Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
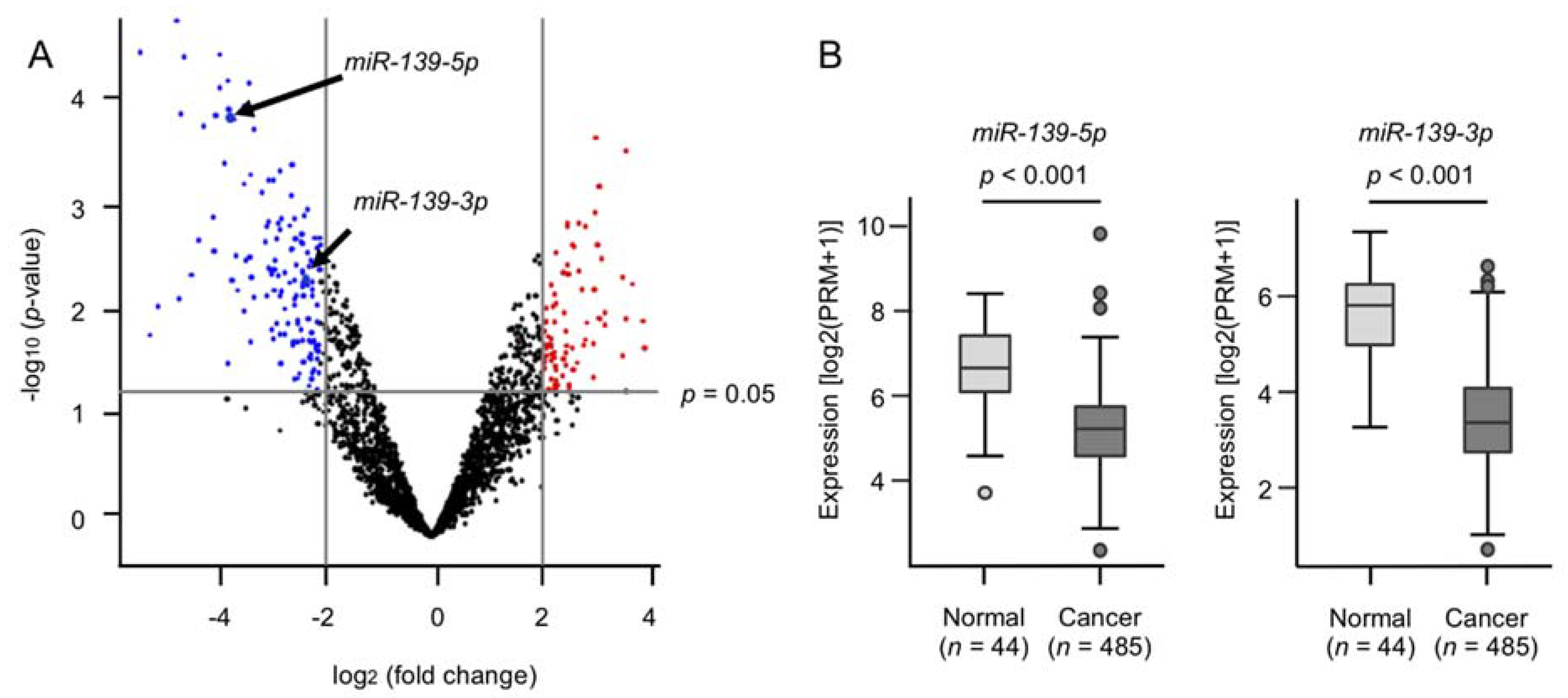
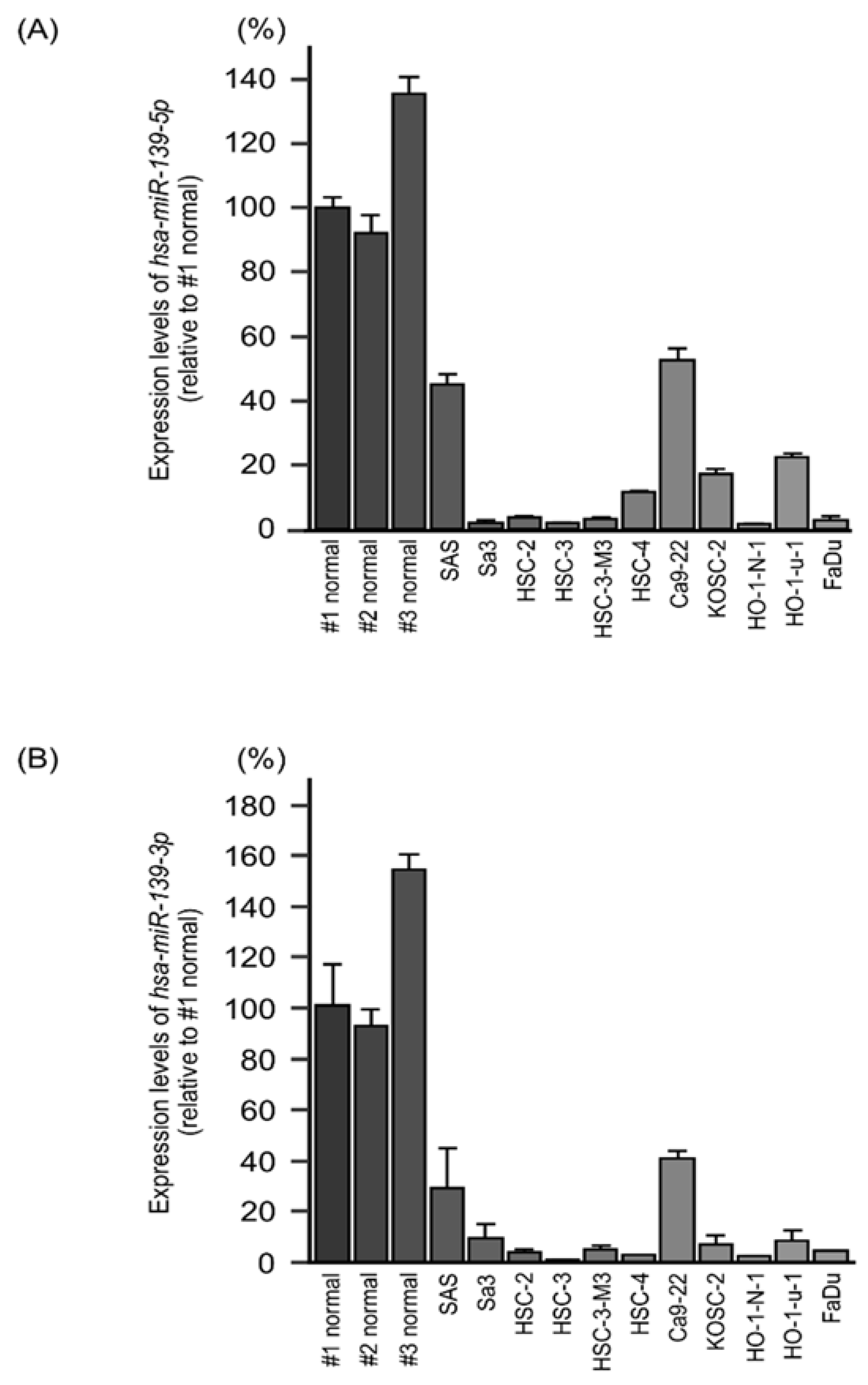
2.1. Generation of a miRNA Expression Signature in HNSCC
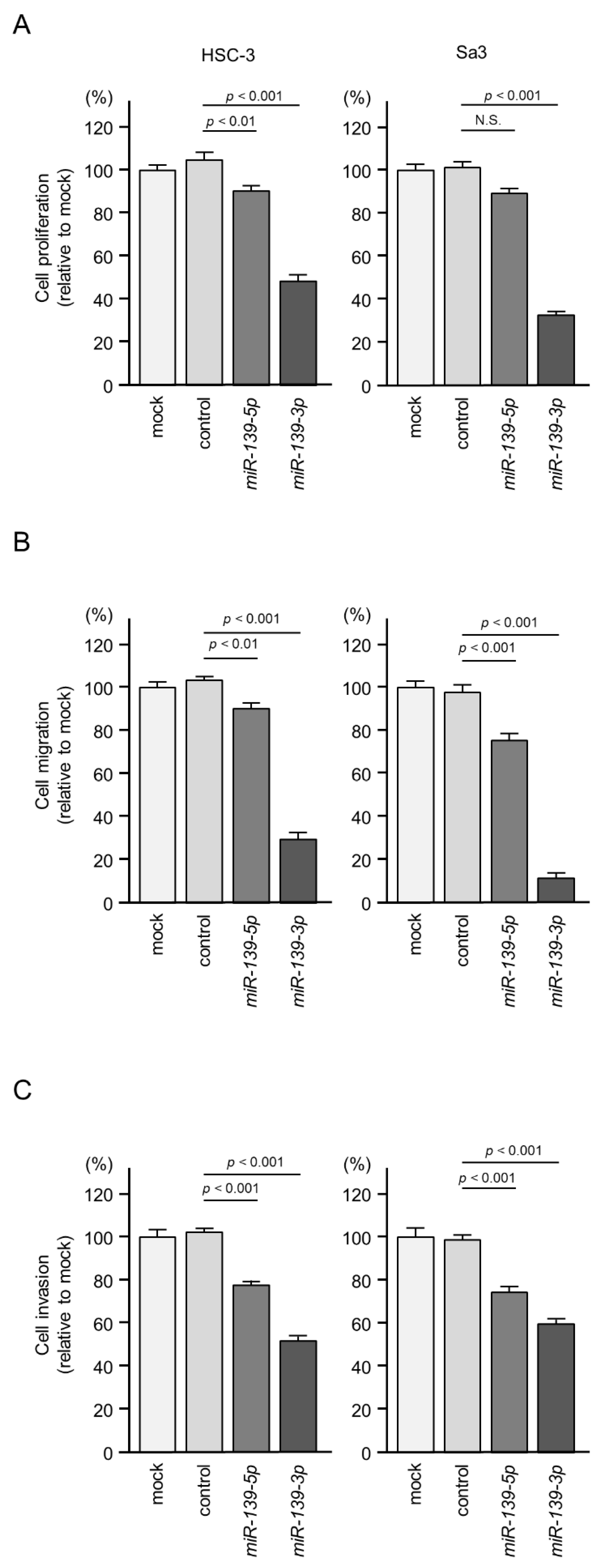
2.2. Effects of Ectopic Expression of miR-139-5p and miR-139-3p on HNSCC Cell Proliferation, Migration, and Invasion
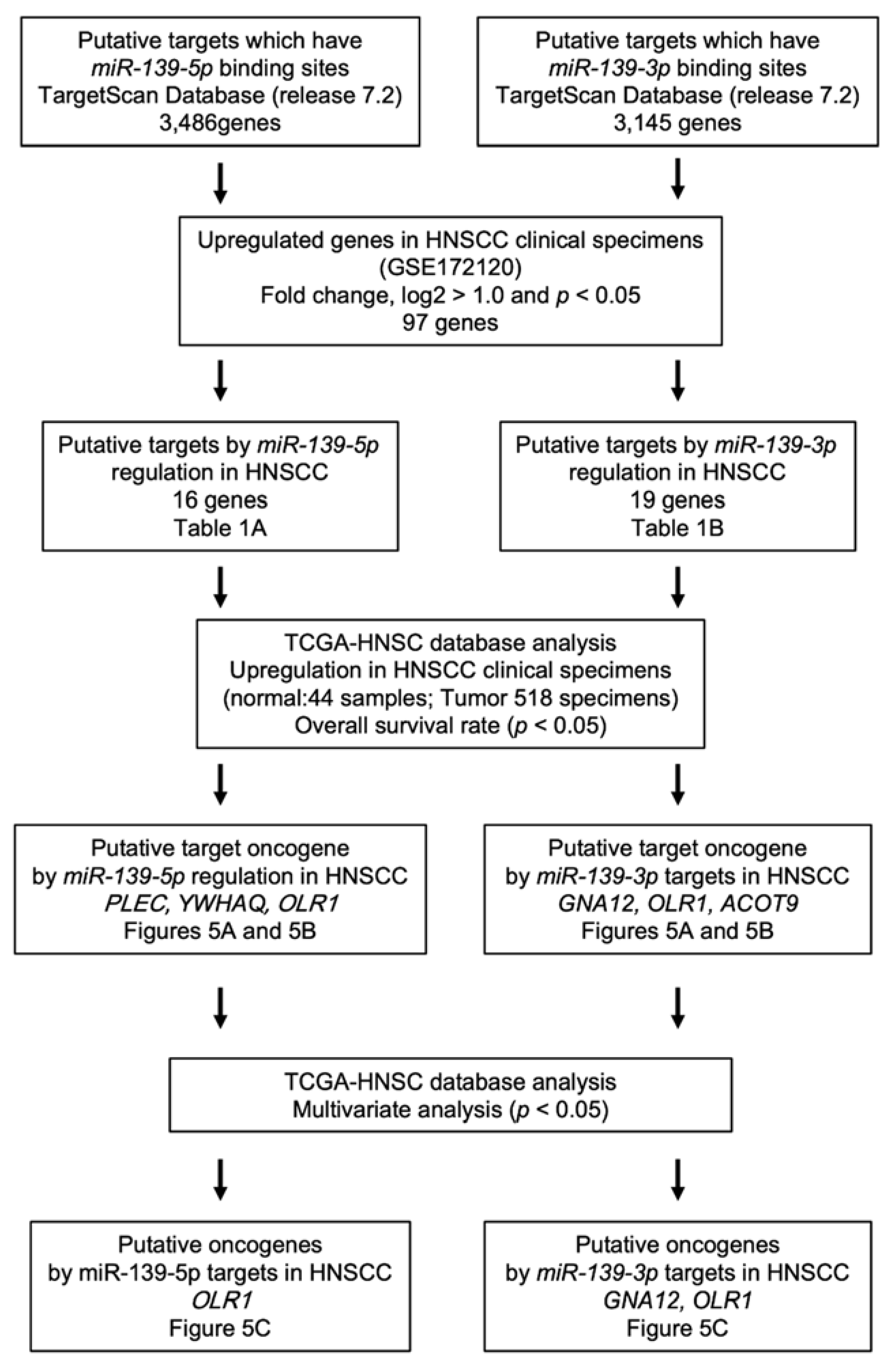
2.3. Identification of Putative Oncogenic Targets through miR-139-5p and miR-139-3p Regulation in HNSCC Cells
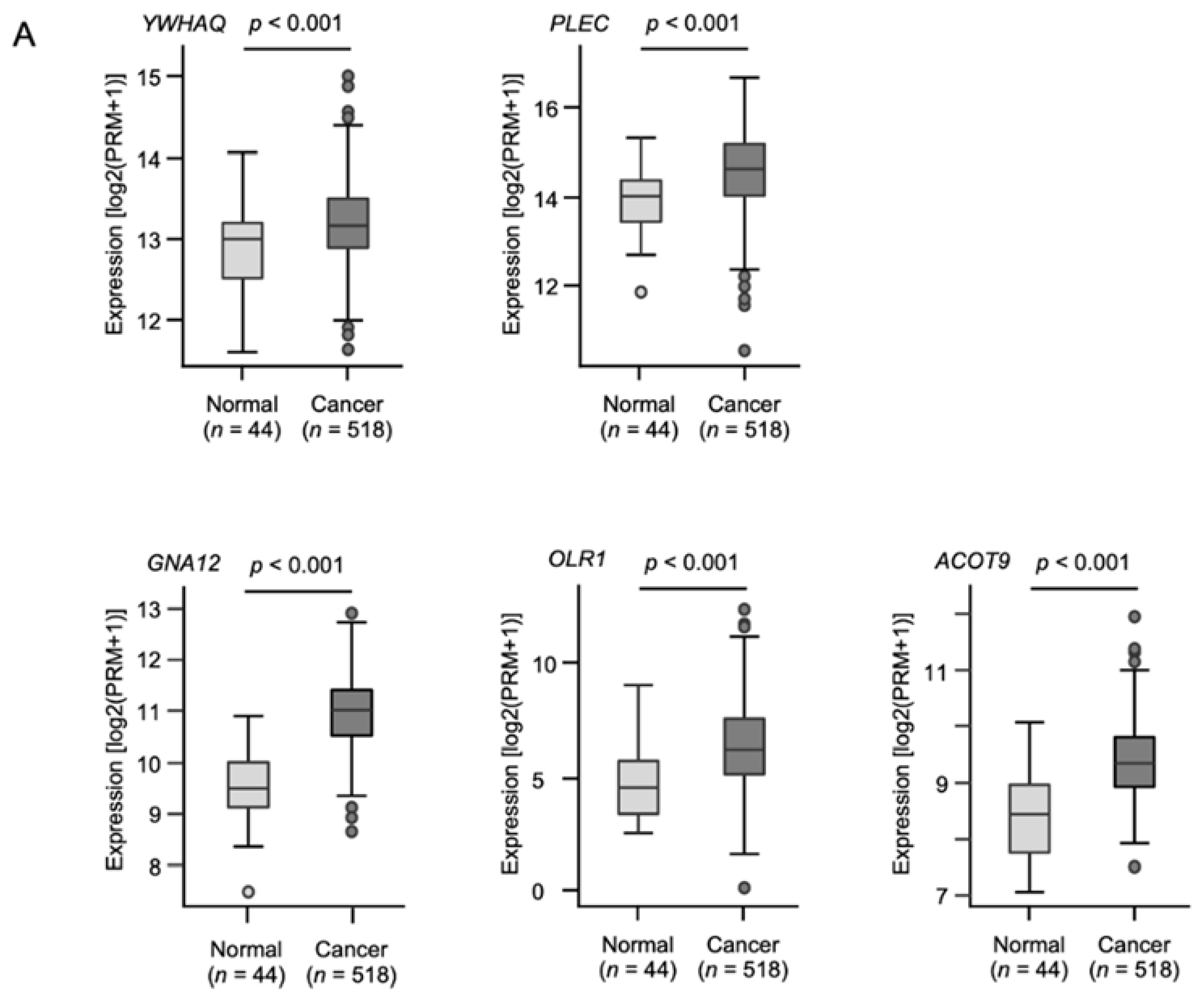
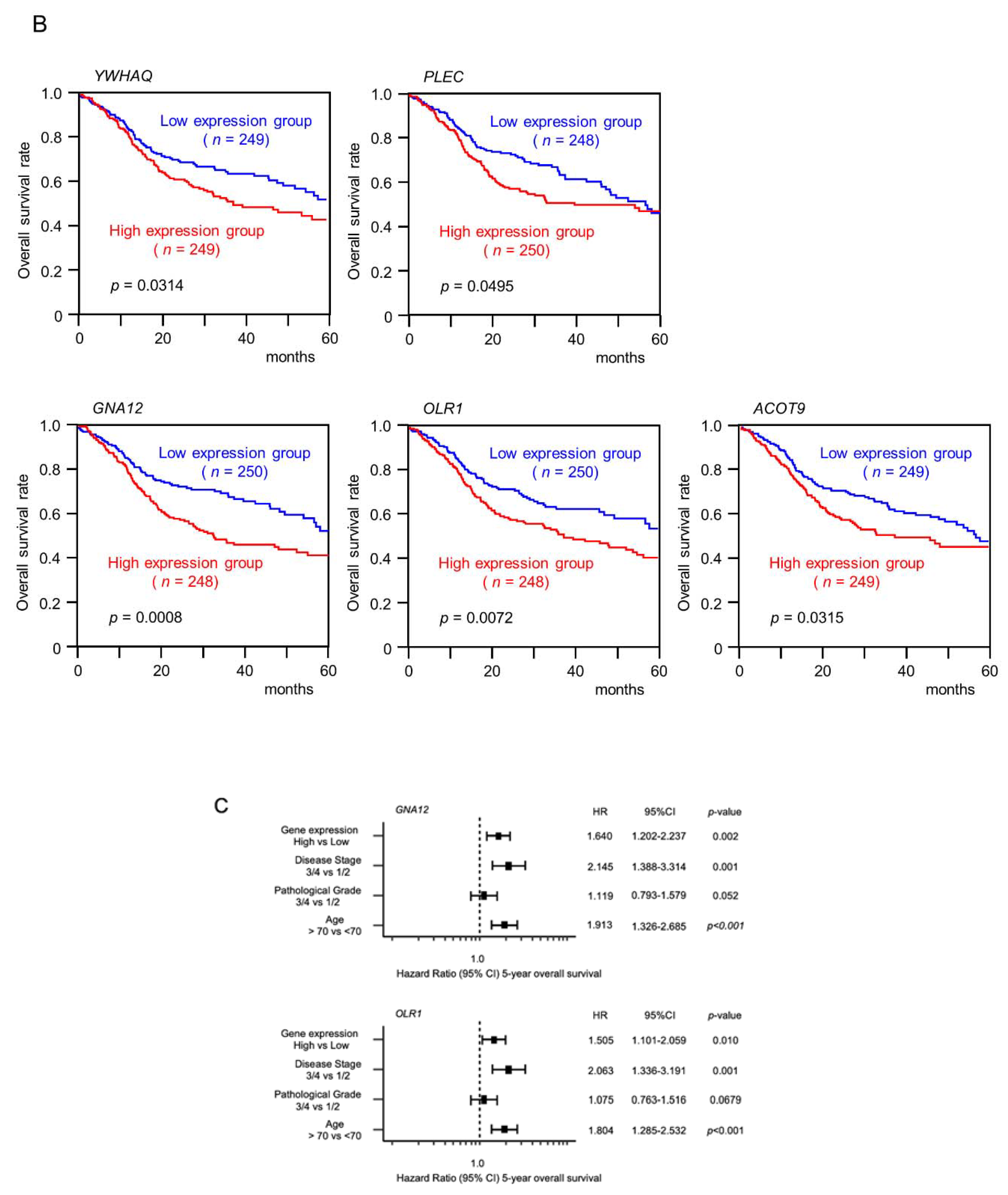
2.4. Clinical Significance of the Putative Target Genes of miR-139-5p and miR-139-3p Determined by the TCGA Analysis
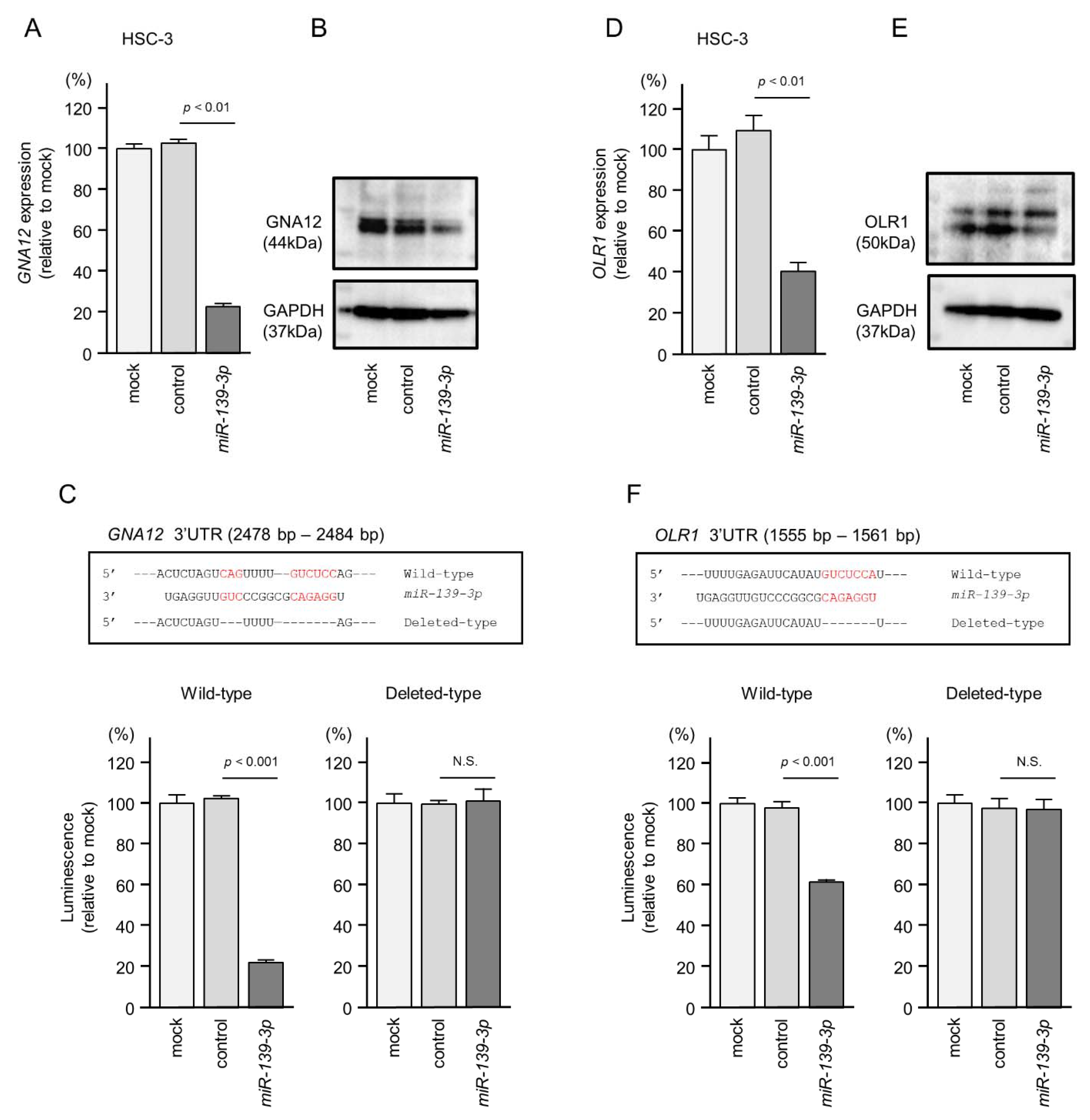
2.5. Direct Regulation of GNA12 and OLR1 by miR-139-3p in HNSCC Cells
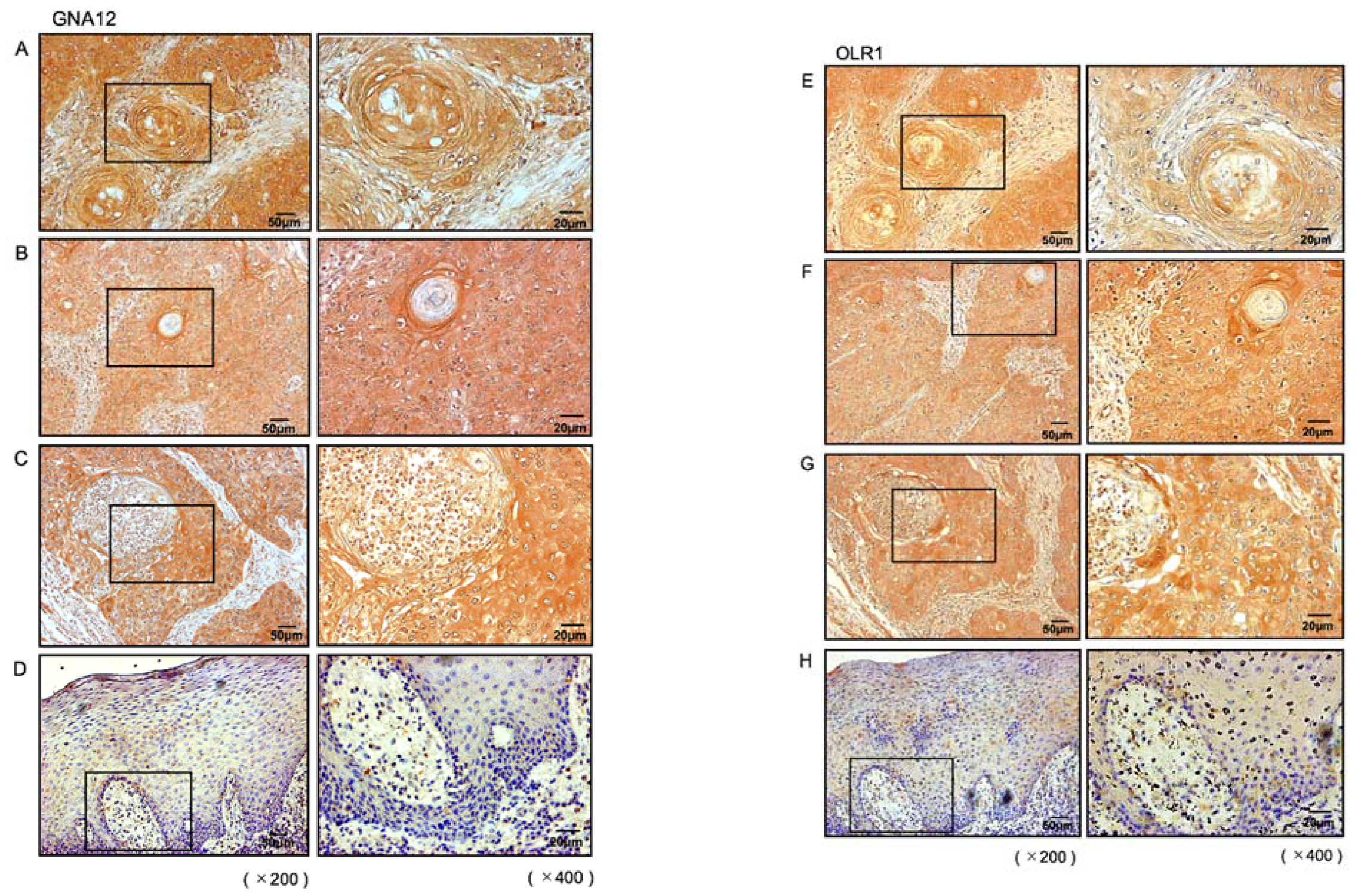
2.6. Overexpression of GNA12 and OLR1 in Clinical Specimens of HNSCC
2.7. GNA12- and OLR1-Mediated Pathways in HNSCC Cells
3. Discussion
4. Materials and Methods
4.1. Clinical HNSCC Specimens, Normal Epithelial Specimens, and HNSCC Cell Lines
4.2. Generation of a miRNA Expression Signature in HNSCC through RNA Sequencing
4.3. Ectopic miRNA Expression Assays of Cell Proliferation, Migration, and Invasion
4.4. Identification of Putative Targets Controlled by miR-139-5p and miR-139-3p in HNSCC Cells
4.5. Direct Regulation of Target Genes by miR-139-3p Using Dual Luciferase Reporter Assays
4.6. Western Blotting and Immunohistochemistry
4.7. Analysis of Molecular Pathways Related to Target Genes Controlled by miR-139-3p
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.E.W.; Bell, R.B.; Bifulco, C.B.; Burtness, B.; Gillison, M.L.; Harrington, K.J.; Le, Q.T.; Lee, N.Y.; Leidner, R.; Lewis, R.L.; et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J. Immunother. Cancer 2019, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, J.; Yu, P.; Guo, L.; Mao, X.; Wang, J.; Miao, S.; Sun, J. The roles of extracellular vesicles in the development, microenvironment, anticancer drug resistance, and therapy of head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 35. