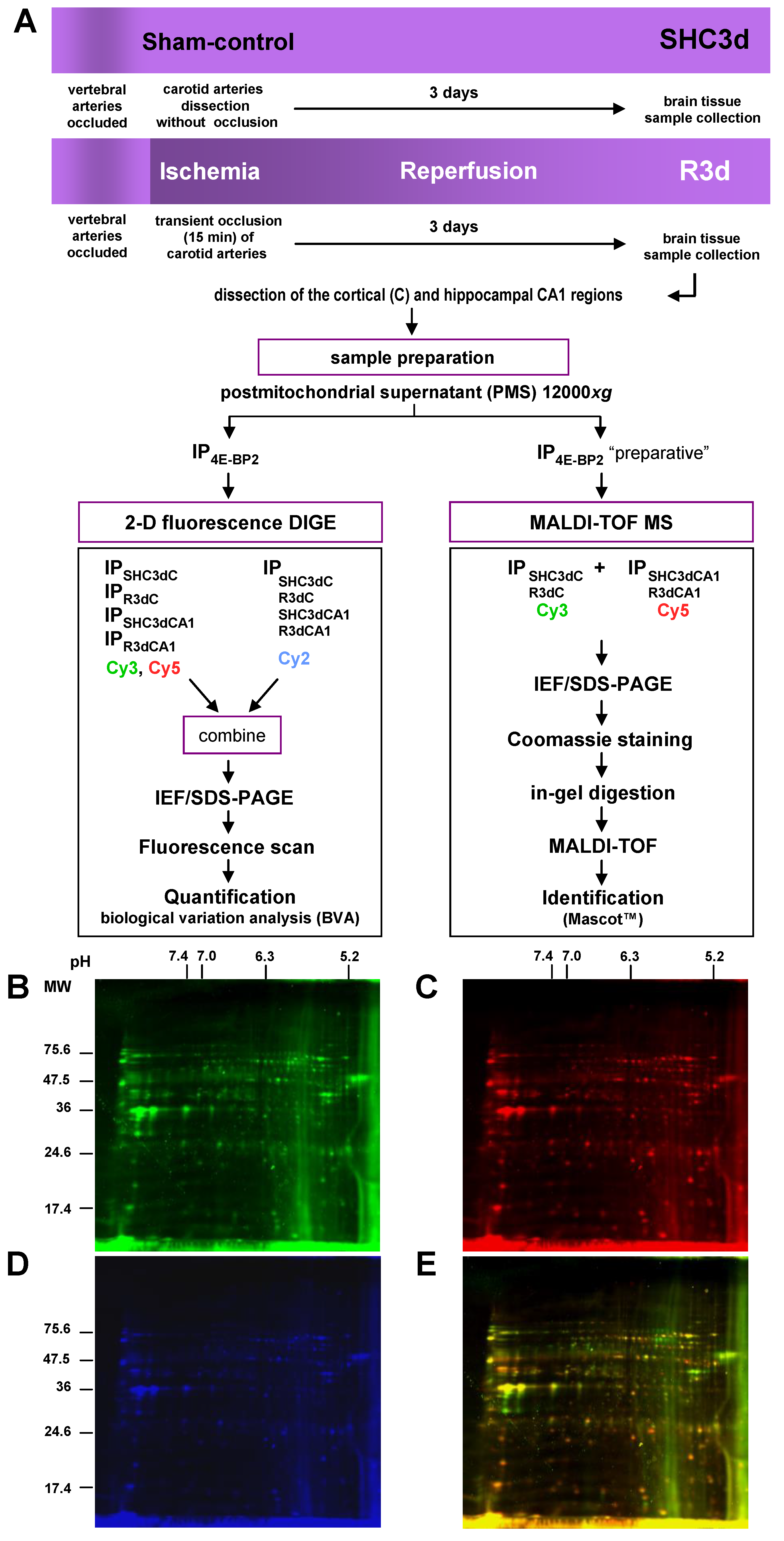
2.1. Differential Detection of eIF4E-Binding Protein 2 (4E-BP2)-Interacting Proteins in Response to Ischemia-Reperfusion (IR)
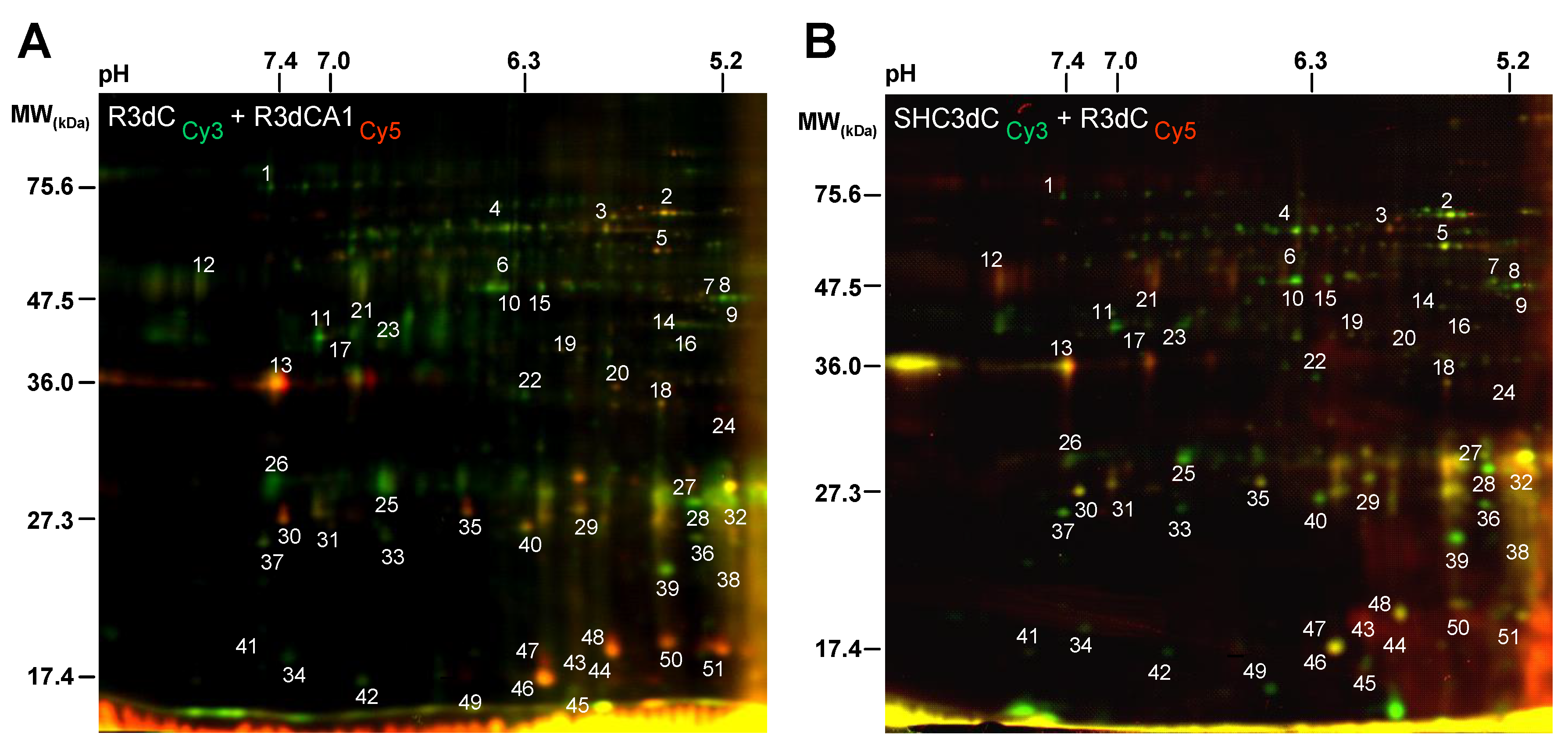
2.2. Biological Variation Analysis of the Differentially Detected 4E-BP2-Interacting Proteins
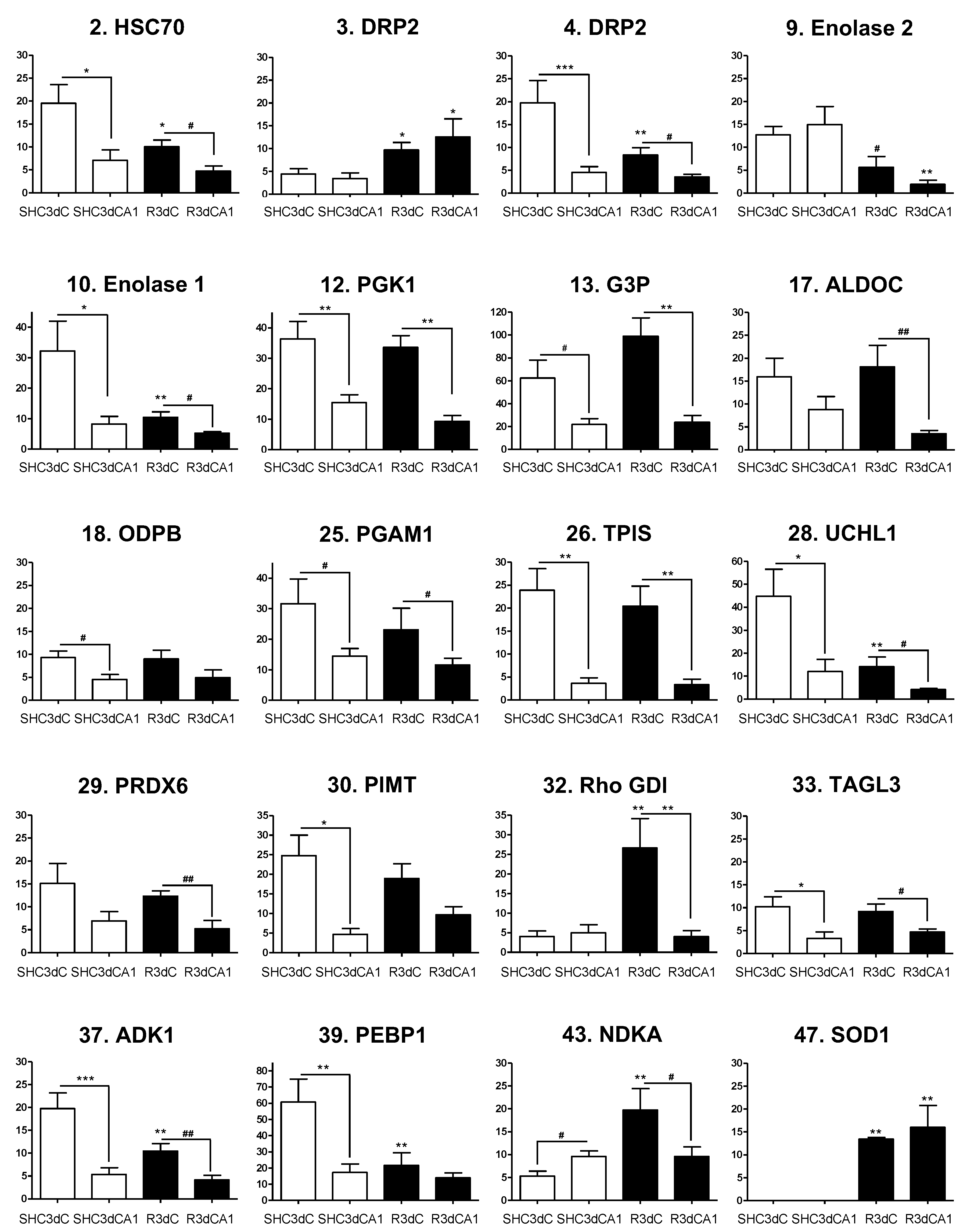
2.3. Identification of 4E-BP2-Interacting Proteins
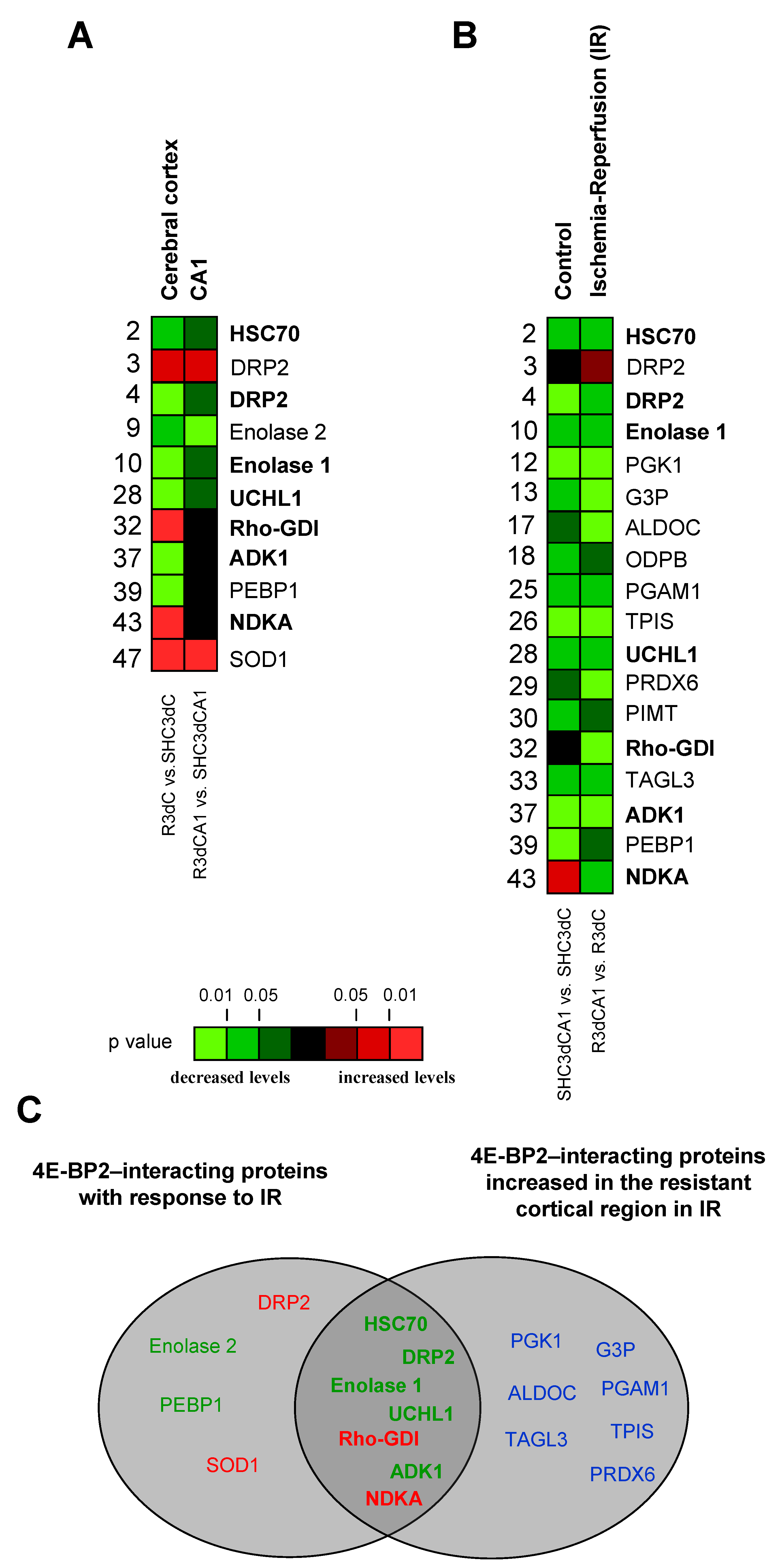
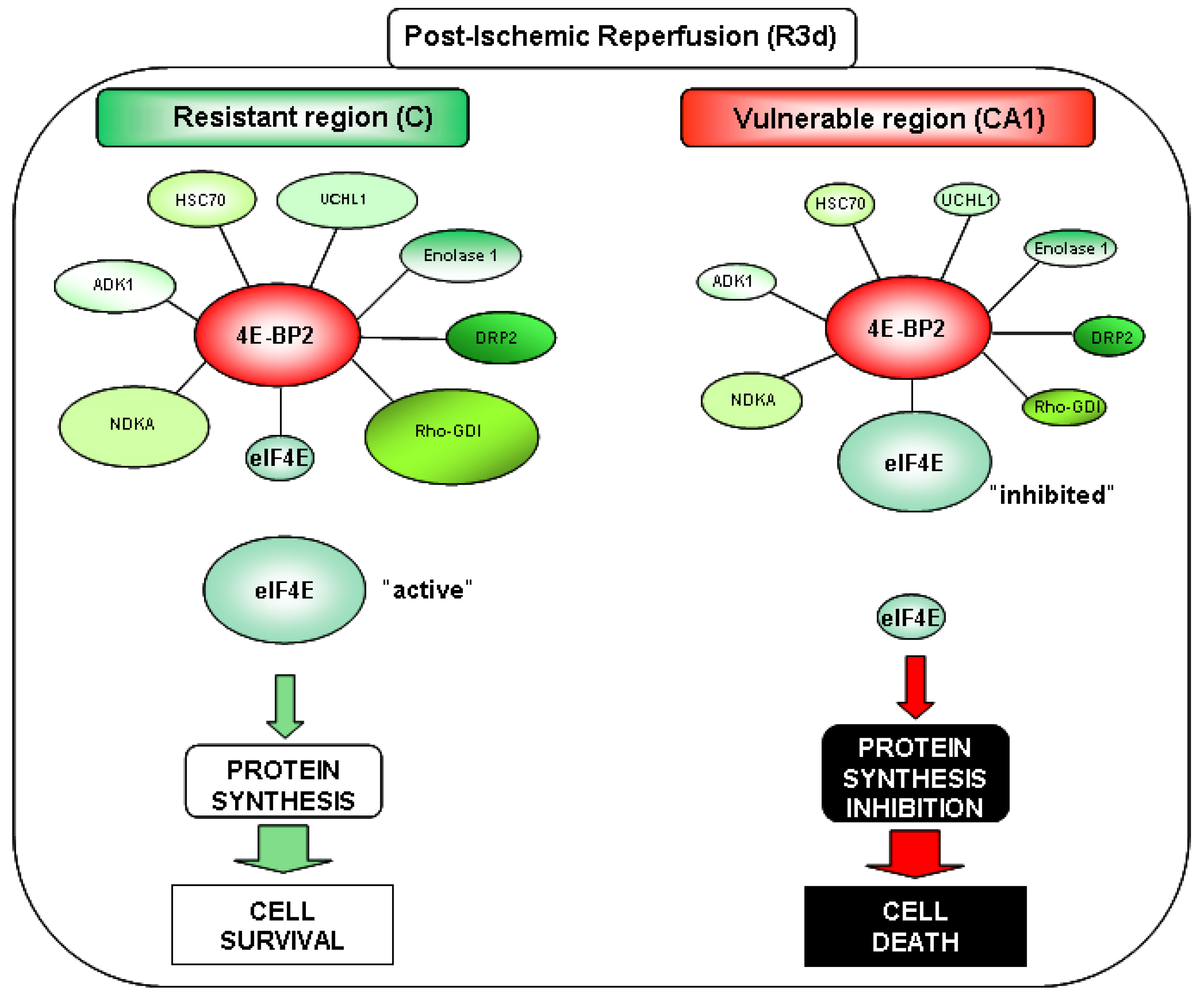
2.4. Changes in the Levels of 4E-BP2-Interacting Proteins in the Cerebral Cortex and Hippocampal CA1 Region under IR. Identification of Biomarkers of Protection or Vulnerability in Cerebral Ischemia
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animal Model of Cerebral Ischemia and Ischemia-Reperfusion
4.3. Brain Tissue Samples
4.4. Immunoprecipitation
4.5. Western Blot Analysis
4.6. Two-Dimensional Fluorescence Difference in Gel Electrophoresis
4.7. Protein Quantification
4.8. Protein Identification by MALDI-TOF Mass Spectrometry
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fukuyo, A.; In, Y.; Ishida, T.; Tomoo, K. Structural scaffold for eIF4E binding selectivity of 4E-BP isoforms: Crystal structure of eIF4E binding region of 4E-BP2 and its comparison with that of 4E-BP1. J. Pept. Sci. 2011, 17, 650–657. [Google Scholar] [CrossRef]
- Haghighat, A.; Mader, S.; Pause, A.; Sonenberg, N. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995, 14, 5701–5709. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Herdy, B.; Costa-Mattioli, M.; Gingras, A.C.; Raught, B.; Sonenberg, N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005, 25, 10556–10565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkogkas, C.G.; Khoutorsky, A.; Ran, I.; Rampakakis, E.; Nevarko, T.; Weatherill, D.B.; Vasuta, C.; Yee, S.; Truitt, M.; Dallaire, P.; et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013, 493, 371–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saran, U.; Foti, M.; Dufour, J.F. Cellular and molecular effects of the mTOR inhibitor everolimus. Clin. Sci. 2015, 129, 895–914. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.Z.; Yao, Q.; Li, H.H. The rationale of targeting mammalian target of rapamycin for ischemic stroke. Cell Signal 2013, 25, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Hartman, N.W.; Lin, T.V.; Zhang, L.; Paquelet, G.E.; Feliciano, D.M.; Bordey, A. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep. 2013, 5, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Raught, B.; Gingras, A.C. Signaling to translation initiation. In Translational Control in Biology and Medicine; Mathews, M.B., Sonenberg, N., Hershey, J.W.B., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2007; pp. 369–401. [Google Scholar]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef]
