A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease
Abstract
:1. Introduction
2. Results
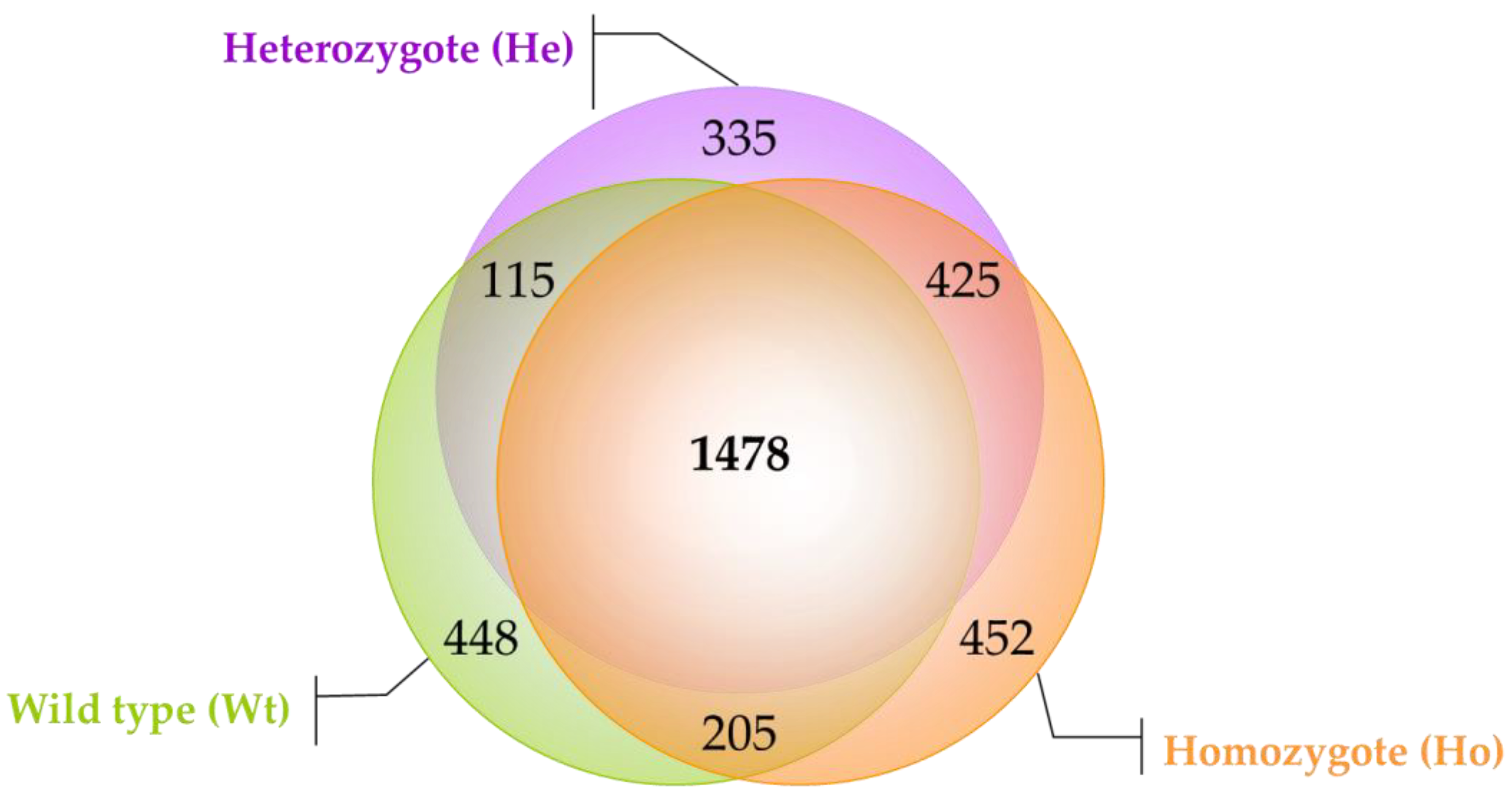
2.1. Protein Profiling
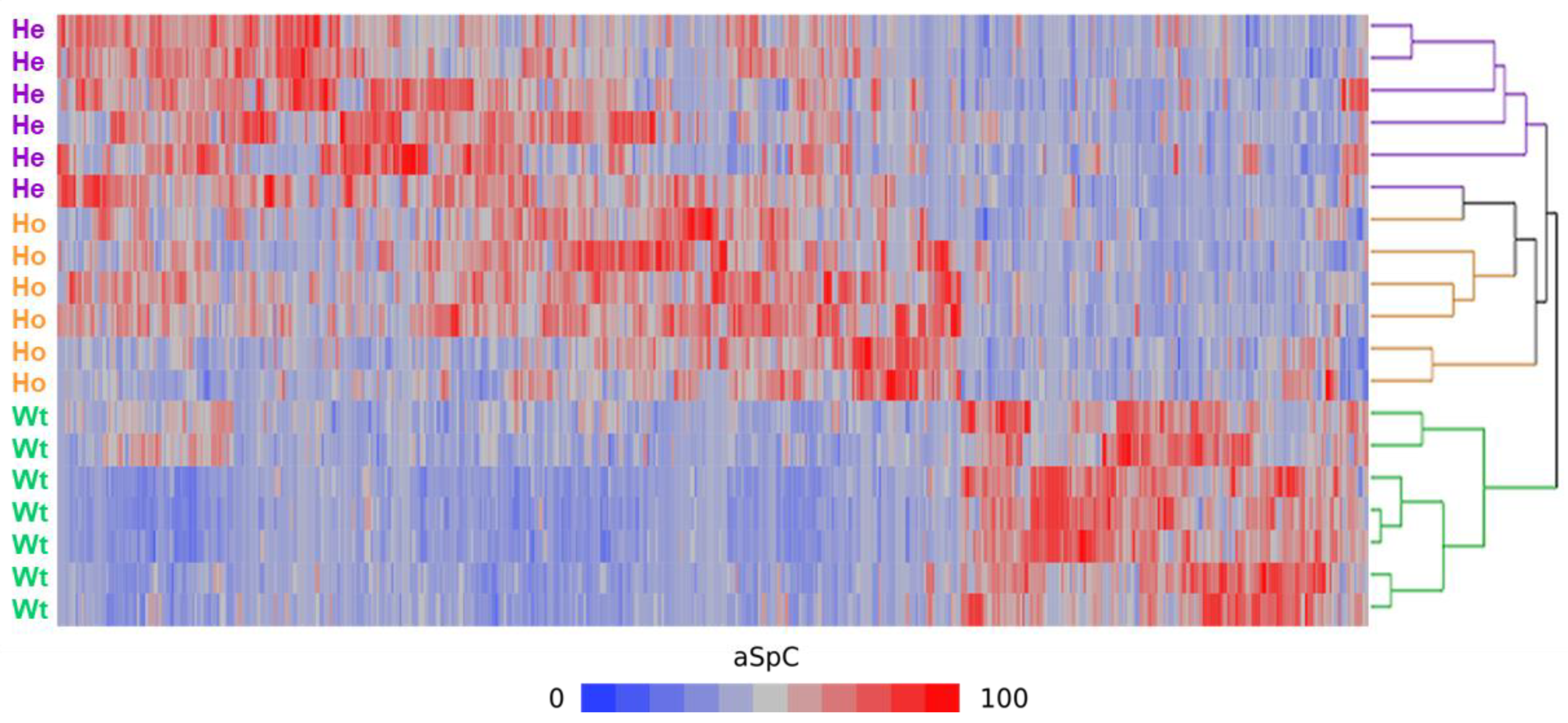
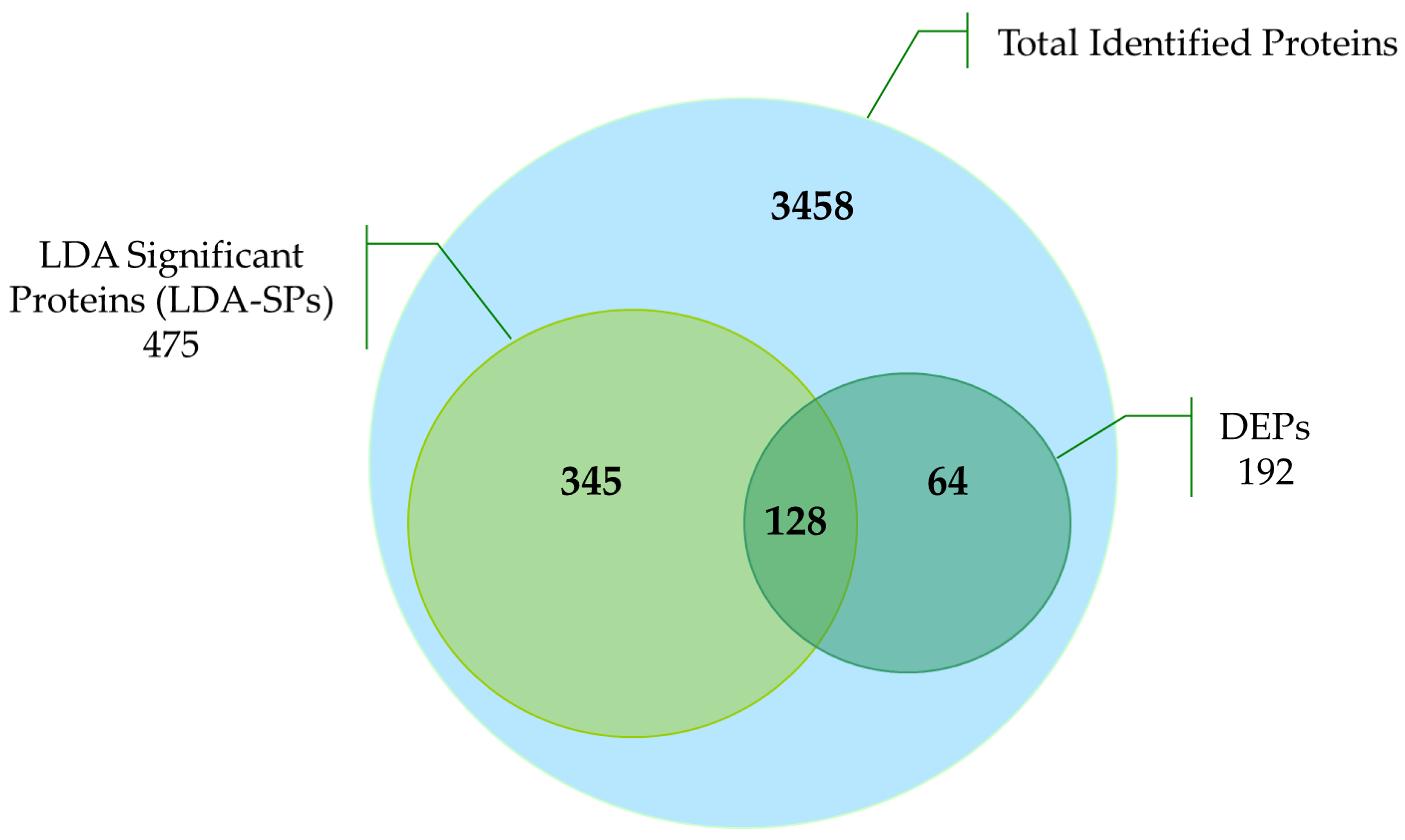
2.2. Clustering and Differential Analysis
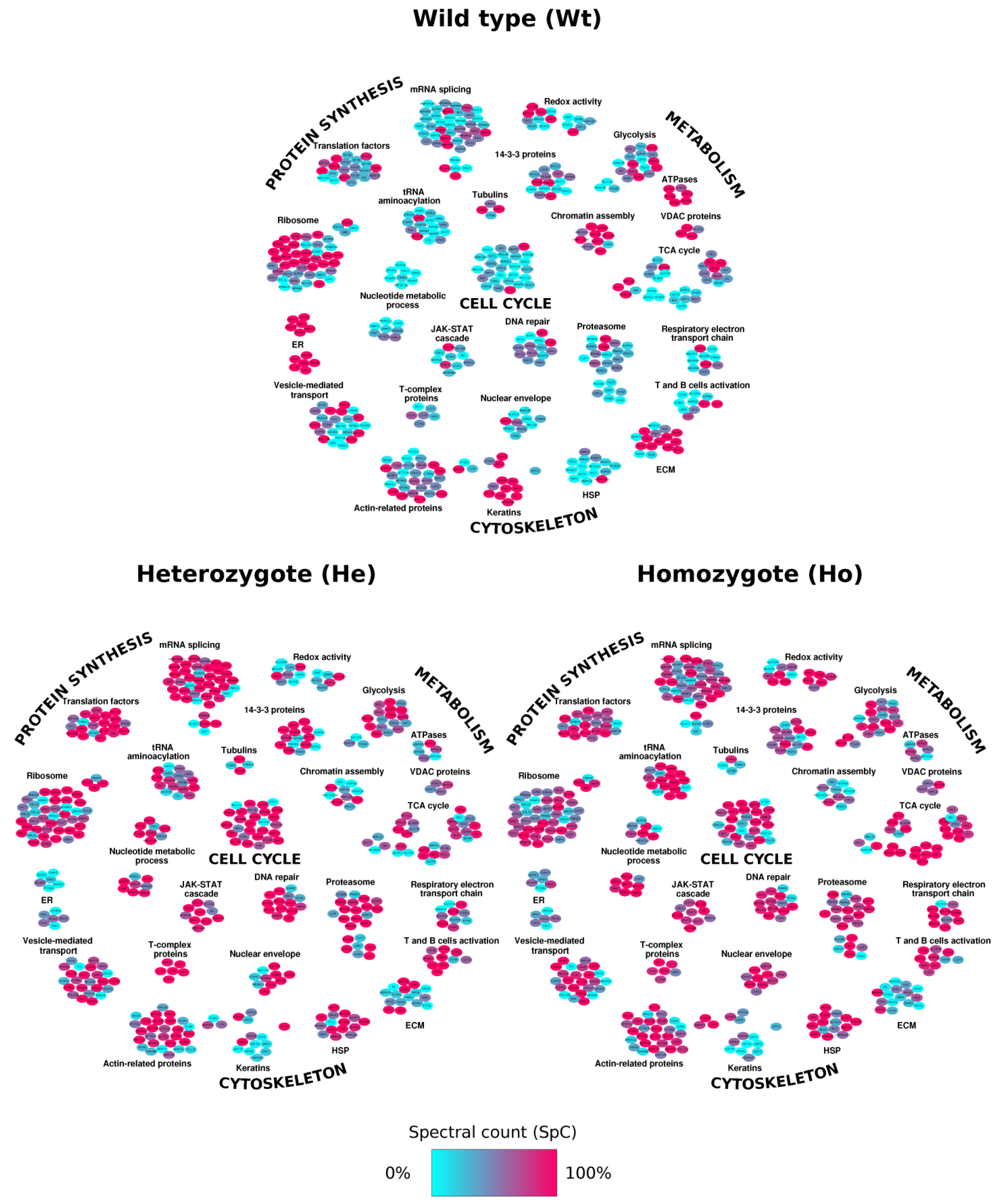
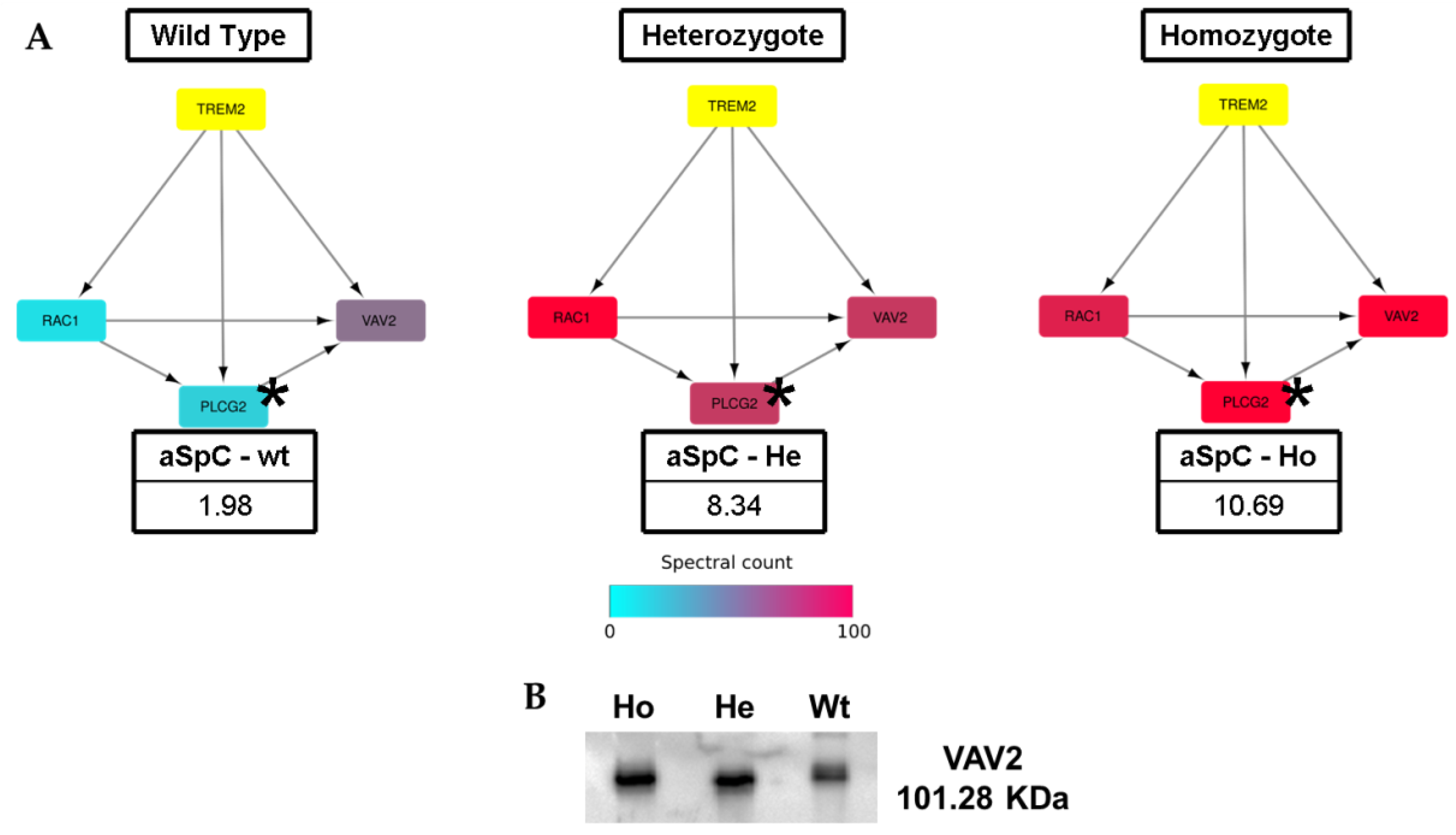
2.3. Network Analysis: Systems Biology Evaluation
3. Discussion
Limitations and Advantages of This Study
4. Materials and Methods
4.1. Subjects
4.2. Lymphoblastoid B-Cell Line and Protein Extraction
4.3. LC-MS/MS Analysis
4.4. Data Handling and Protein Profile of LCLs
4.5. Label-Free Differential Analysis
4.5.1. MAProMa
4.5.2. Linear Discriminant Analysis and Hierarchical Clustering
4.6. Network Analysis
4.7. Western Blotting
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. Report 2017. Alzheimer’s disease facts and figures. Alzheimer’s Dementia 2017, 13, 325–373. [Google Scholar] [CrossRef]
- Sosa-Ortiz, A.L.; Castillo, G.I.A.; Prince, M. Epidemiology of dementias and Alzheimer’s disease. Arch. Med. Res. 2012, 43, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Woollacott, I.; Rohrer, J.D. The clinical spectrum of sporadic and familial forms of frontotemporal dementia. J. Neurochem. 2016, 138 (Suppl. 1), 6–31. [Google Scholar] [CrossRef]
- Bianchin, M.M.; Capella, H.M.; Chaves, D.L.; Steindel, M.; Grisard, E.; Ganev, G.G.; Junior, C.C.; Neto, E.S.; Poffo, M.A.; Walz, R.; et al. Nasu–Hakola Disease (Polycystic Lipomembranous Osteodysplasia with Sclerosing Leukoencephalopathy—PLOSL): A dementia associated with bone cystic lesions. From clinical to genetic and molecular aspects. Cell. Mol. Neurobiol. 