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Cohen, R.B.; Jones, C.U.; Sur, R.K.; Raben, D.; Baselga, J.; Spencer, S.A.; Zhu, J.; et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010, 11, 21–28. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs-An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Koshizuka, K.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Fukumoto, I.; Katada, K.; Okamoto, Y.; Seki, N. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: Dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget 2017, 8, 30288–30304. [Google Scholar] [CrossRef]
- Yonemori, K.; Seki, N.; Idichi, T.; Kurahara, H.; Osako, Y.; Koshizuka, K.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: Anti-tumour functions of the microRNA-216 cluster. Oncotarget 2017, 8, 70097–70115. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Okato, A.; Idichi, T.; Arai, T.; Sugawara, S.; Katada, K.; Okamoto, Y.; et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int. J. Oncol. 2018, 52, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Koshizuka, K.; Yamada, Y.; Moriya, S.; Kikkawa, N.; Kinoshita, T.; Hanazawa, T.; Seki, N. Regulation of Oncogenic Targets by miR-99a-3p (Passenger Strand of miR-99a-Duplex) in Head and Neck Squamous Cell Carcinoma. Cells 2019, 8, 1535. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Kurozumi, A.; Kato, M.; Katada, K.; Okamoto, Y.; Seki, N. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017, 108, 1681–1692. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, H.; Okada, R.; Tanaka, T.; Hozaka, Y.; Wada, M.; Moriya, S.; Idichi, T.; Kita, Y.; Kurahara, H.; Ohtsuka, T.; et al. Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 6459. [Google Scholar] [CrossRef] [PubMed]
