Searching for Biological Function of the Mysterious PA2504 Protein from Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Characteristic of PA2504 Mutants
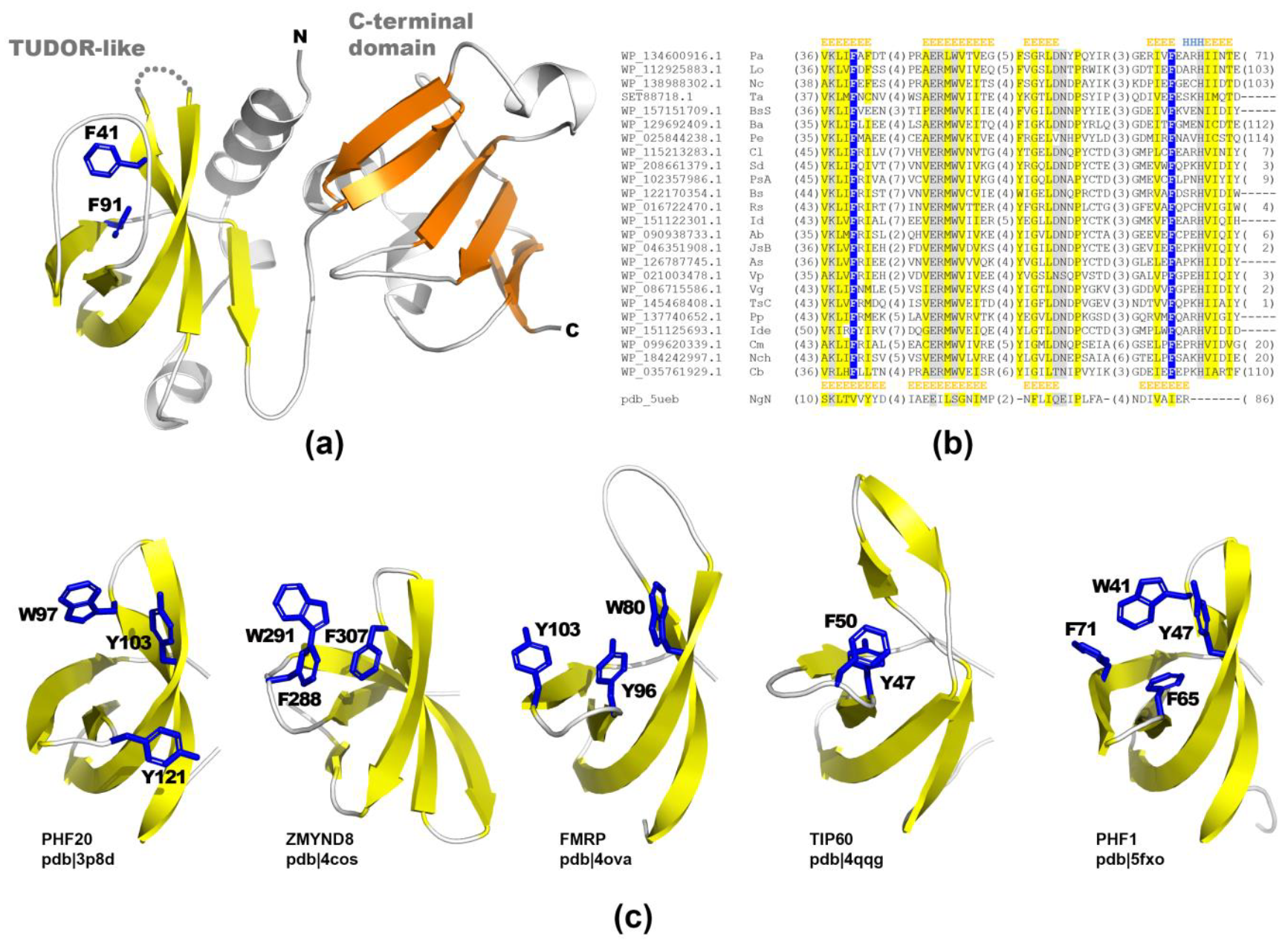
2.2. Structural Studies of PA2504 Protein
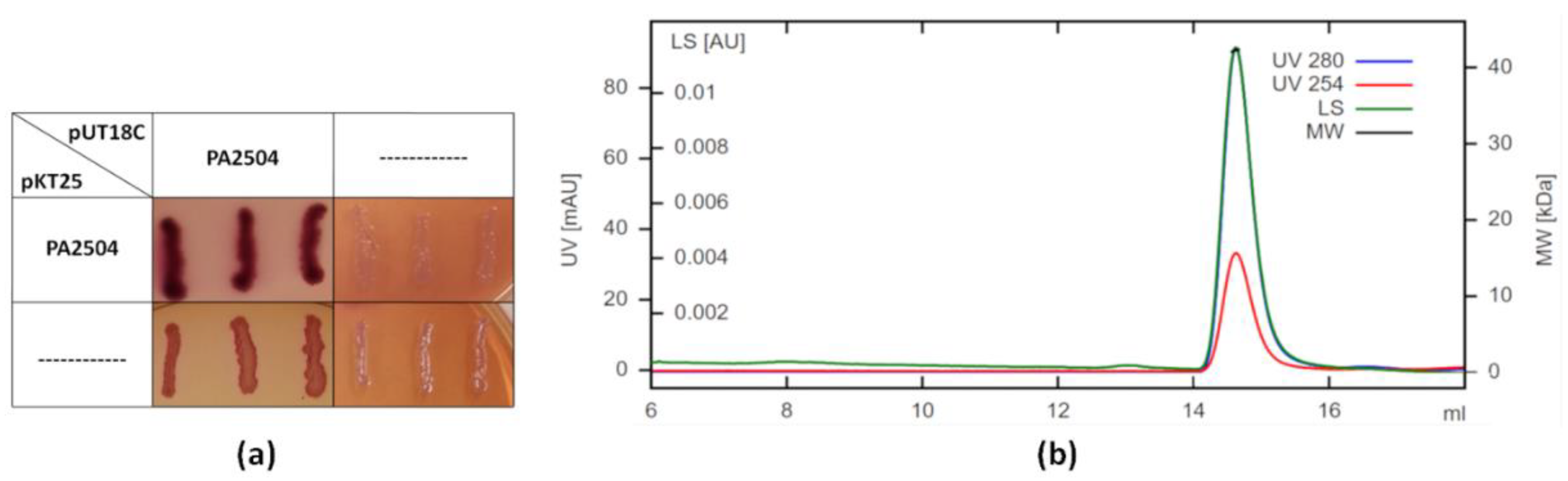
2.3. Oligomerisation of PA2504
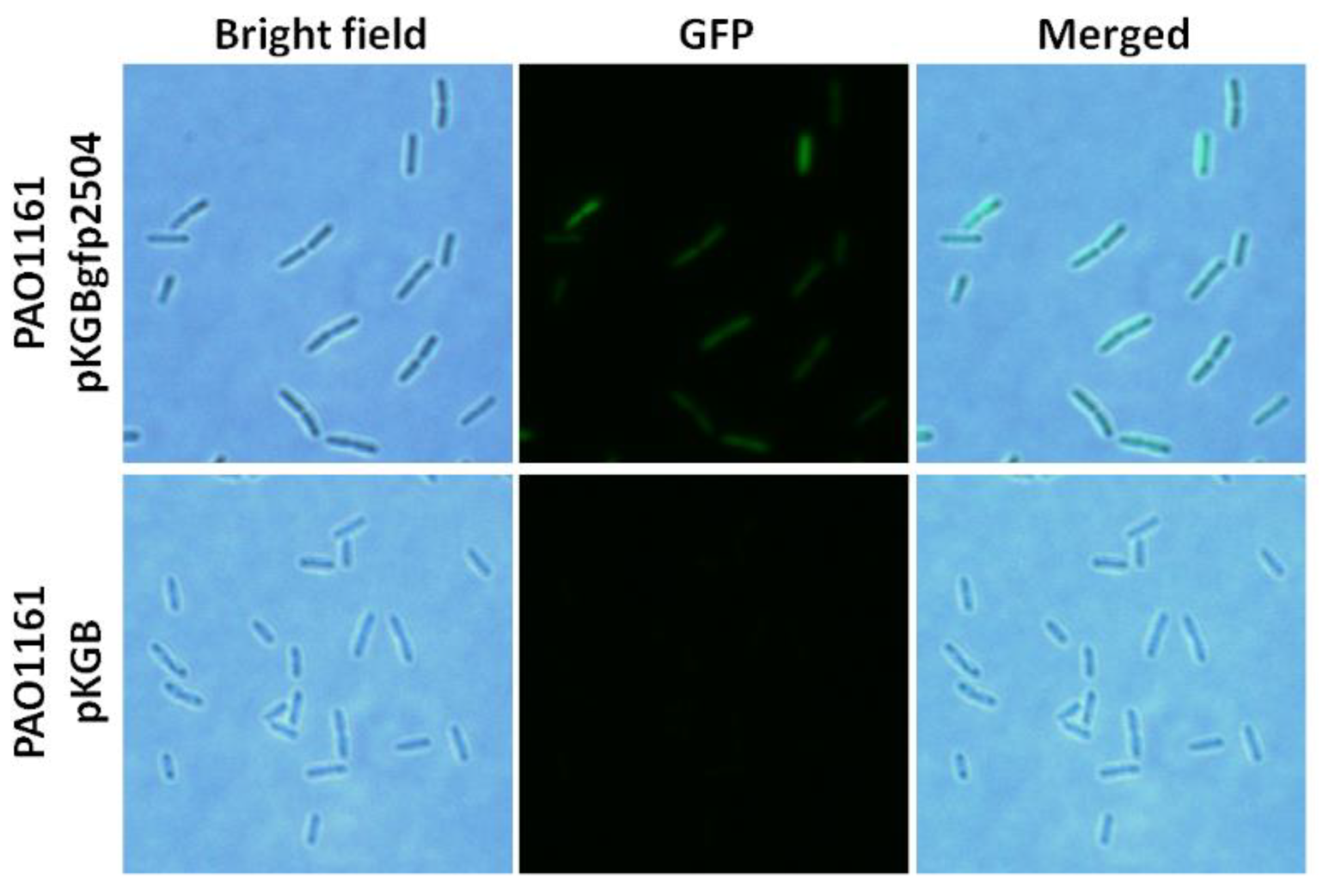
2.4. Cellular Localisation of PA2504
2.5. Transcriptomic Analysis of ΔPA2504 Mutant
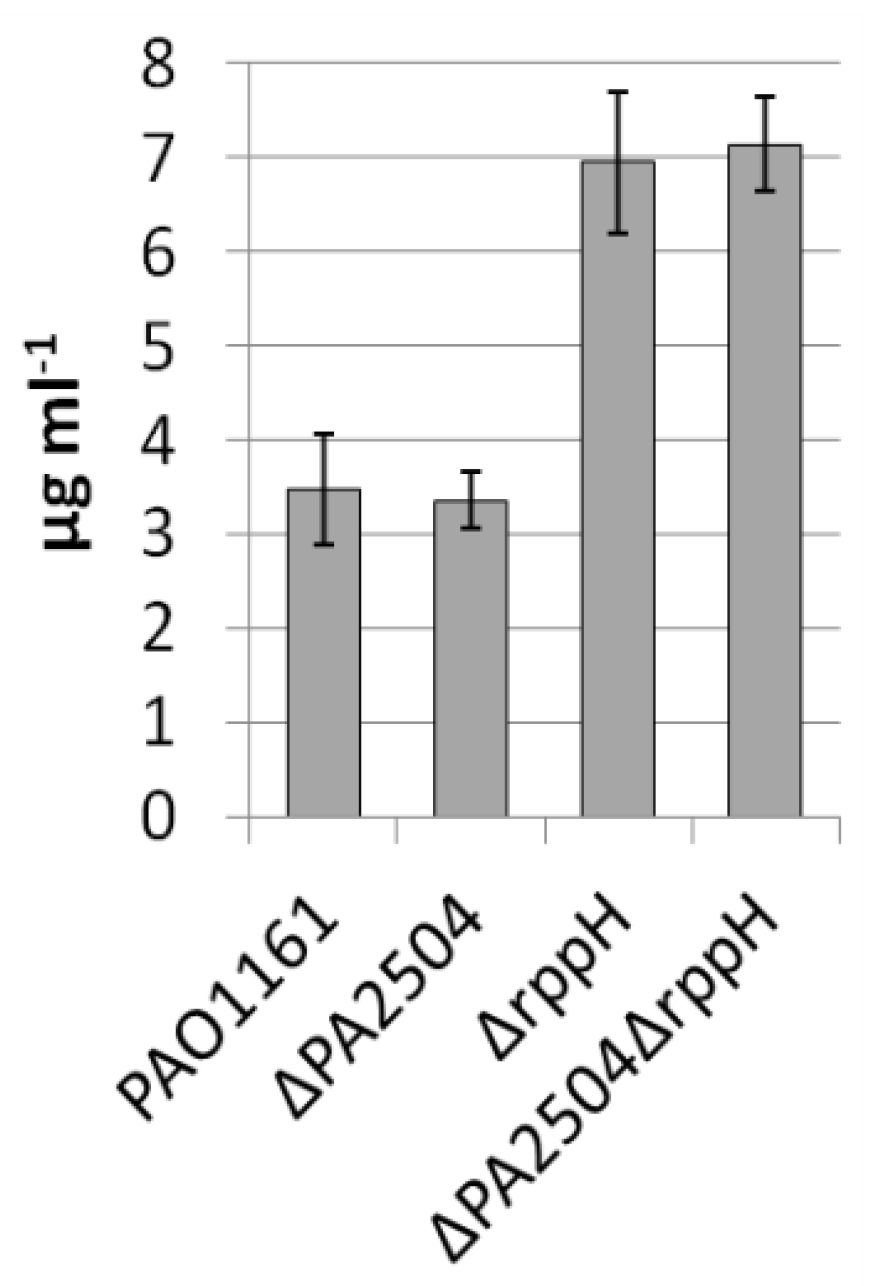
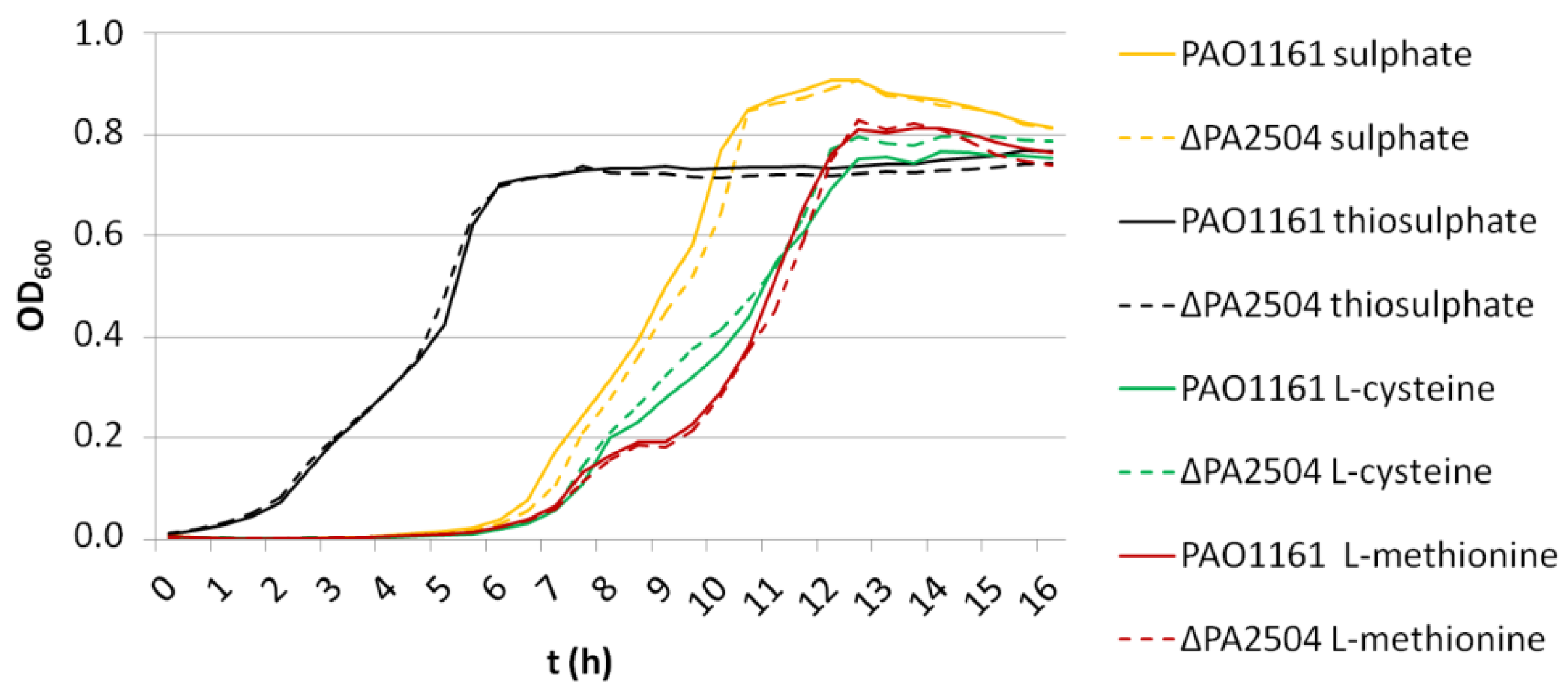
2.6. Growth of ΔPA2504, ΔrppH, and ΔPA2504ΔrppH Mutants on Different Sulphur Sources
2.7. Search for PA2504 Cellular Partners
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Deletion of PA2504
4.3. Overproduction of PA2504
4.4. Pyocyanin Quantification
4.5. Molecular Protein Modelling
4.6. Purification of His6-Tagged PA2504 by Affinity Chromatography
4.7. SEC-MALS Analysis
4.8. Bacterial Two-Hybrid System (BACTH)
4.9. Bacterial Two-Hybrid Library Screening
4.10. Cellular Localisation of PA2504
4.11. In Vivo Protein Crosslinking and Purification of Protein Complexes
4.12. Mass Spectrometry
4.13. RNA Isolation
4.14. RT-qPCR
4.15. RNA-Seq Analysis
4.16. Bioinformatic Analysis of RNA-Seq Results
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BACTH | Bacterial two hybrid |
| DTT | Ditiotreitol |
| EDTA | Ethylenediamine tetraacetic acid |
| FC | Fold change |
| IPTG | Isopropyl ß-D-1-thiogalactopyranoside |
| RNA-Seq | High-throughput RNA sequencing |
| RT-PCR | Reverse transcription PCR |
| RT-qPCR | Reverse transcription quantitative PCR |
| SEC-MALS | Exclusion chromatography combined with multi angle light scattering |
| X-Gal | 5-Bromo-4-Chloro-3-Indolyl β-D-Galactopyranoside |
References
- Spagnolo, A.M.; Sartini, M.; Cristina, M.L. Pseudomonas aeruginosa in the Healthcare Facility Setting. Rev. Med. Microbiol. 2021, 32, 169–175. [Google Scholar] [CrossRef]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Winsor, G.L.; Lam, D.K.W.; Fleming, L.; Lo, R.; Whiteside, M.D.; Yu, N.Y.; Hancock, R.E.W.; Brinkman, F.S.L. Pseudomonas Genome Database: Improved Comparative Analysis and Population Genomics Capability for Pseudomonas Genomes. Nucleic Acids Res. 2011, 39, D596–D600. [Google Scholar] [CrossRef] [PubMed]