- Hidalgo, M.; Le Bouffant, R.; Bello, V.; Buisson, N.; Cormier, P.; Beaudry, M.; Darribere, T. The translational repressor 4E-BP mediates hypoxia-induced defects in myotome cells. J. Cell Sci. 2012, 125, 3989–4000. [Google Scholar] [CrossRef] [Green Version]
- Tsukiyama-Kohara, K.; Vidal, S.M.; Gingras, A.C.; Glover, T.W.; Hanash, S.M.; Heng, H.; Sonenberg, N. Tissue distribution, genomic structure, and chromosome mapping of mouse and human eukaryotic initiation factor 4E-binding proteins 1 and 2. Genomics 1996, 38, 353–363. [Google Scholar] [CrossRef]
- Bidinosti, M.; Ran, I.; Sanchez-Carbente, M.R.; Martineau, Y.; Gingras, A.C.; Gkogkas, C.; Raught, B.; Bramham, C.R.; Sossin, W.S.; Costa-Mattioli, M.; et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol. Cell 2010, 37, 797–808. [Google Scholar] [CrossRef] [Green Version]
- Ayuso, M.I.; Martinez-Alonso, E.; Salvador, N.; Bonova, P.; Regidor, I.; Alcazar, A. Dissociation of eIF4E-binding protein 2 (4E-BP2) from eIF4E independent of Thr37/Thr46 phosphorylation in the ischemic stress response. PLoS ONE 2015, 10, e0121958. [Google Scholar]
- Chien, S. Cerebral blood flow and metabolism. In Principles of Neural Science, 2nd ed.; Kandel, E.R., Schwartz, J.H., Eds.; Elsevier: Amsterdam, The Netherlands, 1985; pp. 845–852. [Google Scholar]
- Lipton, P. Ischemic cell death in brain neurons. Physiol. Rev. 1999, 79, 1431–1568. [Google Scholar] [CrossRef] [PubMed]
- White, B.C.; Sullivan, J.M.; DeGracia, D.J.; O′Neil, B.J.; Neumar, R.W.; Grossman, L.I.; Rafols, J.A.; Krause, G.S. Brain ischemia and reperfusion: Molecular mechanisms of neuronal injury. J. Neurol. Sci. 2000, 179, 1–33. [Google Scholar] [CrossRef]
- Thilmann, R.; Xie, Y.; Kleihues, P.; Kiessling, M. Persistent inhibition of protein synthesis precedes delayed neuronal death in postischemic gerbil hippocampus. Acta Neuropathol. 1986, 71, 88–93. [Google Scholar] [CrossRef]
- Hossmann, K.A. Disturbances of cerebral protein synthesis and ischemic cell death. Prog. Brain Res. 1993, 96, 161–177. [Google Scholar]
- Kirino, T. Delayed neuronal death. Neuropathology 2000, 20, S95–S97. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Lorsch, J.R.; Hellen, C.U.T. The mechanism of translation initation in eucaryotes. In Translational Control in Biology and Medicine; Mathews, M.B., Sonenberg, N., Hershey, J.M.W., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2007; pp. 87–128. [Google Scholar]