2004, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Paloneva, J.; Autti, T.; Hakola, P.; Haltia, M.J. Polycystic Lipomembranous Osteodysplasia with Sclerosing Leukoencephalopathy. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Mirzaa, G., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 2002; pp. 1993–2021. [Google Scholar]
- Yeh, F.L.; Hansen, D.; Sheng, M. TREM2, microglia, and neurodegenerative diseases. Trends Mol. Med. 2017, 23, 512–533. [Google Scholar] [CrossRef]
- Paloneva, J.; Manninen, T.; Christman, G.; Hovanes, K.; Mandelin, J.; Adolfsson, R.; Bianchin, M.; Bird, T.; Miranda, R.; Salmaggi, A.; et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002, 71, 656–662. [Google Scholar] [CrossRef] [Green Version]
- Klunemann, H.H.; Ridha, B.H.; Magy, L.; Wherrett, J.R.; Hemelsoet, D.; Keen, R.W.; De Bleecker, J.L.; Rossor, M.; Marienhagen, J.; Klein, H.E.; et al. The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology 2005, 64, 1502–1507. [Google Scholar] [CrossRef]
- Sessa, G.; Podini, P.; Mariani, M.; Meroni, A.; Spreafico, R.; Sinigaglia, F.; Colonna, M.; Panina, P.; Meldolesi, J. Distribution and signaling of TREM2/DAP12, the receptor system mutated in human polycystic lipomembraneous osteodysplasia with sclerosing leukoencephalopathy dementia. Eur. J. Neurosci. 2004, 20, 2617–2628. [Google Scholar] [CrossRef]
- Paloneva, J.; Mandelin, J.; Kiialainen, A.; Böhling, T.; Prudlo, J.; Hakola, P.; Haltia, M.; Konttinen, Y.T.; Peltonen, L. DAP12/TREM2 Deficiency results in impaired osteoclast differentiation and osteoporotic features. J. Exp. Med. 2003, 198, 669–675. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, S.; Agresta, A.M.; De Palma, A.; Viglio, S.; Mauri, P.; Fumagalli, M.; Iadarola, P.; Montalbetti, L.; Salvini, R.; Bardoni, A. Proteomic Analysis of lymphoblastoid cells from nasu-hakola patients: A step forward in our understanding of this neurodegenerative disorder. PLoS ONE 2014, 9, e110073. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, E.C.; Yates, J.R., III; Gerace, L. MudPIT: A powerful proteomics tool for discovery. Discov. Med. 2003, 3, 38–39. [Google Scholar] [PubMed]
- Comunian, C.; Rusconi, F.; De Palma, A.; Brunetti, P.; Catalucci, D.; Mauri, P. A comparative MudPIT analysis identifies different expression profiles in heart compartments. Proteomics 2011, 11, 2320–2328. [Google Scholar] [CrossRef] [PubMed]
- Mauri, P.; Dehò, G. A proteomic approach to the analysis of RNA degradosome composition in Escherichia coli. Methods Enzymol. 2008, 447, 99–117. [Google Scholar] [CrossRef]
- Roffia, V.; De Palma, A.; Lonati, C.; Di Silvestre, D.; Rossi, R.; Mantero, M.; Gatti, S.; Dondossola, D.; Valenza, F.; Mauri, P.; et al. Proteome investigation of rat lungs subjected to ex vivo perfusion (EVLP). Molecules 2018, 23, 3061. [Google Scholar] [CrossRef] [Green Version]
- Hilario, M.; Kalousis, A. Approaches to dimensionality reduction in proteomic biomarker studies. Brief. Bioinform. 2007, 9, 102–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.; Bader, G.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Martínez, A.; Dalfó, E.; Muntane, G.; Ferrer, I. Glycolitic enzymes are targets of oxidation in aged human frontal cortex and oxidative damage of these proteins is increased in progressive supranuclear palsy. J. Neural Transm. 2008, 115, 59–66. [Google Scholar] [CrossRef]
- Castegna, A.; Aksenov, M.; Aksenova, M.; Thongboonkerd, V.; Klein, J.B.; Pierce, W.M.; Booze, R.; Markesbery, W.R.; Butterfield, D. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. part I: Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 2002, 33, 562–571. [Google Scholar] [CrossRef]