- Sanada, H.; Seki, N.; Mizuno, K.; Misono, S.; Uchida, A.; Yamada, Y.; Moriya, S.; Kikkawa, N.; Machida, K.; Kumamoto, T.; et al. Involvement of Dual Strands of miR-143 (miR-143-5p and miR-143-3p) and Their Target Oncogenes in the Molecular Pathogenesis of Lung Adenocarcinoma. Int. J. Mol. Sci. 2019, 20, 4482. [Google Scholar] [CrossRef]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016, 7, 72084–72098. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Soulières, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Arai, T.; Okato, A.; Kikkawa, N.; Seki, N. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev. 2017, 36, 525–545. [Google Scholar] [CrossRef]
- Koshizuka, K.; Kikkawa, N.; Hanazawa, T.; Yamada, Y.; Okato, A.; Arai, T.; Katada, K.; Okamoto, Y.; Seki, N. Inhibition of integrin β1-mediated oncogenic signalling by the antitumor microRNA-29 family in head and neck squamous cell carcinoma. Oncotarget 2018, 9, 3663–3676. [Google Scholar] [CrossRef]
- Toda, H.; Kurozumi, S.; Kijima, Y.; Idichi, T.; Shinden, Y.; Yamada, Y.; Arai, T.; Maemura, K.; Fujii, T.; Horiguchi, J.; et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: Antitumor miR-204-5p targets AP1S3. J. Hum. Genet. 2018, 63, 1197–1210. [Google Scholar] [CrossRef]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; et al. RNA-sequence-based microRNA expression signature in breast cancer: Tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Goto, Y.; Tanaka, T.; Okada, R.; Moriya, S.; Idichi, T.; Noda, M.; Sasaki, K.; Kita, Y.; Kurahara, H.; et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: Oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J. Hum. Genet. 2020, 65, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kurozumi, A.; Arai, T.; Nohata, N.; Kojima, S.; Okato, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y.; et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br. J. Cancer 2017, 117, 409–420. [Google Scholar] [CrossRef]