- Deana, A.; Celesnik, H.; Belasco, J.G. The Bacterial Enzyme RppH Triggers Messenger RNA Degradation by 5′ Pyrophosphate Removal. Nature 2008, 451, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Kujawa, M.; Lirski, M.; Ziecina, M.; Drabinska, J.; Modzelan, M.; Kraszewska, E. Nudix-Type RNA Pyrophosphohydrolase Provides Homeostasis of Virulence Factor Pyocyanin and Functions as a Global Regulator in Pseudomonas aeruginosa: Nudix RNA Hydrolase Regulates, P. Aeruginosa Virulence. Mol. Microbiol. 2017, 106, 381–394. [Google Scholar] [CrossRef] [Green Version]
- Bielecki, P.; Komor, U.; Bielecka, A.; Müsken, M.; Puchałka, J.; Pletz, M.W.; Ballmann, M.; Martins dos Santos, V.A.; Weiss, S.; Häussler, S. Ex Vivo Transcriptional Profiling Reveals a Common Set of Genes Important for the Adaptation of Pseudomonas aeruginosa to Chronically Infected Host Sites: Ex Vivo Transcriptional Profiling. Environ. Microbiol. 2013, 15, 570–587. [Google Scholar] [CrossRef]
- Juhas, M.; Wiehlmann, L.; Huber, B.; Jordan, D.; Lauber, J.; Salunkhe, P.; Limpert, A.S.; von Götz, F.; Steinmetz, I.; Eberl, L.; et al. Global Regulation of Quorum Sensing and Virulence by VqsR in Pseudomonas aeruginosa. Microbiology 2004, 150, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Raneri, M.; Pinatel, E.; Peano, C.; Rampioni, G.; Leoni, L.; Bianconi, I.; Jousson, O.; Dalmasio, C.; Ferrante, P.; Briani, F. Pseudomonas aeruginosa Mutants Defective in Glucose Uptake Have Pleiotropic Phenotype and Altered Virulence in Non-Mammal Infection Models. Sci. Rep. 2018, 8, 16912. [Google Scholar] [CrossRef]
- Kusiak, M.B. Molecular and Structural Analysis of parB Gene of Pseudomonas aeruginosa. Ph.D. Thesis, Department of Microbial Biochemistry IBB PAS, Warsaw, Poland, 5 October 2010. [Google Scholar]