- Korolainen, M.A.; Goldsteins, G.; Nyman, T.; Alafuzoff, I.; Koistinaho, J.; Pirttilä, T. Oxidative modification of proteins in the frontal cortex of Alzheimer’s disease brain. Neurobiol. Aging 2006, 27, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Bigl, M.; Apelt, J.; Eschrich, K.; Schliebs, R. Cortical glucose metabolism is altered in aged transgenic Tg2576 mice that demonstrate Alzheimer plaque pathology. J. Neural Transm. 2003, 110, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Boyd-Kimball, D.; Poon, H.F.; Lynn, B.C.; Cai, J.; Pierce, W.M., Jr.; Klein, J.B.; Ferguson, J.; Link, C.D.; Butterfield, D.A. Proteomic identification of proteins specifically oxidized in caenorhabditis elegans expressing human Aβ(1–42): Implications for Alzheimer’s disease. Neurobiol. Aging 2006, 27, 1239–1249. [Google Scholar] [CrossRef]
- Sultana, R.; Newman, S.F.; Abdul, H.M.; Cai, J.; Pierce, W.M.; Klein, J.B.; Merchant, M.; Butterfield, D.A. Protective effect of D609 against amyloid-beta1–42-induced oxidative modification of neuronal proteins: Redox proteomics study. J. Neurosci. Res. 2006, 84, 409–417. [Google Scholar] [CrossRef]
- Croteau, E.; Castellano, C.-A.; Fortier, M.; Bocti, C.; Fulop, T.; Paquet, N.; Cunnane, S. A cross-sectional comparison of brain glucose and ketone metabolism in cognitively healthy older adults, mild cognitive impairment and early Alzheimer’s disease. Exp. Gerontol. 2018, 107, 18–26. [Google Scholar] [CrossRef]
- Jensen, N.J.; Wodschow, H.Z.; Nilsson, M.; Rungby, J. Effects of ketone bodies on brain metabolism and function in neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 8767. [Google Scholar] [CrossRef] [PubMed]