- Yamada, Y.; Arai, T.; Kojima, S.; Sugawara, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018, 109, 2919–2936. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Katada, K.; Okato, A.; Arai, T.; Idichi, T.; Osako, Y.; Okamoto, Y.; Seki, N. Antitumor miR-150-5p and miR-150-3p inhibit cancer cell aggressiveness by targeting SPOCK1 in head and neck squamous cell carcinoma. Auris Nasus Larynx 2018, 45, 854–865. [Google Scholar] [CrossRef]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, M.; Seki, N.; Yoshino, H.; Matsushita, R.; Miyamoto, K.; Nakagawa, M.; Enokida, H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016, 107, 1233–1242. [Google Scholar] [CrossRef]
- Okada, R.; Goto, Y.; Yamada, Y.; Kato, M.; Asai, S.; Moriya, S.; Ichikawa, T.; Seki, N. Regulation of Oncogenic Targets by the Tumor-Suppressive miR-139 Duplex (miR-139-5p and miR-139-3p) in Renal Cell Carcinoma. Biomedicines 2020, 8, 599. [Google Scholar] [CrossRef]
- Huang, L.L.; Huang, L.W.; Wang, L.; Tong, B.D.; Wei, Q.; Ding, X.S. Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncol. Lett. 2017, 14, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jin, J.; Ma, T.; Zhai, H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J. Cell. Mol. Med. 2017, 21, 3730–3740. [Google Scholar] [CrossRef]
- Jiang, Q.; Cao, Y.; Qiu, Y.; Li, C.; Liu, L.; Xu, G. Progression of squamous cell carcinoma is regulated by miR-139-5p/CXCR4. Front Biosci 2020, 25, 1732–1745. [Google Scholar]
- Yu, Y.; Yang, J.; Li, Q.; Xu, B.; Lian, Y.; Miao, L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017, 50, 4. [Google Scholar] [CrossRef]
- Li, M.; Ning, J.; Li, Z.; Wang, J.; Zhao, C.; Wang, L. LINC00152 promotes the growth and invasion of oral squamous cell carcinoma by regulating miR-139-5p. Onco. Targets Ther. 2018, 11, 6295–6304. [Google Scholar] [CrossRef] [PubMed]