- Ayuso, M.I.; Martinez-Alonso, E.; Cid, C.; Alonso de Lecinana, M.; Alcazar, A. The translational repressor eIF4E-binding protein 2 (4E-BP2) correlates with selective delayed neuronal death after ischemia. J. Cereb. Blood Flow Metab. 2013, 33, 1173–1181. [Google Scholar] [CrossRef]
- Pulsinelli, W.A.; Brierley, J.B.; Plum, F. Temporal profile of neuronal damage in a model of transient forebrain ischemia. Ann. Neurol. 1982, 11, 491–498. [Google Scholar] [CrossRef]
- Wollenhaupt, K.; Brussow, K.P.; Albrecht, D.; Tomek, W. The eIF4E repressor protein 4E-BP2 is merely truncated, despite 4E-BP1 degradation in the porcine uterine tissue during implantation. Mol. Reprod. Dev. 2012, 79, 767–776. [Google Scholar] [CrossRef]
- Schaller, B.; Graf, R. Cerebral ischemia and reperfusion: The pathophysiologic concept as a basis for clinical therapy. J. Cereb. Blood Flow Metab. 2004, 24, 351–371. [Google Scholar] [CrossRef] [Green Version]
- Burda, J.; Martin, M.E.; Garcia, A.; Alcazar, A.; Fando, J.L.; Salinas, M. Phosphorylation of the alpha subunit of initiation factor 2 correlates with the inhibition of translation following transient cerebral ischaemia in the rat. Biochem. J. 1994, 302, 335–338. [Google Scholar] [CrossRef]
- DeGracia, D.J.; Neumar, R.W.; White, B.C.; Krause, G.S. Global brain ischemia and reperfusion: Modifications in eukaryotic initiation factors associated with inhibition of translation initiation. J. Neurochem. 1996, 67, 2005–2012. [Google Scholar] [CrossRef] [Green Version]
- DeGracia, D.J.; Hu, B.R. Irreversible translation arrest in the reperfused brain. J. Cereb. Blood Flow Metab. 2007, 27, 875–893. [Google Scholar] [CrossRef] [Green Version]
- Cid, C.; Garcia-Bonilla, L.; Camafeita, E.; Burda, J.; Salinas, M.; Alcazar, A. Proteomic characterization of protein phosphatase 1 complexes in ischemia-reperfusion and ischemic tolerance. Proteomics 2007, 7, 3207–3218. [Google Scholar] [CrossRef]
- Chen, A.; Liao, W.P.; Lu, Q.; Wong, W.S.; Wong, P.T. Upregulation of dihydropyrimidinase-related protein 2, spectrin alpha II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats—A proteomics approach. Neurochem. Int. 2007, 50, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, E.; Rosell, A.; Colome, N.; Hernandez-Guillamon, M.; Garcia-Berrocoso, T.; Ribo, M.; Alcazar, A.; Ortega-Aznar, A.; Salinas, M.; Canals, F.; et al. The proteome of human brain after ischemic stroke. J. Neuropathol. Exp. Neurol. 