- Błaszczyk, J.W. Energy metabolism decline in the aging brain—Pathogenesis of neurodegenerative disorders. Metabolites 2020, 10, 450. [Google Scholar] [CrossRef] [PubMed]
- English, J.A.; Pennington, K.; Dunn, M.J.; Cotter, D.R. The neuroproteomics of schizophrenia. Biol. Psychiatry 2011, 69, 163–172. [Google Scholar] [CrossRef]
- Vercauteren, F.G.; Bergeron, J.J.; Vandesande, F.; Arckens, L.; Quirion, R. Proteomic approaches in brain research and neuropharmacology. Eur. J. Pharmacol. 2004, 500, 385–398. [Google Scholar] [CrossRef]
- Kazuno, A.-A.; Ohtawa, K.; Otsuki, K.; Usui, M.; Sugawara, H.; Okazaki, Y.; Kato, T. Proteomic analysis of lymphoblastoid cells derived from monozygotic twins discordant for bipolar disorder: A preliminary study. PLoS ONE 2013, 8, e53855. [Google Scholar] [CrossRef]
- Novikova, S.I.; He, F.; Cutrufello, N.J.; Lidow, M.S. Identification of protein biomarkers for schizophrenia and bipolar disorder in the postmortem prefrontal cortex using SELDI-TOF-MS ProteinChip profiling combined with MALDI-TOF-PSD-MS analysis. Neurobiol. Dis. 2006, 23, 61–76. [Google Scholar] [CrossRef]
- Johnston-Wilson, N.L.; Sims, C.D.; Hofmann, J.-P.; Anderson, L.; Shore, A.D.; Torrey, E.F.; Yolken, R.H. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. Mol. Psychiatry 2000, 5, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Chen, J.; Wang, L.; Ivashkiv, L.B. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J. Leukoc. Biol. 2007, 82, 237–243. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-Q.; Tao, B.-B.; Li, B.; Wang, X.-H.; Zhang, W.-C.; Wan, L.; Wen-Chuan, Z.; Li, S.-T. Overexpression of TREM2 enhances glioma cell proliferation and invasion: A therapeutic target in human glioma. Oncotarget 2016, 7, 2354–2366. [Google Scholar] [CrossRef] [Green Version]