- Savitsky, P.; Krojer, T.; Fujisawa, T.; Lambert, J.-P.; Picaud, S.; Wang, C.-Y.; Shanle, E.K.; Krajewski, K.; Friedrichsen, H.; Kanapin, A.; et al. Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin. Cell Rep. 2016, 17, 2724–2737. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like Proteins Link the PRC2 Complex to CpG Islands. Nature 2017, 549, 287–291. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Park, S.; Badeaux, A.I.; Kim, D.; Lee, J.; Thompson, J.R.; Yan, F.; Kaneko, S.; Yuan, Z.; Botuyan, M.V.; et al. PHF20 Is an Effector Protein of P53 Double Lysine Methylation That Stabilizes and Activates P53. Nat. Struct. Mol. Biol. 2012, 19, 916–924. [Google Scholar] [CrossRef]
- Ramos, A.; Hollingworth, D.; Adinolfi, S.; Castets, M.; Kelly, G.; Frenkiel, T.A.; Bardoni, B.; Pastore, A. The Structure of the N-Terminal Domain of the Fragile X Mental Retardation Protein: A Platform for Protein-Protein Interaction. Structure 2006, 14, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, Z.; Fu, Y.; He, Q.; Jiang, L.; Zheng, J.; Gao, Y.; Mei, P.; Chen, Z.; Ren, X. The Amino-Terminal Structure of Human Fragile X Mental Retardation Protein Obtained Using Precipitant-Immobilized Imprinted Polymers. Nat. Commun. 2015, 6, 6634. [Google Scholar] [CrossRef] [PubMed]
- Vermeij, P.; Kertesz, M.A. Pathways of Assimilative Sulfur Metabolism in Pseudomonas Putida. J. Bacteriol. 1999, 181, 5833–5837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, D.H. Escherichia Coli Contains a Protein That Is Homologous in Function and N-Terminal Sequence to the Protein Encoded by the NifS Gene of AzotobacterVinelandii and That Can Participate in the Synthesis of the Fe-S Cluster of Dihydroxy-Acid Dehydratase. J. Biol. Chem. 1996, 271, 16068–16074. [Google Scholar] [CrossRef]
- Morigasaki, S.; Umeyama, A.; Kawano, Y.; Aizawa, Y.; Ohtsu, I. Defect of RNA PyrophosphohydrolaseRppH Enhances Fermentative Production of L-Cysteine in Escherichia Coli. J. Gen. Appl. Microbiol. 2020, 66, 307–314. [Google Scholar] [CrossRef]
- Culver, G.M. Assembly of the 30S Ribosomal Subunit. Biopolymers 2003, 68, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, A.; Rydén-Aulin, M. A Novel Mutation in Ribosomal Protein S4 Thataffects the Function of a Mutated RF1. Biochimie 2000, 82, 683–691. [Google Scholar] [CrossRef]