2010, 69, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, P.O. Proteomic analysis of focal cerebral ischemic injury in male rats. J. Vet. Med. Sci. 2010, 72, 181–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, P.O. Identification of proteins differentially expressed in cerebral cortexes of Ginkgo biloba extract (EGb761)-treated rats in a middle cerebral artery occlusion model—A proteomics approach. Am. J. Chin. Med. 2011, 39, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Hori, M.; Nakamachi, T.; Rakwal, R.; Shibato, J.; Ogawa, T.; Aiuchi, T.; Tsuruyama, T.; Tamaki, K.; Shioda, S. Transcriptomics and proteomics analyses of the PACAP38 influenced ischemic brain in permanent middle cerebral artery occlusion model mice. J. Neuroinflamm. 2012, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos-Martorell, M.; Salvador, N.; Monge, M.; Canals, F.; Garcia-Bonilla, L.; Hernandez-Guillamon, M.; Ayuso, M.I.; Chacon, P.; Rosell, A.; Alcazar, A.; et al. Brain proteomics identifies potential simvastatin targets in acute phase of stroke in a rat embolic model. J. Neurochem. 2014, 130, 301–312. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Q.; Lin, H.; Li, S.; Sun, L.; Yang, Y. HSP72 and gp96 in gastroenterological cancers. Clin. Chim. Acta 2013, 417, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Xilouri, M.; Stefanis, L. Chaperone mediated autophagy in aging: Starve to prosper. Ageing Res. Rev. 2016, 32, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kawagoe, J.; Abe, K.; Sato, S.; Nagano, I.; Nakamura, S.; Kogure, K. Distributions of heat shock protein (HSP) 70 and heat shock cognate protein (HSC) 70 mRNAs after transient focal ischemia in rat brain. Brain Res. 1992, 587, 195–202. [Google Scholar] [CrossRef]
- Aoki, M.; Abe, K.; Kawagoe, J.; Sato, S.; Nakamura, S.; Kogure, K. Temporal profile of the induction of heat shock protein 70 and heat shock cognate protein 70 mRNAs after transient ischemia in gerbil brain. Brain Res. 1993, 601, 185–192. [Google Scholar] [CrossRef]
- Brea, D.; Agulla, J.; Staes, A.; Gevaert, K.; Campos, F.; Sobrino, T.; Blanco, M.; Davalos, A.; Castillo, J.; Ramos-Cabrer, P. Study of Protein Expression in Peri-Infarct Tissue after Cerebral Ischemia. Sci. Rep. 2015, 5, 12030. [Google Scholar] [CrossRef] [Green Version]
- Pancholi, V. Multifunctional alpha-enolase: Its role in diseases. Cell Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef]
- Poon, H.F.; Vaishnav, R.A.; Getchell, T.V.; Getchell, M.L.; Butterfield, D.A. Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol. Aging 2006, 27, 1010–1019. [Google Scholar] [CrossRef]
