A Human Pan-Cancer System Analysis of Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 3 (PLOD3)
Abstract
1. Introduction
2. Results
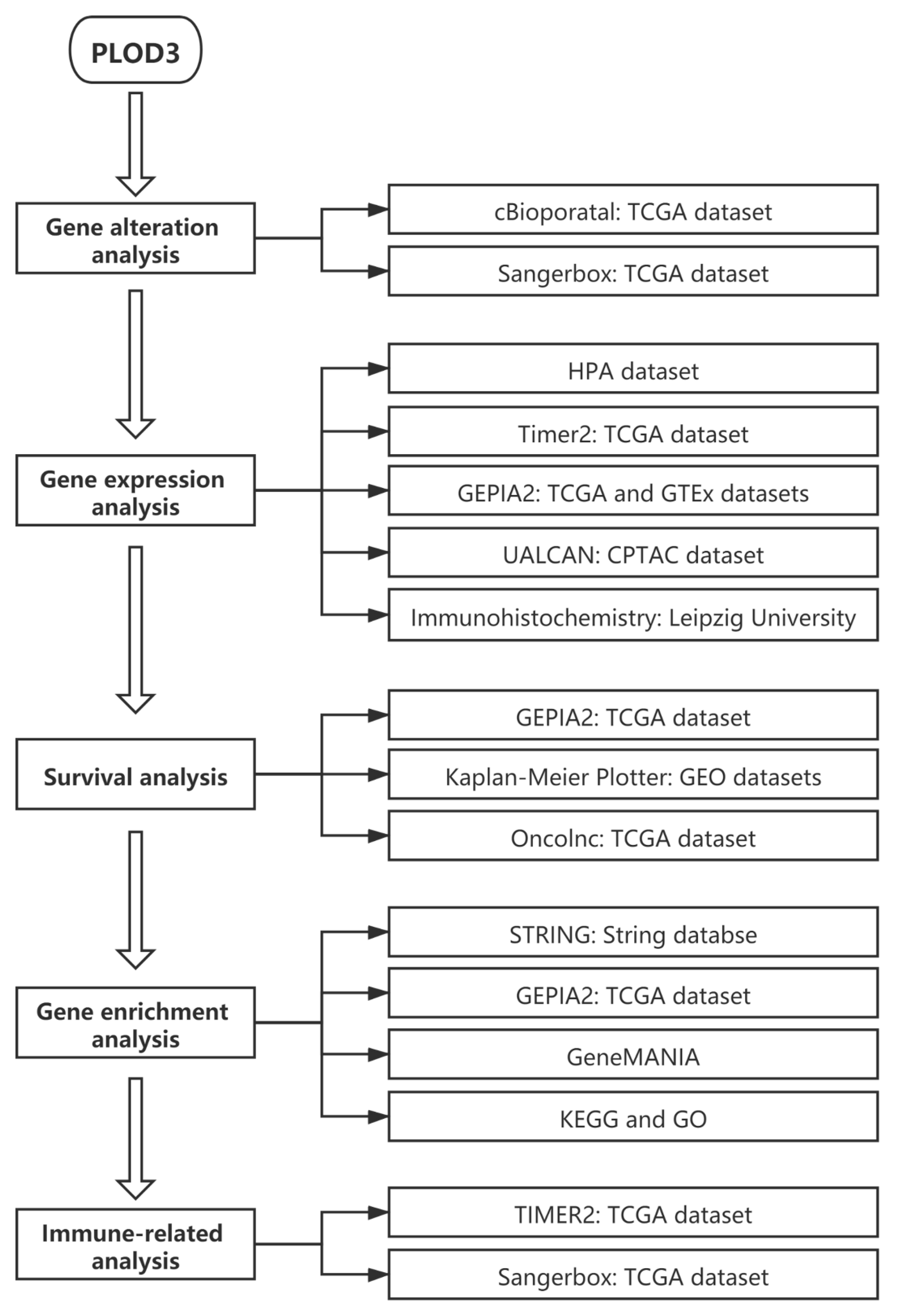
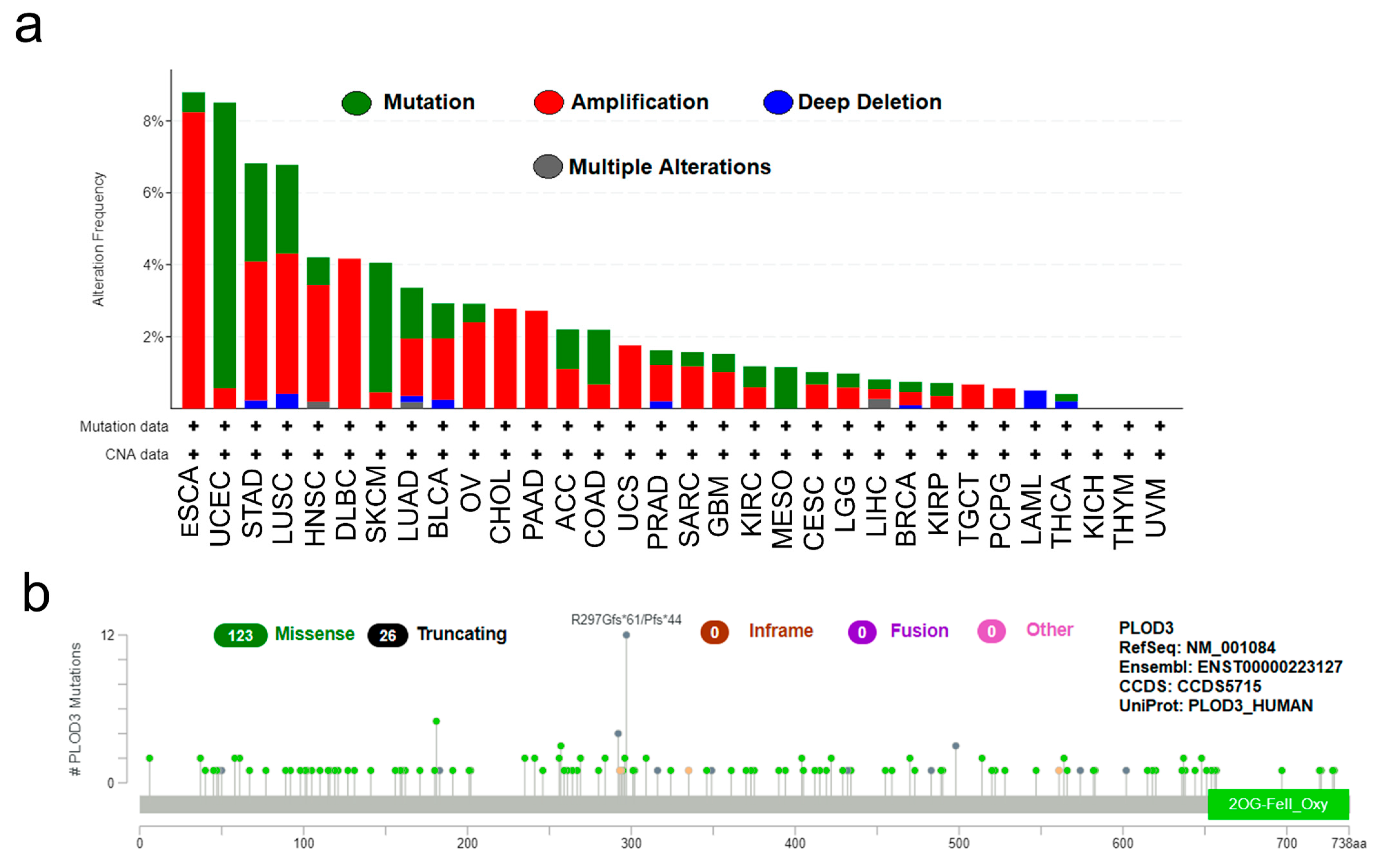
2.1. Experiment Setup and Genetic Alteration Analysis Data
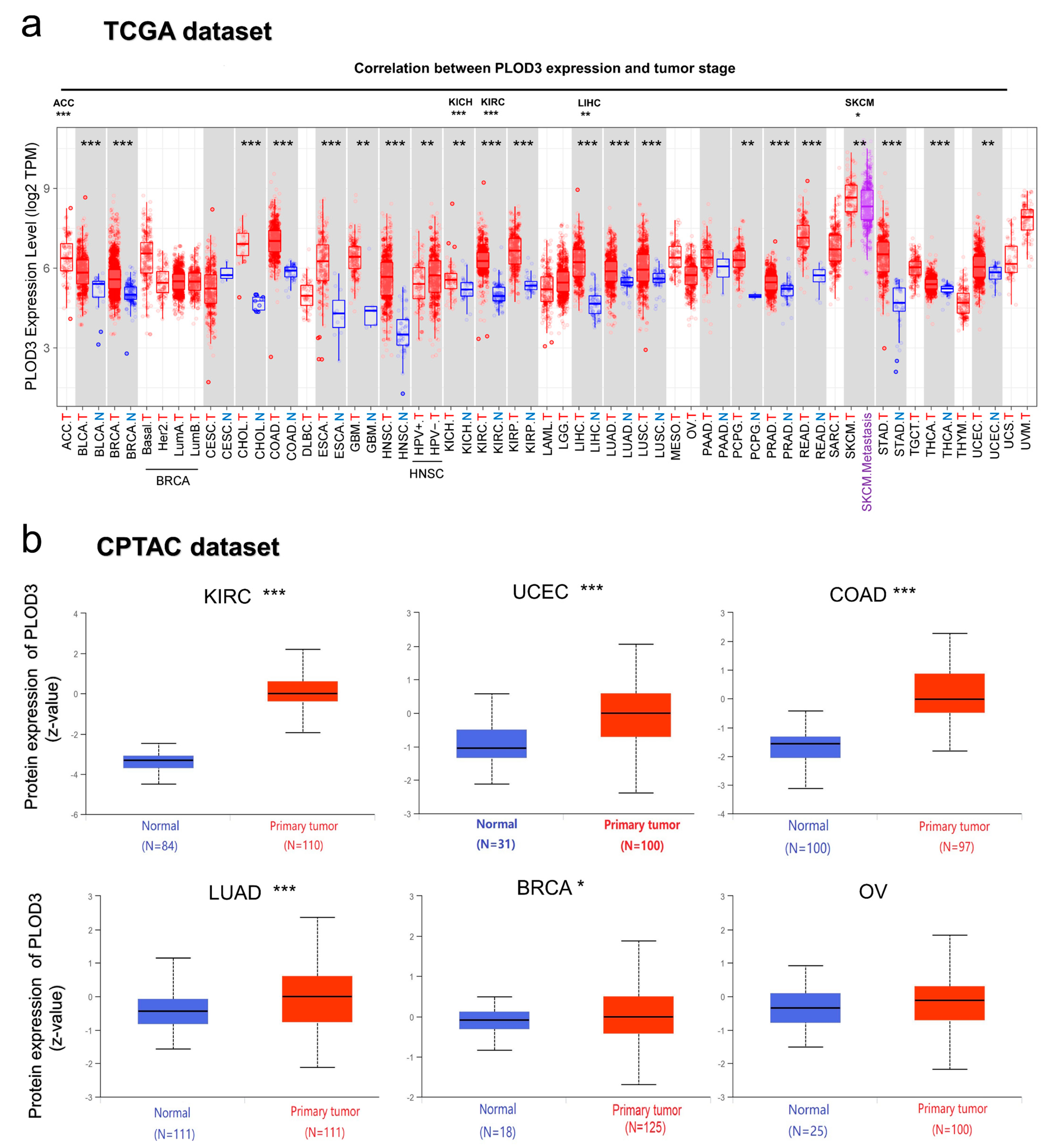
2.2. Gene Expression Analysis Data
2.3. Survival Analysis Data
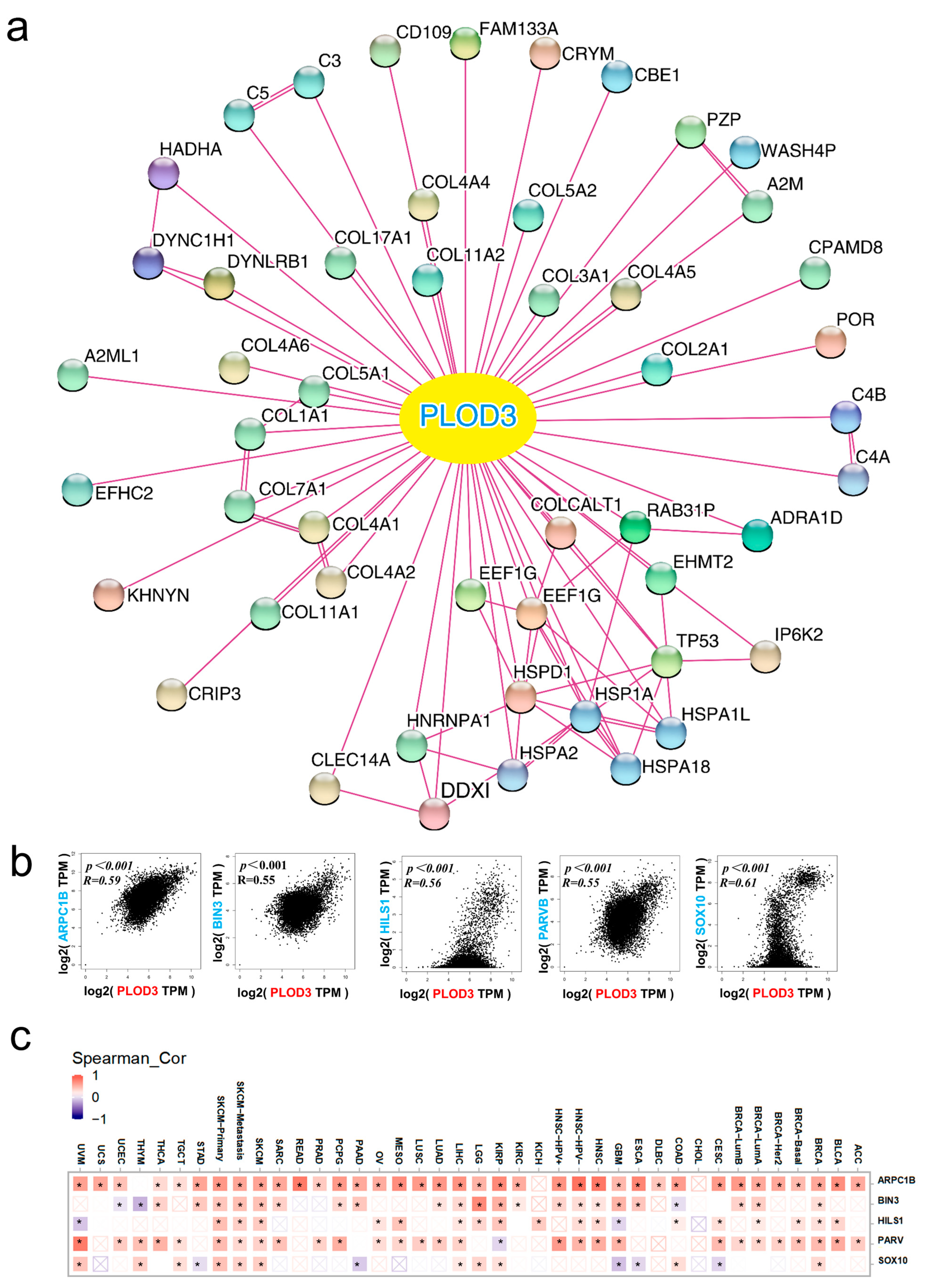
2.4. Analysis of Related Genes Network
2.5. KEGG Pathway and GO Enrichment Analysis Data
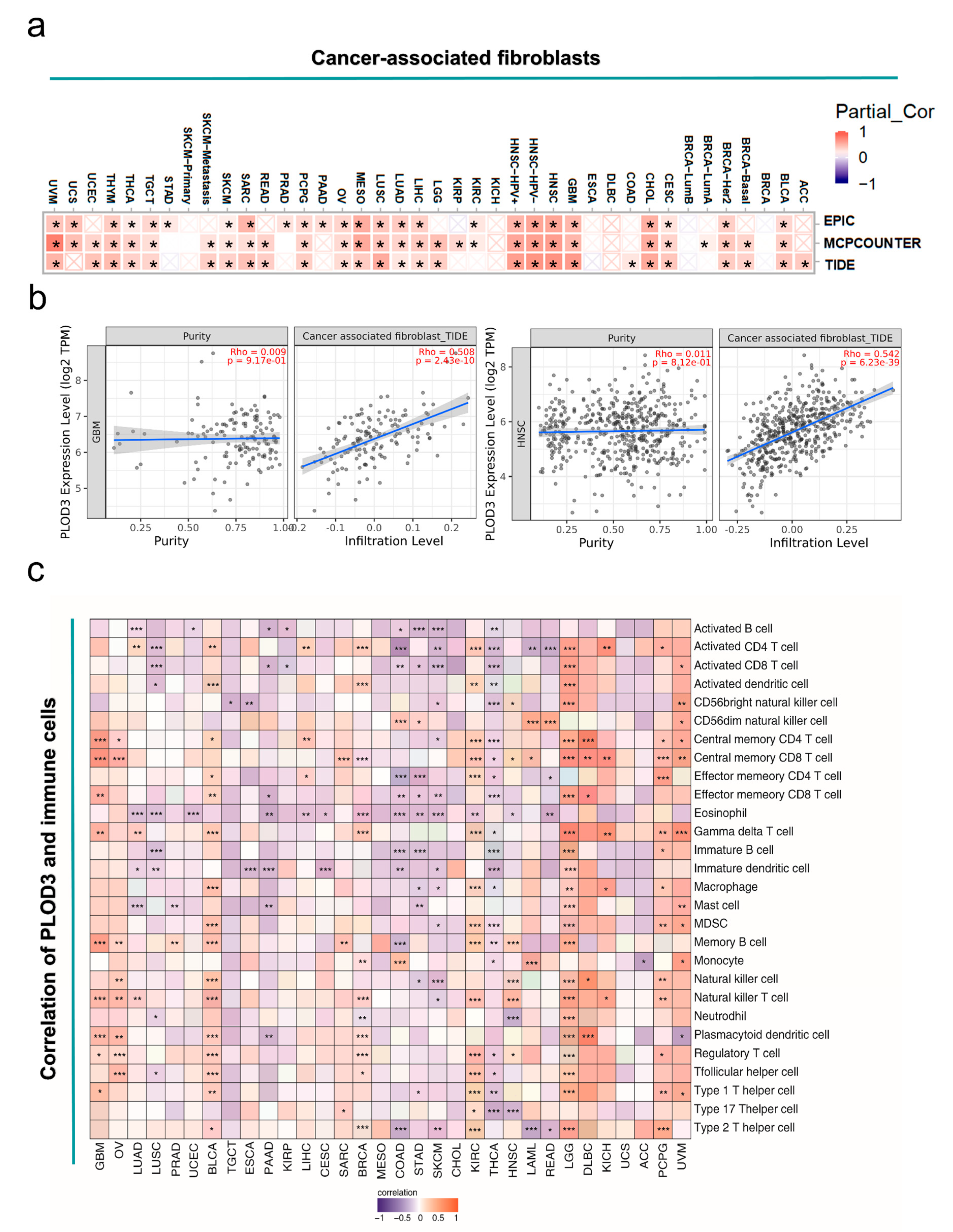
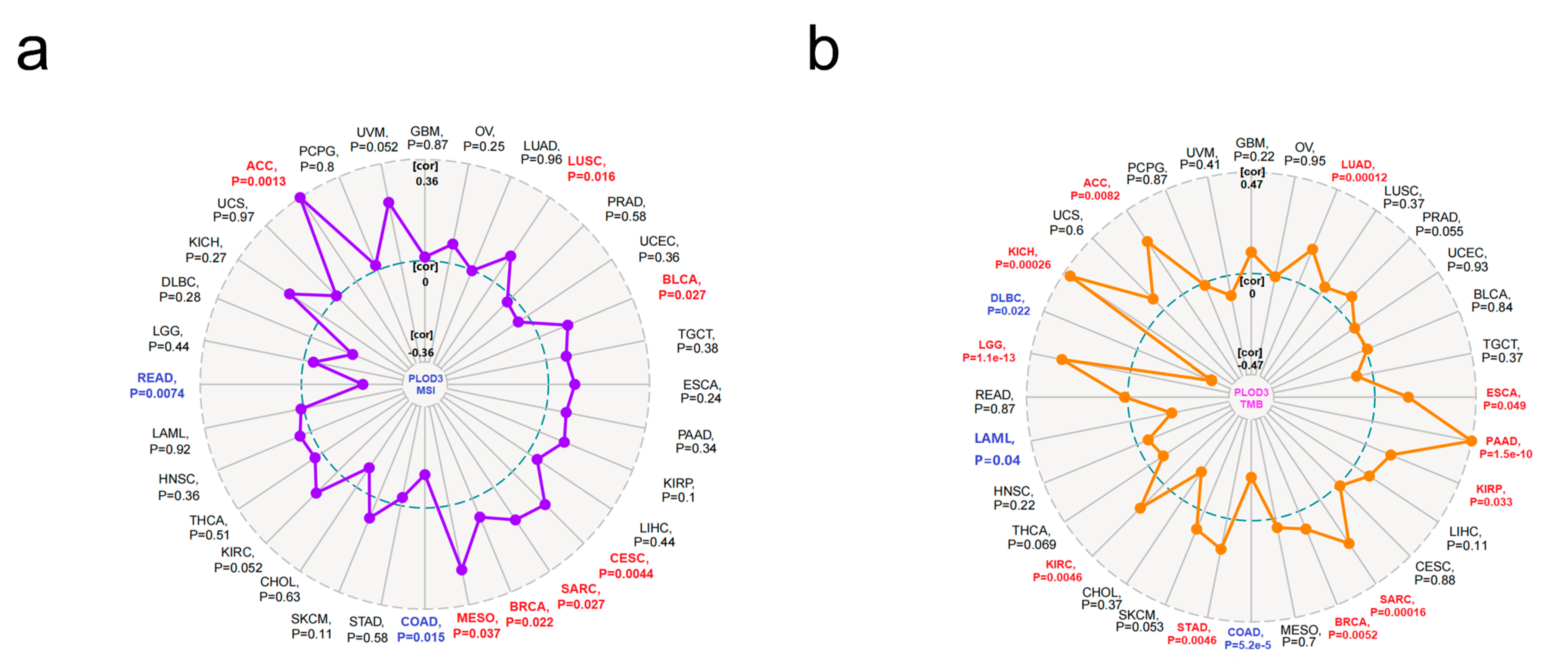
2.6. Immune Infiltration Analysis Data
3. Discussion
4. Materials and Methods
4.1. Genetic Alteration Analysis
4.2. Gene expression Analysis
4.3. Survival Prognosis Analysis
4.4. Construction of Related Genes Network
4.5. KEGG Pathway and GO Enrichment Analysis
4.6. Immune Infiltration Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salo, A.M.; Cox, H.; Farndon, P.; Moss, C.; Grindulis, H.; Risteli, M.; Robins, S.P.; Myllylä, R. A Connective Tissue Disorder Caused by Mutations of the Lysyl Hydroxylase 3 Gene. Am. J. Hum. Genet. 2008, 83, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Vahidnezhad, H.; Youssefian, L.; Saeidian, A.H.; Touati, A.; Pajouhanfar, S.; Baghdadi, T.; Shadmehri, A.A.; Giunta, C.; Kraenzlin, M.; Syx, D.; et al. Mutations in PLOD3, encoding lysyl hydroxylase 3, cause a complex connective tissue disorder including recessive dystrophic epidermolysis bullosa-like blistering phenotype with abnormal anchoring fibrils and type VII collagen deficiency. Matrix Biol. 2019, 81, 91–106. [Google Scholar] [CrossRef]