- Torres, M. Ribosomal Protein S4 Is a Transcription Factor with Properties Remarkably Similar to NusA, a Protein Involved in Both Non-Ribosomal and Ribosomal RNA Antitermination. EMBO J. 2001, 20, 3811–3820. [Google Scholar] [CrossRef] [Green Version]
- Sprinzl, M. Elongation Factor Tu: A Regulatory GTPase with an Integrated Effector. Trends Biochem. Sci. 1994, 19, 245–250. [Google Scholar] [CrossRef]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- López-Ochoa, J.; Montes-García, J.F.; Vázquez, C.; Sánchez-Alonso, P.; Pérez-Márquez, V.M.; Blackall, P.J.; Vaca, S.; Negrete-Abascal, E. Gallibacterium Elongation Factor-Tu Possesses Amyloid-like Protein Characteristics, Participates in Cell Adhesion, and Is Present in Biofilms. J. Microbiol. 2017, 55, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, D.S.; Zegarra, V.; Maksimova, E.; Nakamoto, J.A.; Kasatsky, P.; Paleskava, A.; Konevega, A.L.; Milón, P. How the Initiating Ribosome Copes with PpGpp to Translate MRNAs. PLoS Biol. 2020, 18, e3000593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeb, S.; Haas, D. Regulatory Roles of the GacS/GacA Two-Component System in Plant-Associated and Other Gram-Negative Bacteria. MPMI 2001, 14, 1351–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Huang, X.; Tang, L.; Wu, D.; Xu, Y. Global Control of GacA in Secondary Metabolism, Primary Metabolism, Secretion Systems, and Motility in the Rhizobacterium Pseudomonas aeruginosa M18. J. Bacteriol. 2013, 195, 3387–3400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pek, J.W.; Anand, A.; Kai, T. Tudor Domain Proteins in Development. Development 2012, 139, 2255–2266. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Dutta, D.; Thakur, K.G. Crystal Structure of Mycobacterium Tuberculosis CarD, an Essential RNA Polymerase Binding Protein, reveals a Quasidomain-Swapped Dimeric Structural Architecture: Crystal Structure of M. Tuberculosis CarD. Proteins 2014, 82, 879–884. [Google Scholar] [CrossRef]
- Ortiz, C.; Kureisaite-Ciziene, D.; Schmitz, F.; McLaughlin, S.H.; Vicente, M.; Löwe, J. Crystal Structure of the Z-Ring Associated Cell Division Protein ZapC from Escherichia Coli. FEBS Lett. 2015, 589, 3822–3828. [Google Scholar] [CrossRef] [Green Version]