- Doran, J.F.; Jackson, P.; Kynoch, P.A.; Thompson, R.J. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J. Neurochem. 1983, 40, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Osaka, H.; Wang, Y.L.; Takada, K.; Takizawa, S.; Setsuie, R.; Li, H.; Sato, Y.; Nishikawa, K.; Sun, Y.J.; Sakurai, M.; et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 2003, 12, 1945–1958. [Google Scholar] [CrossRef]
- Ahmad, M.; Dar, N.J.; Bhat, Z.S.; Hussain, A.; Shah, A.; Liu, H.; Graham, S.H. Inflammation in ischemic stroke: Mechanisms, consequences and possible drug targets. CNS Neurol. Disord. Drug Targets 2014, 13, 1378–1396. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo-Pereira, M.E.; Rockwell, P.; Schmidt-Glenewinkel, T.; Serrano, P. Neuroinflammation and J2 prostaglandins: Linking impairment of the ubiquitin-proteasome pathway and mitochondria to neurodegeneration. Front Mol. Neurosci. 2015, 7, 104. [Google Scholar] [CrossRef] [Green Version]
- Kondo, M.; Shibata, T.; Kumagai, T.; Osawa, T.; Shibata, N.; Kobayashi, M.; Sasaki, S.; Iwata, M.; Noguchi, N.; Uchida, K. 15-Deoxy-Delta (12,14)-prostaglandin J (2): The endogenous electrophile that induces neuronal apoptosis. Proc. Natl. Acad. Sci. USA 2002, 99, 7367–7372. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, W.; Rose, M.E.; Hickey, R.W.; Chen, J.; Uechi, G.T.; Balasubramani, M.; Day, B.W.; Patel, K.V.; Graham, S.H. The point mutation UCH-L1 C152A protects primary neurons against cyclopentenone prostaglandin-induced cytotoxicity: Implications for post-ischemic neuronal injury. Cell Death Dis. 2015, 6, e1966. [Google Scholar] [CrossRef] [Green Version]
- Graham, S.H.; Liu, H. Life and death in the trash heap: The ubiquitin proteasome pathway and UCHL1 in brain aging, neurodegenerative disease and cerebral Ischemia. Ageing Res. Rev. 2017, 34, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, W.; Ahmad, M.; Miller, T.M.; Rose, M.E.; Poloyac, S.M.; Uechi, G.; Balasubramani, M.; Hickey, R.W.; Graham, S.H. Modification of ubiquitin-C-terminal hydrolase-L1 by cyclopentenone prostaglandins exacerbates hypoxic injury. Neurobiol. Dis. 2011, 41, 318–328. [Google Scholar] [CrossRef] [Green Version]
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef]
- Dzeja, P.P.; Bast, P.; Pucar, D.; Wieringa, B.; Terzic, A. Defective metabolic signaling in adenylate kinase AK1 gene knock-out hearts compromises post-ischemic coronary reflow. J. Biol. Chem. 2007, 282, 31366–31372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teoh, J.; Boulos, S.; Chieng, J.; Knuckey, N.W.; Meloni, B.P. Erythropoietin increases neuronal NDPKA expression, and NDPKA up-regulation as well as exogenous application protects cortical neurons from in vitro ischemia-related insults. Cell Mol. Neurobiol. 2014, 34, 379–392. [Google Scholar] [CrossRef]
