Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango
Abstract
:1. Introduction
2. Results
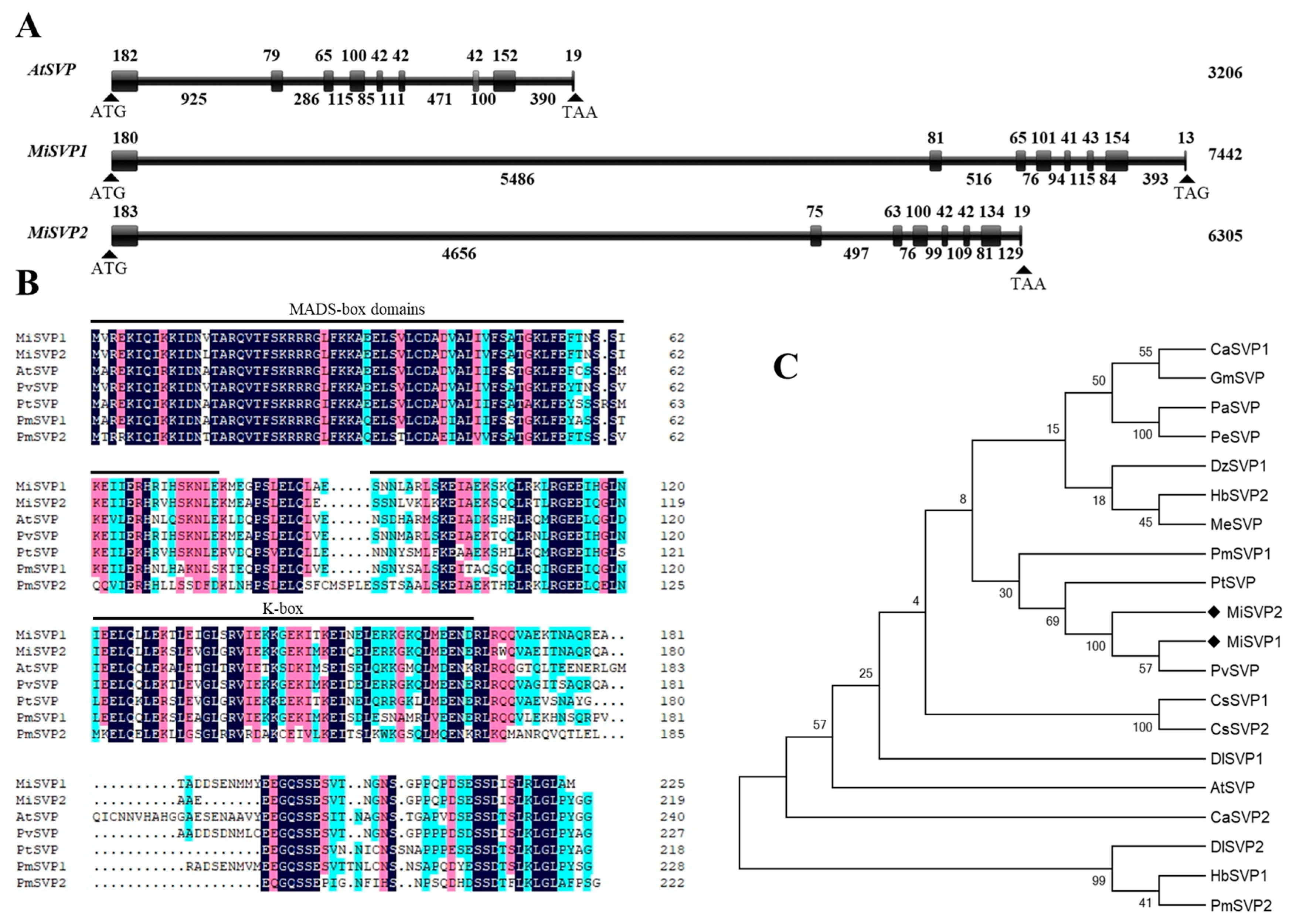
2.1. Cloning and Bioinformatics Analysis of MiSVPs