- Forcada-Nadal, A.; Palomino-Schätzlein, M.; Neira, J.L.; Pineda-Lucena, A.; Rubio, V. The PipX Protein, When Not Bound to Its Targets, Has Its Signaling C-Terminal Helix in a Flexed Conformation. Biochemistry 2017, 56, 3211–3224. [Google Scholar] [CrossRef]
- Kawale, A.A.; Burmann, B.M. UvrD Helicase–RNA Polymerase Interactions Are Governed by UvrD’s Carboxy-Terminal Tudor Domain. Commun. Biol. 2020, 3, 607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, J.-Y.; Hu, H.; Ye, B.-C.; Tan, M. Systematic Proteomic Analysis of Protein Methylation in Prokaryotes and Eukaryotes Revealed Distinct Substrate Specificity. Proteomics 2018, 18, 1700300. [Google Scholar] [CrossRef]
- Singhal, A.; Virmani, R.; Naz, S.; Arora, G.; Gaur, M.; Kundu, P.; Sajid, A.; Misra, R.; Dabla, A.; Kumar, S.; et al. Methylation of Two-Component Response Regulator MtrA in Mycobacteria Negatively Modulates Its DNA Binding and Transcriptional Activation. Biochem. J. 2020, 477, 4473–4489. [Google Scholar] [CrossRef]
- Widjaja, M.; Harvey, K.L.; Hagemann, L.; Berry, I.J.; Jarocki, V.M.; Raymond, B.B.A.; Tacchi, J.L.; Gründel, A.; Steele, J.R.; Padula, M.P.; et al. Elongation Factor Tu Is a Multifunctional and Processed Moonlighting Protein. Sci. Rep. 2017, 7, 11227. [Google Scholar] [CrossRef] [Green Version]
- Prezioso, S.M.; Duong, D.M.; Kuiper, E.G.; Deng, Q.; Albertí, S.; Conn, G.L.; Goldberg, J.B. Trimethylation of Elongation Factor-Tu by the Dual Thermoregulated Methyltransferase EftM Does Not Impact Its Canonical Function in Translation. Sci. Rep. 2019, 9, 3553. [Google Scholar] [CrossRef] [PubMed]
- Głąbski, K.T. Role of Putative Partners of Par Proteins in Chromosome Segregation, Cell Division and Motility of Pseudomonas aeruginosa. Ph.D. Thesis, Department of Microbial Biochemistry IBB PAS, Warsaw, Poland, 6 June 2014. [Google Scholar]