- Otero, K.; Shinohara, M.; Zhao, H.; Cella, M.; Gilfillan, S.; Colucci, A.; Faccio, R.; Ross, F.P.; Teitelbaum, S.; Takayanagi, H.; et al. TREM2 and β-Catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 2012, 188, 2612–2621. [Google Scholar] [CrossRef] [PubMed]
- Stefano, L.; Racchetti, G.; Bianco, F.; Passini, N.; Gupta, R.S.; Bordignon, P.P.; Meldolesi, J. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J. Neurochem. 2009, 110, 284–294. [Google Scholar] [CrossRef]
- Koh, J.-M.; Lee, Y.-S.; Kim, Y.S.; Park, S.-H.; Lee, S.H.; Kim, H.-H.; Lee, M.-S.; Lee, K.-U.; Kim, G.S. Heat shock protein 60 causes osteoclastic bone resorption via toll-like receptor-2 in estrogen deficiency. Bone 2009, 45, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Forabosco, P.; Vandrovcova, J.; Botía, J.A.; Guelfi, S.; Warren, J.D.; UK Brain Expression Consortium (UKBEC); Momeni, P.; Weale, M.E.; Ryten, M.; et al. Frontotemporal dementia: Insights into the biological underpinnings of disease through gene co-expression network analysis. Mol. Neurodegener. 2016, 11, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novellino, F.; Saccà, V.; Donato, A.; Zaffino, P.; Spadea, M.F.; Vismara, M.; Arcidiacono, B.; Malara, N.; Presta, I.; Donato, G. Innate immunity: A common denominator between neurodegenerative and neuropsychiatric diseases. Int. J. Mol. Sci. 2020, 21, 1115. [Google Scholar] [CrossRef] [Green Version]