- Li, S.S.; Lian, Y.F.; Huang, Y.L.; Huang, Y.H.; Xiao, J. Overexpressing PLOD family genes predict poor prognosis in gastric cancer. J. Cancer 2020, 11, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Heikkinen, J.; Hautala, T.; Kivirikko, K.I.; Myllylä, R. Structure and Expression of the Human Lysyl Hydroxylase Gene (PLOD): Introns 9 and 16 Contain Alu Sequences at the Sites of Recombination in Ehlers-Danlos Syndrome Type VI Patients. Genomics 1994, 24, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Schneider, V.A.; Granato, M. Genomic structure and embryonic expression of zebrafish lysyl hydroxylase 1 and lysyl hydroxylase 2. Matrix Biol. 2007, 26, 12–19. [Google Scholar] [CrossRef][Green Version]
- Scietti, L.; Campioni, M.; Forneris, F. SiMPLOD, a Structure-Integrated Database of Collagen Lysyl Hydroxylase (LH/PLOD) Enzyme Variants. J. Bone Miner. Res. 2019, 34, 1376–1382. [Google Scholar] [CrossRef]
- Wang, B.; Xu, L.; Ge, Y.; Cai, X.; Li, Q.; Yu, Z.; Wang, J.; Wang, Y.; Lu, C.; Wang, D.; et al. PLOD3 is Upregulated in Gastric Cancer and Correlated with Clinicopathologic Characteristics. Clin. Lab. 2019, 65, 25–30. [Google Scholar] [CrossRef]
- Stone, R.C.; Pastar, I.; Ojeh, N.; Chen, V.; Liu, S.; Garzon, K.I.; Tomic-Canic, M. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016, 365, 495–506. [Google Scholar] [CrossRef]
- Lee, C.H. Reversal of Epithelial–Mesenchymal Transition by Natural Anti-Inflammatory and Pro-Resolving Lipids. Cancers 2019, 11, 1841. [Google Scholar] [CrossRef]
- Roversi, G.; Pfundt, R.; Moroni, R.F.; Magnani, I.; Van Reijmersdal, S.; Pollo, B.; Straatman, H.; Larizza, L.; Schoenmakers, E.F.P.M. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene 2006, 25, 1571–1583. [Google Scholar] [CrossRef]
- Hui, D.; Paiva, C.E.; Del Fabbro, E.G.; Steer, C.; Naberhuis, J.; Van De Wetering, M.; Fernandez-Ortega, P.; Morita, T.; Suh, S.Y.; Bruera, E.; et al. Prognostication in advanced cancer: Update and directions for future research. Support. Care Cancer 2019, 27, 1973–1984. [Google Scholar] [CrossRef]