- Sannigrahi, M.K.; Sharma, R.; Singh, V.; Panda, N.K.; Rattan, V.; Khullar, M. Role of Host miRNA Hsa-miR-139-3p in HPV-16-Induced Carcinomas. Clin. Cancer Res. 2017, 23, 3884–3895. [Google Scholar] [CrossRef]
- Ma, Y.; Gong, Z.; Wang, H.; Liang, Y.; Huang, X.; Yu, G. Anti-cancer effect of miR-139-3p on laryngeal squamous cell carcinoma by targeting rab5a: In vitro and in vivo studies. Pathol. Res. Pract. 2020, 216, 153194. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J.; Wang, K.; Tang, X.; He, J. miR-139-3p suppresses the invasion and migration properties of breast cancer cells by targeting RAB1A. Oncol. Rep. 2019, 42, 1699–1708. [Google Scholar] [CrossRef] [PubMed]
- Siehler, S. Regulation of RhoGEF proteins by G12/13-coupled receptors. Br. J. Pharmacol. 2009, 158, 41–49. [Google Scholar] [CrossRef]
- Kozasa, T.; Hajicek, N.; Chow, C.R.; Suzuki, N. Signalling mechanisms of RhoGTPase regulation by the heterotrimeric G proteins G12 and G13. J. Biochem. 2011, 150, 357–369. [Google Scholar] [CrossRef]
- Jian, S.L.; Hsieh, H.Y.; Liao, C.T.; Yen, T.C.; Nien, S.W.; Cheng, A.J.; Juang, J.L. Gα₁₂ drives invasion of oral squamous cell carcinoma through up-regulation of proinflammatory cytokines. PLoS ONE 2013, 8, e66133. [Google Scholar] [CrossRef]
- Gan, C.P.; Patel, V.; Mikelis, C.M.; Zain, R.B.; Molinolo, A.A.; Abraham, M.T.; Teo, S.H.; Abdul Rahman, Z.A.; Gutkind, J.S.; Cheong, S.C. Heterotrimeric G-protein alpha-12 (Gα12) subunit promotes oral cancer metastasis. Oncotarget 2014, 5, 9626–9640. [Google Scholar] [CrossRef][Green Version]
- Yang, G.; Xiong, G.; Feng, M.; Zhao, F.; Qiu, J.; Liu, Y.; Cao, Z.; Wang, H.; Yang, J.; You, L.; et al. OLR1 Promotes Pancreatic Cancer Metastasis via Increased c-Myc Expression and Transcription of HMGA2. Mol. Cancer Res. 2020, 18, 685–697. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, H.; Zhao, L.; Zhang, Y.; Wan, Q.; Shen, Y.; Bu, X.; Wan, M.; Shen, C. Up-regulation of OLR1 expression by TBC1D3 through activation of TNFα/NF-κB pathway promotes the migration of human breast cancer cells. Cancer Lett. 2017, 408, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J. Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