- Tobin, R.P.; Mukherjee, S.; Kain, J.M.; Rogers, S.K.; Henderson, S.K.; Motal, H.L.; Rogers, M.K.N.; Shapiro, L.A. Traumatic brain injury causes selective, CD74-dependent peripheral lymphocyte activation that exacerbates neurodegeneration. Acta Neuropathol. Commun. 2014, 2, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Lin, J.; Schlotterer, A.; Wu, L.; Fleming, T.; Busch, S.; Dietrich, N.; Hammes, H.-P. CD74 indicates microglial activation in experimental diabetic retinopathy and exogenous methylglyoxal mimics the response in normoglycemic retina. Acta Diabetol. 2014, 51, 813–821. [Google Scholar] [CrossRef]
- Blacher, E.; Dadali, T.; Bespalko, A.; Msc, V.J.H.; Grimm, M.; Hartmann, T.; Lund, F.E.; Stein, R.; Levy, A. Alzheimer’s disease pathology is attenuated in a CD38-deficient mouse model. Ann. Neurol. 2015, 78, 88–103. [Google Scholar] [CrossRef]
- Mao, D.; Epple, H.; Uthgenannt, B.; Veis, D.; Faccio, R. PLCγ2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J. Clin. Investig. 2006, 116, 2869–2879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalbetti, L.; Soragna, D.; Ratti, M.T.; Bini, P.; Buscone, S.; Moglia, A. Nasu-Hakola disease: A rare entity in Italy. Critical review of the literature. Funct. Neurol. 2004, 19, 171–179. [Google Scholar] [PubMed]
- Gurwitz, D. Human iPSC-derived neurons and lymphoblastoid cells for personalized medicine research in neuropsychiatric disorders. Dialogues Clin. Neurosci. 2016, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Montalbetti, L.; Ratti, M.T.; Greco, B.; Aprile, C.; Moglia, A.; Soragna, D. Neuropsychological tests and functional nuclear neuroimaging provide evidence of subclinical impairment in Nasu-Hakola disease heterozygotes. Funct. Neurol. 2005, 20, 71–75. [Google Scholar]
- Soragna, D.; Tupler, R.; Ratti, M.T.; Montalbetti, L.; Papi, L.; Sestini, R. An Italian family affected by Nasu-Hakola disease with a novel genetic mutation in the TREM2 gene. J. Neurol. Neurosurg. Psychiatry 2003, 74, 825–826. [Google Scholar] [CrossRef] [Green Version]