- Baek, J.H.; Yun, H.S.; Kwon, G.T.; Kim, J.Y.; Lee, C.W.; Song, J.Y.; Um, H.D.; Kang, C.M.; Park, J.K.; Kim, J.S.; et al. PLOD3 promotes lung metastasis via regulation of STAT3. Cell Death Dis. 2018, 9, 1138. [Google Scholar] [CrossRef]
- Tsai, C.K.; Huang, L.C.; Tsai, W.C.; Huang, S.M.; Lee, J.T.; Hueng, D.Y. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget 2018, 9, 15705–15720. [Google Scholar] [CrossRef]
- Saxena, A.K.; Tiwari, M.; Agarwal, M.; Kumar, A.A. Prediction of 3D Protein Structure Based on The Mutation of AKAP3 and PLOD3 Genes in The Case of Non-Obstructive Azoospermia. Int. J. Fertil. Steril. 2020, 14, 102–109. [Google Scholar] [CrossRef]
- Ewans, L.J.; Colley, A.; Gaston-Massuet, C.; Gualtieri, A.; Cowley, M.J.; McCabe, M.J.; Anand, D.; Lachke, S.A.; Scietti, L.; Forneris, F.; et al. Pathogenic variants in PLOD3 result in a Stickler syndrome-like connective tissue disorder with vascular complications. J. Med. Genet. 2019, 56, 629–638. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Balmain, A. The critical roles of somatic mutations and environmental tumor-promoting agents in cancer risk. Nat. Genet. 2020, 52, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Visconti, L.; Nelissen, K.; Deckx, L.; Van Den Akker, M.; Adriaensen, W.; Daniels, L.; Matheï, C.; Linsen, L.; Hellings, N.; Stinissen, P.; et al. Prognostic value of circulating cytokines on overall survival and disease-free survival in cancer patients. Biomark. Med. 2014, 8, 297–306. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, R. Roles of PLODs in Collagen Synthesis and Cancer Progression. Front. Cell Dev. Biol. 2018, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Deng, L.; Zhu, J.; Rychahou, P.G.; Xu, R. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014, 14, 1. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Oudin, M.J.; Jonas, O.; Kosciuk, T.; Broye, L.C.; Guido, B.C.; Wyckoff, J.; Riquelme, D.; Lamar, J.M.; Asokan, S.B.; Whittaker, C.; et al. Tumor Cell–Driven Extracellular Matrix Remodeling Drives Haptotaxis during Metastatic Progression. Cancer Discov. 2016, 6, 516–531. [Google Scholar] [CrossRef]
- Cheon, D.J.; Tong, Y.; Sim, M.S.; Dering, J.; Berel, D.; Cui, X.; Lester, J.; Beach, J.A.; Tighiouart, M.; Walts, A.E.; et al. A Collagen-Remodeling Gene Signature Regulated by TGF-β Signaling Is Associated with Metastasis and Poor Survival in Serous Ovarian Cancer. Clin. Cancer Res. 2014, 20, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Gournier, H.; Goley, E.D.; Niederstrasser, H.; Trinh, T.; Welch, M.D. Reconstitution of Human Arp2/3 Complex Reveals Critical Roles of Individual Subunits in Complex Structure and Activity. Mol. Cell 2001, 8, 1041–1052. [Google Scholar] [CrossRef]
- Welch, M.D.; DePace, A.H.; Verma, S.; Iwamatsu, A.; Mitchison, T.J. The Human Arp2/3 Complex Is Composed of Evolutionarily Conserved Subunits and Is Localized to Cellular Regions of Dynamic Actin Filament Assembly. J. Cell Biol. 1997, 138, 375–384. [Google Scholar] [CrossRef]
- Matsuda, C.; Kameyama, K.; Suzuki, A.; Mishima, W.; Yamaji, S.; Okamoto, H.; Nishino, I.; Hayashi, Y.K. Affixin activates Rac1 via βPIX in C2C12 myoblast. FEBS Lett. 2008, 582, 1189–1196. [Google Scholar] [CrossRef]