| A. Sixteen Candidate Target Genes Regulated by miR-139-5p | |||||
| Entrez Gene ID | Gene Symbol | Gene Name | Fold Change (log2 > 1.0) | p-Value | Total Sites |
| 19 | ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 2.065 | 0.043 | 1 |
| 64919 | BCL11B | BAF chromatin remodeling complex subunit BCL11B | 2.620 | 0.030 | 0 |
| 80005 | DOCK5 | dedicator of cytokinesis 5 | 1.940 | 0.040 | 0 |
| 1993 | ELAVL2 | ELAV-like neuron-specific RNA-binding protein 2 | 2.606 | 0.013 | 2 |
| 4973 | OLR1 | oxidized low-density lipoprotein (lectin-like) receptor 1 | 4.620 | 0.035 | 1 |
| 26031 | OSBPL3 | oxysterol-binding protein-like 3 | 1.799 | 0.038 | 1 |
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 | 1.270 | 0.015 | 1 |
| 5209 | PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1.840 | 0.040 | 0 |
| 5339 | PLEC | plectin | 1.020 | 0.040 | 0 |
| 10154 | PLXNC1 | plexin C1 | 1.463 | 0.038 | 1 |
| 51312 | SLC25A37 | solute carrier family 25 (mitochondrial iron transporter), member 37 | 1.740 | 0.040 | 0 |
| 6733 | SRPK2 | SRSF protein kinase 2 | 1.550 | 0.050 | 0 |
| 91937 | TIMD4 | T cell immunoglobulin and mucin domain containing 4 | 2.140 | 0.010 | 0 |
| 25829 | TMEM184B | transmembrane protein 184B | 1.120 | 0.046 | 2 |
| 8743 | TNFSF10 | TNF superfamily member 10 | 3.540 | 0.040 | 0 |
| 10971 | YWHAQ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta | 1.059 | 0.047 | 1 |
| B. Nineteen Candidate Target Genes Regulated by miR-139-3p | |||||
| Entrez Gene ID | Gene Symbol | Gene Name | Fold Change (log2 > 1.0) | p-value | Total sites |
| 2768 | GNA12 | guanine nucleotide-binding protein (G protein) alpha 12 | 1.114 | 0.039 | 1 |
| 440689 | HIST2H2BF | histone cluster 2, H2bf | 2.816 | 0.035 | 2 |
| 124935 | SLC43A2 | solute carrier family 43 (amino acid system L transporter), member 2 | 1.962 | 0.045 | 1 |