- Okabe-Kado, J.; Kasukabe, T.; Honma, Y.; Kobayashi, H.; Maseki, N.; Kaneko, Y. Extracellular NM23 protein promotes the growth and survival of primary cultured human acute myelogenous leukemia cells. Cancer Sci. 2009, 100, 1885–1894. [Google Scholar] [CrossRef]
- Huang, C.Y.; Liou, Y.F.; Chung, S.Y.; Lin, W.Y.; Jong, G.P.; Kuo, C.H.; Tsai, F.J.; Cheng, Y.C.; Cheng, F.C.; Lin, J.Y. Role of ERK signaling in the neuroprotective efficacy of magnesium sulfate treatment during focal cerebral ischemia in the gerbil cortex. Chin. J. Physiol. 2010, 53, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Karmarkar, S.W.; Bottum, K.M.; Krager, S.L.; Tischkau, S.A. ERK/MAPK is essential for endogenous neuroprotection in SCN2.2 cells. PLoS ONE 2011, 6, e23493. [Google Scholar] [CrossRef]
- Wang, S.; Guo, S.X.; Dai, Z.G.; Dong, X.W.; Liu, Y.; Jiang, S.; Wang, Z.P. Dual isoflurane-induced preconditioning improves neuroprotection in rat brain in vitro and the role of extracellular signal—Regulated protein kinase. Chin. Med. Sci. J. 2011, 26, 36–42. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Zou, Y.Y.; Wang, J.T. p38 mitogen-activated protein kinase-induced nuclear factor kappa-light-chain-enhancer of activated B cell activity is required for neuroprotection in retinal ischemia/reperfusion injury. Mol. Vis. 2012, 18, 2096–2106. [Google Scholar] [PubMed]
- Zhou, Y.; Bhatia, I.; Cai, Z.; He, Q.Y.; Cheung, P.T.; Chiu, J.F. Proteomic analysis of neonatal mouse brain: Evidence for hypoxia- and ischemia-induced dephosphorylation of collapsin response mediator proteins. J. Proteome Res. 2008, 7, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Teilum, M.; Krogh, M.; Wieloch, T.; Mattiasson, G. Hypothermia affects translocation of numerous cytoplasmic proteins following global cerebral ischemia. J. Proteome Res. 2007, 6, 2822–2832. [Google Scholar] [CrossRef] [PubMed]
- Kobeissy, F.H.; Ottens, A.K.; Zhang, Z.; Liu, M.C.; Denslow, N.D.; Dave, J.R.; Tortella, F.C.; Hayes, R.L.; Wang, K.K. Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol. Cell Proteomics 2006, 5, 1887–1898. [Google Scholar] [CrossRef] [Green Version]
- Kwan, J.A.; Schulze, C.J.; Wang, W.; Leon, H.; Sariahmetoglu, M.; Sung, M.; Sawicka, J.; Sims, D.E.; Sawicki, G.; Schulz, R. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J. 2004, 18, 690–692. [Google Scholar] [CrossRef]
- Martin de la Vega, C.; Burda, J.; Nemethova, M.; Quevedo, C.; Alcazar, A.; Martin, M.E.; Danielisova, V.; Fando, J.L.; Salinas, M. Possible mechanisms involved in the down-regulation of translation during transient global ischaemia in the rat brain. Biochem. J. 2001, 357, 819–826. [Google Scholar] [CrossRef]