- Ye, X.; Feng, C.; Gao, T.; Mu, G.; Zhu, W.; Yang, Y. Linker Histone in Diseases. Int. J. Biol. Sci. 2017, 13, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Gulfidan, G.; Turanli, B.; Beklen, H.; Sinha, R.; Arga, K.Y. Pan-cancer mapping of differential protein-protein interactions. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Yeger-Lotem, E.; Sharan, R. Human protein interaction networks across tissues and diseases. Front. Genet. 2015, 6, 257. [Google Scholar] [CrossRef] [PubMed]
- Patil, C.; Walter, P. Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001, 13, 349–355. [Google Scholar] [CrossRef]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef]
- Schröder, M.; Kaufman, R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005, 74, 739–789. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016, 529, 326–335. [Google Scholar] [CrossRef]
- Wu, C.; Duan, Y.; Gong, S.; Kallendrusch, S.; Schopow, N.; Osterhoff, G. Integrative and Comprehensive Pancancer Analysis of Regulator of Chromatin Condensation 1 (RCC1). Int. J. Mol. Sci. 2021, 22, 7374. [Google Scholar] [CrossRef]
- Fridman, W.H.; Galon, J.; Dieu-Nosjean, M.-C.; Cremer, I.; Fisson, S.; Damotte, D.; Pagès, F.; Tartour, E.; Sautès-Fridman, C. Immune Infiltration in Human Cancer: Prognostic Significance and Disease Control. Curr. Top. Microbiol. Immunol. 2010, 344, 1–24. [Google Scholar] [CrossRef]
- Fu, J.; Yu, A.; Tang, J.; He, B.; Chen, W. Adoptive CD8 + T cell therapy generates immunological memory to inhibit melanoma metastasis. Am. J. Transl. Res. 2020, 12, 7262–7274. [Google Scholar] [PubMed]
- Wu, Z.; Chen, H.; Luo, W.; Zhang, H.; Li, G.; Zeng, F.; Deng, F. The Landscape of Immune Cells Infiltrating in Prostate Cancer. Front. Oncol. 2020, 10, 517637. [Google Scholar] [CrossRef] [PubMed]