- El-Sayed, A.K.; Hothersall, J.; Thomas, C.M. Quorum-Sensing-Dependent Regulation of Biosynthesis of the Polyketide Antibiotic Mupirocin in Pseudomonas Fluorescens NCIMB 10586 The GenBank Accession Numbers for the Sequences Determined in This Work Are AF318063 (MupA), AF318064 (MupR) and AF318065 (MupI). Microbiology 2001, 147, 2127–2139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A Bacterial Two-Hybrid System Based on a Reconstituted Signal Transduction Pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malagon, F. RNase III Is Required for Localization to the Nucleoid of the 5′ Pre-RRNA Leader and for Optimal Induction of RRNA Synthesis in E. Coli. RNA 2013, 19, 1200–1207. [Google Scholar] [CrossRef] [Green Version]
- Bartosik, A.A.; Mierzejewska, J.; Thomas, C.M.; Jagura-Burdzy, G. ParB Deficiency in Pseudomonas aeruginosa Destabilizes the Partner Protein ParA and Affects a Variety of Physiological Parameters. Microbiology 2009, 155, 1080–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and Characterization of Genes for a Second Anthranilate Synthase in Pseudomonas aeruginosa: Interchangeability of the Two Anthranilate Synthases and Evolutionary Implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [Green Version]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-Suite3 for Fast Remote Homology Detection and Deep Protein Annotation. BMC Bioinform. 2019, 20, 473. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved Protein Structure Prediction Using Predicted Interresidue Orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T. Protein Secondary Structure Prediction Based on Position-Specific Scoring Matrices 1 1Edited by G. von Heijne. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, L. DALI and the Persistence of Protein Shape. Protein Sci. 2020, 29, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lougheed, K.E.A.; Bennett, M.H.; Williams, H.D. An in Vivo Crosslinking System for Identifying Mycobacterial Protein–Protein Interactions. J. Microbiol. Methods 2014, 105, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Klockenbusch, C.; Kast, J. Optimization of Formaldehyde Cross-Linking for Protein Interaction Analysis of Non-Tagged Integrin β 1. J. Biomed. Biotechnol. 2010, 2010, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 8 June 2020).