| 25829 | TMEM184B | transmembrane protein 184B | 1.120 | 0.046 | 2 |
| 57464 | STRIP2 | striatin interacting protein 2 | 2.572 | 0.047 | 1 |
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 | 1.270 | 0.015 | 1 |
| 10154 | PLXNC1 | plexin C1 | 1.463 | 0.038 | 1 |
| 4973 | OLR1 | oxidized low-density lipoprotein (lectin-like) receptor 1 | 4.620 | 0.035 | 1 |
| 55784 | MCTP2 | multiple C2 domains, transmembrane 2 | 1.274 | 0.035 | 2 |
| 89796 | NAV1 | neuron navigator 1 | 1.998 | 0.036 | 1 |
| 4288 | MKI67 | marker of proliferation Ki-67 | 1.005 | 0.048 | 1 |
| 51312 | SLC25A37 | solute carrier family 25 (mitochondrial iron transporter), member 37 | 1.739 | 0.044 | 1 |
| 26031 | OSBPL3 | oxysterol-binding protein-like 3 | 1.799 | 0.038 | 1 |
| 55502 | HES6 | hes family bHLH transcription factor 6 | 1.782 | 0.048 | 1 |
| 19 | ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 2.065 | 0.043 | 1 |
| 23597 | ACOT9 | acyl-CoA thioesterase 9 | 1.121 | 0.034 | 1 |
| 85407 | NKD1 | naked cuticle homolog 1 (Drosophila) | 1.215 | 0.042 | 1 |
| 351 | APP | amyloid beta (A4) precursor protein | 1.412 | 0.046 | 1 |
| 9123 | SLC16A3 | solute carrier family 16 (monocarboxylate transporter), member 3 | 2.763 | 0.046 | 1 |
| A. Gene Sets in the High GNA12 Expression Group | ||
| Name | Normalized Enrichment Score | FDR q-Value |
| epithelial–mesenchymal transition | 3.685 | q < 0.001 |
| myogenesis | 3.268 | q < 0.001 |
| UV response dn | 2.605 | q < 0.001 |
| angiogenesis | 2.561 | q < 0.001 |
| KRAS signaling up | 2.543 | q < 0.001 |
| inflammatory response | 2.434 | q < 0.001 |
| apical junction | 2.411 | q < 0.001 |
| coagulation | 2.407 | q < 0.001 |
| TNF-alpha signaling via NFkb | 2.309 | q < 0.001 |
| hypoxia | 2.119 | q < 0.001 |
| TGF-beta signaling | 2.096 | q < 0.001 |
| IL6/JAK/STAT3 signaling | 2.059 | q < 0.001 |
| hedgehog signaling | 2.005 | q < 0.001 |