- Steven, A.; Seliger, B. The Role of Immune Escape and Immune Cell Infiltration in Breast Cancer. Breast Care 2018, 13, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Kwa, M.Q.; Herum, K.M.; Brakebusch, C. Cancer-associated fibroblasts: How do they contribute to metastasis? Clin. Exp. Metastasis 2019, 36, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- He, Q.F.; Xu, Y.; Li, J.; Huang, Z.M.; Li, X.H.; Wang, X. CD81 T-cell exhaustion in cancer: Mechanisms and new area for cancer immunotherapy. Brief. Funct. Genom. 2019, 18, 99–106. [Google Scholar] [CrossRef]
- Zhang, L.; Romero, P. Metabolic Control of CD8+ T Cell Fate Decisions and Antitumor Immunity. Trends Mol. Med. 2018, 24, 30–48. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef]
- Uchihara, T.; Miyake, K.; Yonemura, A.; Komohara, Y.; Itoyama, R.; Koiwa, M.; Yasuda, T.; Arima, K.; Harada, K.; Eto, K.; et al. Extracellular Vesicles from Cancer-Associated Fibroblasts Containing Annexin A6 Induces FAK-YAP Activation by Stabilizing b1 Integrin, Enhancing Drug Resistance. Cancer Res. 2020, 80, 3222–3235. [Google Scholar] [CrossRef]
- Seton-Rogers, S. Fibroblasts orchestrate tumour initiation. Nat. Rev. Cancer 2020, 20, 301. [Google Scholar] [CrossRef] [PubMed]
- Pein, M.; Insua-Rodríguez, J.; Hongu, T.; Riedel, A.; Meier, J.; Wiedmann, L.; Decker, K.; Essers, M.A.G.; Sinn, H.P.; Spaich, S.; et al. Metastasis-initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat. Commun. 2020, 11, 1494. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blaženović, I.; et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Murciano-Goroff, Y.R.; Warner, A.B.; Wolchok, J.D. The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell Res. 2020, 30, 507–519. [Google Scholar] [CrossRef]
- Mattox, A.K.; Lee, J.; Westra, W.H.; Pierce, R.H.; Ghossein, R.; Faquin, W.C.; Diefenbach, T.J.; Morris, L.G.; Lin, D.T.; Wirth, L.J.; et al. PD-1 Expression in Head and Neck Squamous Cell Carcinomas Derives Primarily from Functionally Anergic CD4+ TILs in the Presence of PD-L1+ TAMs. Cancer Res. 2017, 77, 6365–6374. [Google Scholar] [CrossRef]