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Analyzing RNA-seq Data with DESeq2. Available online: https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html (accessed on 8 June 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Edgar, R. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comparison of the Pseudomonas aeruginosa PA2504 Deficient Strain and the PAO1161 Wild Type Strain. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179150 (accessed on 1 July 2021).
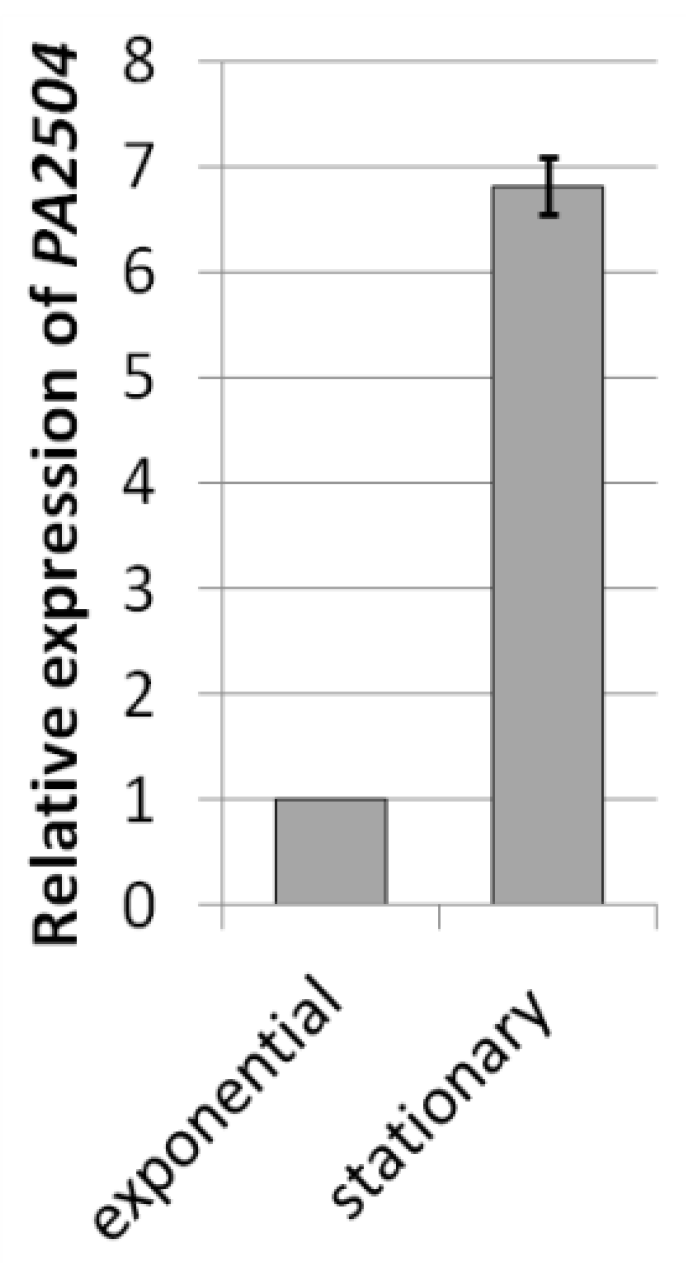


| Conditions | Strain | FC | Source |
|---|---|---|---|
| Burn wound isolate (human) vs. stationary growth in rich liquid medium | clinical isolate | −11.0 | [6] |
| Cystic fibrosis patient lung isolate vs. stationary growth in rich liquid medium | clinical isolate | −9.0 | [6] |
| ΔvqsR vs. WT, ABC minimal medium | PAO1 | +8.0 | [7] |
| Murine tumour isolate (mouse) vs. stationary growth phase in rich liquid medium | veterinary isolate | −6.3 | [6] |
| GUN (glucose uptake null) + glucose vs. WT | PAO1 | +3.6 | [8] |
| GUN (glucose uptake null) + glucose vs. WT + glucose | PAO1 | +2.8 | [8] |
| Gene ID | Name | Log2 FC | Product Description |
|---|---|---|---|
| PA0280 | cysA | −2.70 | Sulphatethiosulphate ABC transporter ATP-binding protein CysA |
| PA0281 | cysW | −2.76 | Sulphate transporter CysW |
| PA0282 | cysT | −2.79 | Sulphate transporter CysT |
| PA0283 | sbp | −3.13 | Sulphate-binding protein |
| PA0284 | oscA | −3.11 | Hypothetical protein/sulphur starvation response protein |
| PA0399 | PA0399 | −0.73 | Cystathionine beta-synthase |
| PA0400 | PA0400 | −0.54 | Cystathionine gamma-lyase |
| PA1245 | aprX | −0.92 | Extracellular protease AprX |
| PA1246 | aprD | −0.70 | Alkaline protease secretion ATP-binding protein AprD |
| PA1247 | aprE | −0.64 | Alkaline protease secretion protein AprE |
| PA1248 | aprF | −0.73 | Alkaline protease secretion protein AprF |
| PA1249 | aprA | −0.83 | Alkaline metalloproteinase |
| PA1493 | cysP | −0.55 | Sulphate ABC transporter substrate-binding protein |
| PA1756 | cysH | −0.98 | Phosphoadenosinephosphosulphate reductase |
| PA1837 | PA1837 | −1.52 | Hypothetical protein/oxidoreductase probably involved in sulphite reduction |
| PA1838 | cysI | −1.29 | Sulphite reductase |
| PA1912 | femI | −0.39 | ECF sigma factor FemI |
| PA2062 | PA2062 | −1.00 | Probable pyridoxal-phosphate dependent protein/IscS subfamily cysteine desulphurase |
| PA2086 | PA2086 | −1.12 | Epoxide hydrolase |
| PA2202 | PA2202 | −1.79 | Amino acid permease |
| PA2203 | PA2203 | −2.32 | Amino acid permease |
| PA2204 | PA2204 | −3.02 | ABC transporter |
| PA2328 | PA2328 | −0.86 | Hypothetical protein/nitrate transport protein NrtA precursor |
| PA2329 | PA2329 | −0.72 | ABC transporter ATP-binding protein/nitrate/sulphonate/bicarbonate ABC transporter ATPase |
| PA2330 | PA2330 | −0.74 | Hypothetical protein/acyl-CoA/acyl-ACP dehydrogenase |
| PA2426 | pvdS | −0.43 | Extracytoplasmic-function sigma-70 factor |
| PA2481 | PA2481 | +0.46 | Hypothetical protein/thiosulphate dehydrogenase |
| PA2594 | PA2594 | −1.40 | Putative periplasmic aliphatic sulphonate binding protein |
| PA2598 | PA2598 | −0.70 | Hypothetical protein/methanesulphonate monooxygenase |
| PA2786 | PA2786 | −1.10 | Hypothetical protein/GAF domain-containing protein |