- Nomura, E.; Katsuta, K.; Ueda, T.; Toriyama, M.; Mori, T.; Inagaki, N. Acid-labile surfactant improves in-sodium dodecyl sulfate polyacrylamide gel protein digestion for matrix-assisted laser desorption/ionization mass spectrometric peptide mapping. J. Mass Spectrom. 2004, 39, 202–207. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.K.; Murty, M.N.; Flynn, P.J. Data clustering. ACM Comput. Surv. 1999, 31, 264–323. [Google Scholar] [CrossRef]
- Zhao, Y.; Karypis, G. Data clustering in life sciences. Mol. Biotechnol. 2005, 31, 55–80. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Palma, A.; Agresta, A.M.; Viglio, S.; Rossi, R.; D’Amato, M.; Di Silvestre, D.; Mauri, P.; Iadarola, P. A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease. Int. J. Mol. Sci. 2021, 22, 9959. https://doi.org/10.3390/ijms22189959
De Palma A, Agresta AM, Viglio S, Rossi R, D’Amato M, Di Silvestre D, Mauri P, Iadarola P. A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease. International Journal of Molecular Sciences. 2021; 22(18):9959. https://doi.org/10.3390/ijms22189959
Chicago/Turabian StyleDe Palma, Antonella, Anna Maria Agresta, Simona Viglio, Rossana Rossi, Maura D’Amato, Dario Di Silvestre, Pierluigi Mauri, and Paolo Iadarola. 2021. "A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease" International Journal of Molecular Sciences 22, no. 18: 9959. https://doi.org/10.3390/ijms22189959
APA StyleDe Palma, A., Agresta, A. M., Viglio, S., Rossi, R., D’Amato, M., Di Silvestre, D., Mauri, P., & Iadarola, P. (2021). A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease. International Journal of Molecular Sciences, 22(18), 9959. https://doi.org/10.3390/ijms22189959