- Haanen, J.B.A.G.; Robert, C. Immune Checkpoint Inhibitors. Prog. Tumor Res. 2015, 42, 55–66. [Google Scholar]
- Syn, N.L.; Teng, M.W.L.; Mok, T.S.K.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Yang, B.; Zhao, Y.; Wang, L.; Zhao, Y.; Wei, L.; Chen, D.; Chen, Z. Identification of PLOD Family Genes as Novel Prognostic Biomarkers for Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 1695. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front. Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Postow, M.A.; Callahan, M.K.; Wolchok, J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015, 33, 1974–1982. [Google Scholar] [CrossRef]
- Shekarian, T.; Valsesia-Wittmann, S.; Brody, J.; Michallet, M.C.; Depil, S.; Caux, C.; Marabelle, A. Pattern recognition receptors: Immune targets to enhance cancer immunotherapy. Ann. Oncol. 2017, 28, 1756–1766. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yao, J.; Song, L.; Zhang, S.; Huang, T.; Li, Y. Local and abscopal responses in advanced intrahepatic cholangiocarcinoma with low TMB, MSS, pMMR and negative PD-L1 expression following combined therapy of SBRT with PD-1 blockade. J. Immunother. Cancer 2019, 7, 204. [Google Scholar] [CrossRef]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.H.; Yun, H.S.; Kwon, G.T.; Lee, J.; Kim, J.Y.; Jo, Y.; Cho, J.M.; Lee, C.W.; Song, J.Y.; Ahn, J.; et al. PLOD3 suppression exerts an anti-tumor effect on human lung cancer cells by modulating the PKC-delta signaling pathway. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Li, J.; Wei, S.; Qi, P.; Ji, H.; Su, J.; Du, N.; Zhang, X. Knockdown of PLOD3 suppresses the malignant progression of renal cell carcinoma via reducing TWIST1 expression. Mol. Cell. Probes 2020, 53, 101608. [Google Scholar] [CrossRef]