- Garcia-Bonilla, L.; Cid, C.; Alcazar, A.; Burda, J.; Ayuso, I.; Salinas, M. Regulatory proteins of eukaryotic initiation factor 2-alpha subunit (eIF2 alpha) phosphatase, under ischemic reperfusion and tolerance. J. Neurochem. 2007, 103, 1368–1380. [Google Scholar] [CrossRef]
- Alban, A.; David, S.O.; Bjorkesten, L.; Andersson, C.; Sloge, E.; Lewis, S.; Currie, I. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003, 3, 36–44. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Alonso, E.; Alcazar, P.; Camafeita, E.; Fernandez-Lucas, M.; Ruiz-Roso, G.; Alcazar, A. Proteomic analysis of plasma proteins of high-flux haemodialysis and on-line haemodiafiltration patients reveals differences in transthyretin levels related with anaemia. Sci. Rep. 2020, 10, 16029. [Google Scholar] [CrossRef] [PubMed]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel maldi lift-tof/tof mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef] [PubMed]

| No. | Protein Identification/Synonyms | Abbreviation a | Accession No. b | Gene Name | Theoretical/Apparent Mass (Da) | Theoretical/Apparent pI | Score c | Coverage (%) | LIFT (Score) c,d |
|---|---|---|---|---|---|---|---|---|---|
| 2 | Heat shock 70 kDa protein 8 /HSP7C | HSC70 | P63018 | Hspa8 | 71,055/ 72,612 | 5.37/ 5.5 | 234 | 39 | 2774.32 (126) |
| 3 | Dihydropyrimidinase-related protein 2/DRP2 /CRMP2 /TOAD64 | DRP2 | P47942 | Dpysl2 | 62,638/ 68,584 | 5.95/ 5.8 | 180 | 38 | 2377.30 (146) |
| 4 | Dihydropyrimidinase-related protein 2 /DRP2 /CRMP2 /TOAD64 | DRP2 | P47942 | Dpysl2 | 62,638/ 68,306 | 5.95/ 6.4 | 239 | 56 | 2070.89 (150) |
| 9 | Gamma-enolase /Enolase 2 /NSE/ENOG | Enolase 2 | P07323 | Eno2 | 47,510/ 47,553 | 5.03/ 5.2 | 232 | 61 | 1358.68 (77) |
| 10 | Alpha-enolase/Enolase 1 /NNE /ENOA | Enolase 1 | P04764 | Eno1 | 47,440/ 50,094 | 6.16/ 6.4 | 189 | 61 | 1541.74 (85) |
| 12 | Phosphoglycerate kinase 1/PGK1 | PGK1 | P16617 | Pgk1 | 44,909/ 44,365 | 8.02/ 7.6 | 72 | 33 | 1634.80 (75) |
| 13 | Glyceraldehyde-3-phosphate dehydrogenase/GAPDH/G3P | G3P | P04797 | Gapdh | 36,090/ 36,027 | 8.14/ 7.4 | 53 | 21 | 1779.78 (107) |
| 17 | Fructose-bisphosphate aldolase C/ALDOC | ALDOC | P09117 | Aldoc | 39,658/ 39,979 | 6.67/ 7.1 | 234 | 77 | 2100.91 (153) |
| 18 | Pyruvate dehydrogenase E1 component subunit beta/ODPB | ODPB | P49432 | Pdhb | 39,299/ 36,027 | 6.2/ 5.5 | 85 | 31 | 1747.87 (59) |
| 25 | Phosphoglycerate mutase 1 /PGAM-B/PGAM1 | PGAM1 | P25113 | Pgam1 | 28,928/ 29,767 | 6.67/ 6.7 | 257 | 72 | 2135.97 (160) |
| 26 | Triose-phosphate isomerase/TPIS | TPIS | P48500 | Tpi1 | 27,345/ 27,772 | 6.89/ 7.4 | 165 | 55 | 2206.11 (133) |