| PA3441 | ssuF | −3.07 | Molybdopterin-binding protein/organosulphonate utilisation protein SsuF |
| PA3931 | PA3931 | −2.58 | Putative methionine-binding protein |
| PA3932 | PA3932 | −0.97 | Transcriptional regulator |
| PA4067 | oprG | −0.49 | Outer membrane protein OprG |
| PA4195 | PA4195 | −1.64 | Putative amino acid ABC transporter substrate-binding protein/ABC transporter glutamine-binding protein GlnH precursor |
| PA4442 | cysN | −1.87 | Bifunctional sulphate adenylyltransferase subunit 1/adenylylsulphate kinase |
| PA4443 | cysD | −2.22 | Sulphate adenylyltransferase subunit 2 |
| PA4470 | fumC1 | −0.29 | Fumarate hydratase |
| PA4471 | PA4471 | −0.58 | Hypothetical protein |
| PA5024 | ytnM | −0.44 | Hypothetical protein/sulphite exporter TauE/SafE |
| PA5025 | metY | −0.62 | O-acetylhomoserineaminocarboxypropyltransferase |
| PA5103 | puuR | −1.03 | Hypothetical protein/PhnD/SsuA/transferrin family substrate-binding protein |
| Plasmid | Relevant Features | Source |
|---|---|---|
| pKGB | pKGB8.0.2 vector, in this work referred to as pKGB; araBADp, araC, CmR, broad-host-range expression vector | [36] |
| pQE-80L | oriColE1ApR T5p lacOlacIq His6 tag, expression vector | Qiagen |
| pAKE600 | oriMB1 oriRK2ApRsacB | [37] |
| pKT25 | ori P15A, KmR, lacp–cyaT25 | [38] |
| pKNT25 | orip15, KmR, lacp–cyaT25 | [38] |
| pUT18C | oriColE1, ApR, lacp–cyaT18 | [38] |
| pBAD24-sfGFPx1 | araBADp, araC, ApR, Superfolder GFP ORF cloned into pBAD24 for expression in E. coli | [39] |
| pAKE2504 | pAKE600 plasmid with 275 bp upstream region of PA2504 gene with start codon and 253 bp downstream region of PA2504 gene with stop codon, inserted as EcoI-PstI and PstI-BamHI fragments | This work |
| pQE2504 | pQE-80L with PA2504 without start codon, inserted as BamHI-SalI fragment | This work |
| pKGB2504 | pKGB with PA2504Hisx6 inserted as EcoRI-SalI fragment | This work |
| pKGBgfp2504 | pKGB with sfGFP without stop codon, inserted as an EcoRI-HindIII fragment, and PA2504 without start codon, cloned as XbaI-SacI fragment | This work |
| pKT2504 | pKT25 with PA2504 without start codon, inserted as BamHI-EcoRI fragment | This work |
| pNTrppH | pKNT25 with PA2504 without stop codon, inserted as BamHI-EcoRI fragment | This work |
| p18C2504 | pUT18C with PA2504 without start codon, inserted as BamHI-EcoRI fragment | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drabinska, J.; Steczkiewicz, K.; Kujawa, M.; Kraszewska, E. Searching for Biological Function of the Mysterious PA2504 Protein from Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 9833. https://doi.org/10.3390/ijms22189833
Drabinska J, Steczkiewicz K, Kujawa M, Kraszewska E. Searching for Biological Function of the Mysterious PA2504 Protein from Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(18):9833. https://doi.org/10.3390/ijms22189833
Chicago/Turabian StyleDrabinska, Joanna, Kamil Steczkiewicz, Martyna Kujawa, and Elżbieta Kraszewska. 2021. "Searching for Biological Function of the Mysterious PA2504 Protein from Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 18: 9833. https://doi.org/10.3390/ijms22189833
APA StyleDrabinska, J., Steczkiewicz, K., Kujawa, M., & Kraszewska, E. (2021). Searching for Biological Function of the Mysterious PA2504 Protein from Pseudomonas aeruginosa. International Journal of Molecular Sciences, 22(18), 9833. https://doi.org/10.3390/ijms22189833