2.2. Expression Analysis of MiSVPs
2.2.1. Tissue Expression Analysis of MiSVPs
2.2.2. Temporal Expression Analysis of MiSVPs
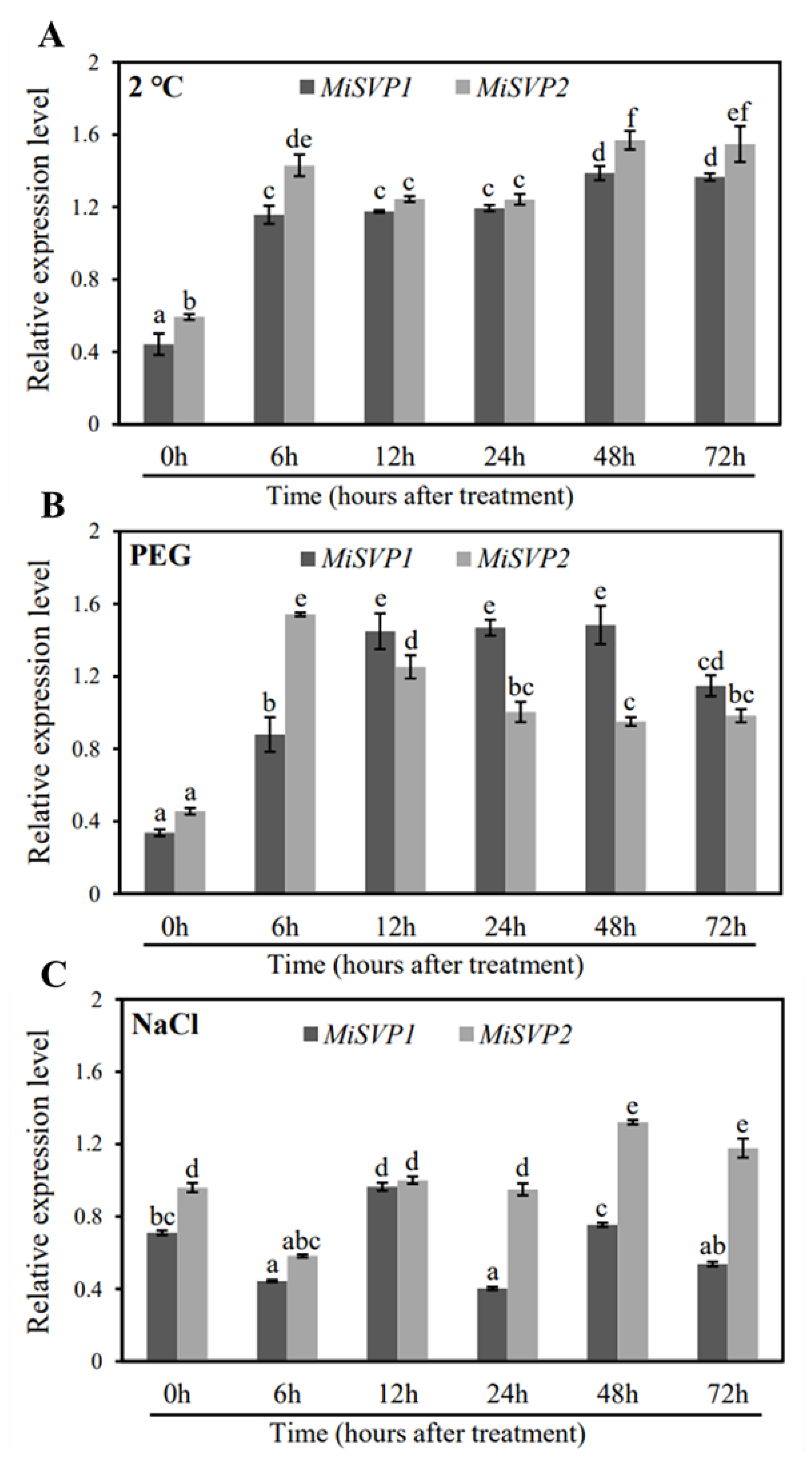