| 28 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 /UCHL1 | UCHL1 | Q00981 | Uchl1 | 25,165/ 28,258 | 5.14/ 5.2 | 188 | 78 | 1678.83 (119) |
| 29 | Peroxiredoxin-6/NSGPx /Thiol-specific antioxidant protein /PRDX6 | PRDX6 | O35244 | Prdx6 | 24,860/ 28,752 | 5.64/ 5.8 | 130 | 49 | 2030.98 (100) |
| 30 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase /PIMT | PIMT | P22062 | Pcmt1 | 24,683/ 27,294 | 7.14/ 7.2 | 103 | 42 | 1694.85 (45) |
| 32 | Rho GDP-dissociation inhibitor 1 /Rho-GDI alpha/GDIR1 | Rho-GDI | Q5XI73 | Arhgdia | 23,450/ 27,294 | 5.12/ 5.2 | 177 | 70 | 1650.92 (122) |
| 33 | Transgelin-3/Neuronal protein NP25/TAGL3 | TAGL3 | P37805 | Tagln3 | 22,657/ 22,164 | 6.84/ 6.7 | 95 | 52 | 1781.72 (82) |
| 37 | Adenylate kinase isoenzyme 1 /KAD1 | ADK1 | P39069 | Ak1 | 21,684/ 24,173 | 7.66/ 7.2 | 125 | 63 | 1495.74 (104) |
| 39 | Phosphatidylethanolamine-binding protein 1 /PEBP1 | PEBP1 | P31044 | Pebp1 | 20,902/ 23,348 | 5.48/ 5.5 | 253 | 66 | 1742.89 (76) |
| 43 | Nucleoside diphosphate kinase A /NDKA | NDKA | Q05982 | Nme1 | 17,296/ 17,385 | 5.96/ 5.8 | 91 | 48 | 1165.62 (39) |
| 47 | Superoxide dismutase [Cu-Zn], SODC | SOD1 | P07632 | Sod1 | 16,073/ 17,998 | 5.88/ 6.1 | 55 | 24 | 1367.79 (108) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Alonso, E.; Guerra-Pérez, N.; Escobar-Peso, A.; Regidor, I.; Masjuan, J.; Alcázar, A. Differential Association of 4E-BP2-Interacting Proteins Is Related to Selective Delayed Neuronal Death after Ischemia. Int. J. Mol. Sci. 2021, 22, 10327. https://doi.org/10.3390/ijms221910327
Martínez-Alonso E, Guerra-Pérez N, Escobar-Peso A, Regidor I, Masjuan J, Alcázar A. Differential Association of 4E-BP2-Interacting Proteins Is Related to Selective Delayed Neuronal Death after Ischemia. International Journal of Molecular Sciences. 2021; 22(19):10327. https://doi.org/10.3390/ijms221910327
Chicago/Turabian StyleMartínez-Alonso, Emma, Natalia Guerra-Pérez, Alejandro Escobar-Peso, Ignacio Regidor, Jaime Masjuan, and Alberto Alcázar. 2021. "Differential Association of 4E-BP2-Interacting Proteins Is Related to Selective Delayed Neuronal Death after Ischemia" International Journal of Molecular Sciences 22, no. 19: 10327. https://doi.org/10.3390/ijms221910327
APA StyleMartínez-Alonso, E., Guerra-Pérez, N., Escobar-Peso, A., Regidor, I., Masjuan, J., & Alcázar, A. (2021). Differential Association of 4E-BP2-Interacting Proteins Is Related to Selective Delayed Neuronal Death after Ischemia. International Journal of Molecular Sciences, 22(19), 10327. https://doi.org/10.3390/ijms221910327