- Jiang, H.; Guo, W.; Yuan, S.; Song, L. PLOD1 Is a Prognostic Biomarker and Mediator of Proliferation and Invasion in Osteosarcoma. BioMed Res. Int. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Brückner, L.; Reinshagen, A.; Hoang, N.A.; Höhn, A.K.; Lordick, F.; Bechmann, I.; Aktas, B.; Nel, I.; Kallendrusch, S. FOXM1 Inhibition in Ovarian Cancer Tissue Cultures Affects Individual Treatment Susceptibility Ex Vivo. Cancers 2021, 13, 956. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, S.; Duan, Y.; Wu, C.; Osterhoff, G.; Schopow, N.; Kallendrusch, S. A Human Pan-Cancer System Analysis of Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 3 (PLOD3). Int. J. Mol. Sci. 2021, 22, 9903. https://doi.org/10.3390/ijms22189903
Gong S, Duan Y, Wu C, Osterhoff G, Schopow N, Kallendrusch S. A Human Pan-Cancer System Analysis of Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 3 (PLOD3). International Journal of Molecular Sciences. 2021; 22(18):9903. https://doi.org/10.3390/ijms22189903
Chicago/Turabian StyleGong, Siming, Yingjuan Duan, Changwu Wu, Georg Osterhoff, Nikolas Schopow, and Sonja Kallendrusch. 2021. "A Human Pan-Cancer System Analysis of Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 3 (PLOD3)" International Journal of Molecular Sciences 22, no. 18: 9903. https://doi.org/10.3390/ijms22189903
APA StyleGong, S., Duan, Y., Wu, C., Osterhoff, G., Schopow, N., & Kallendrusch, S. (2021). A Human Pan-Cancer System Analysis of Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 3 (PLOD3). International Journal of Molecular Sciences, 22(18), 9903. https://doi.org/10.3390/ijms22189903