| complement | 1.891 | 0.001 |
| notch signaling | 1.788 | 0.004 |
| IL2/STAT5 signaling | 1.772 | 0.004 |
| pancreas beta cells | 1.645 | 0.012 |
| mitotic spindle | 1.554 | 0.028 |
| apoptosis | 1.506 | 0.040 |
| B. Gene Sets in the High OLR1 Expression Group | ||
| Name | Normalized Enrichment Score | FDR q-Value |
| epithelial–mesenchymal transition | 2.309 | q < 0.001 |
| inflammatory response | 2.152 | q < 0.001 |
| myogenesis | 2.121 | q < 0.001 |
| angiogenesis | 2.089 | q < 0.001 |
| coagulation | 2.017 | q < 0.001 |
| KRAS signaling up | 1.996 | q < 0.001 |
| allograft rejection | 1.910 | q < 0.001 |
| complement | 1.762 | 0.002 |
| IL2/STAT5 signaling | 1.702 | 0.004 |
| IL6/JAK/STAT3 signaling | 1.696 | 0.004 |
| interferon-gamma response | 1.687 | 0.004 |
| UV response dn | 1.585 | 0.013 |
| TNF-alpha signaling via NFkb | 1.550 | 0.019 |
| apical junction | 1.464 | 0.048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koma, A.; Asai, S.; Minemura, C.; Oshima, S.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 9947. https://doi.org/10.3390/ijms22189947
Koma A, Asai S, Minemura C, Oshima S, Kinoshita T, Kikkawa N, Koshizuka K, Moriya S, Kasamatsu A, Hanazawa T, et al. Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2021; 22(18):9947. https://doi.org/10.3390/ijms22189947
Chicago/Turabian StyleKoma, Ayaka, Shunichi Asai, Chikashi Minemura, Sachi Oshima, Takashi Kinoshita, Naoko Kikkawa, Keiichi Koshizuka, Shogo Moriya, Atsushi Kasamatsu, Toyoyuki Hanazawa, and et al. 2021. "Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma" International Journal of Molecular Sciences 22, no. 18: 9947. https://doi.org/10.3390/ijms22189947
APA StyleKoma, A., Asai, S., Minemura, C., Oshima, S., Kinoshita, T., Kikkawa, N., Koshizuka, K., Moriya, S., Kasamatsu, A., Hanazawa, T., Uzawa, K., & Seki, N. (2021). Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences, 22(18), 9947. https://doi.org/10.3390/ijms22189947