2.2.3. Effects of Adversity Treatments on the Alteration of MiSVPs Expression Levels
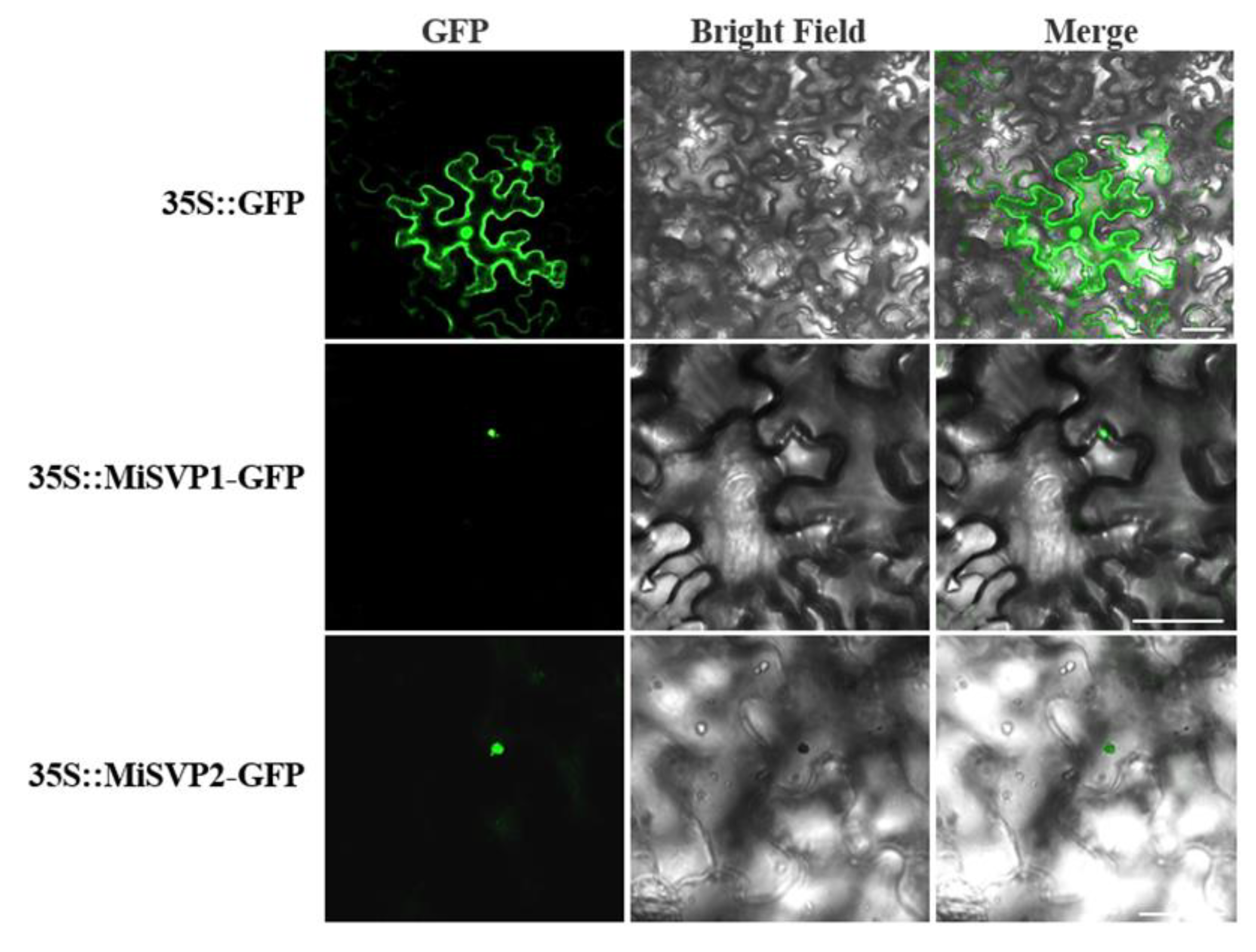
2.3. Subcellular Localization of MiSVPs
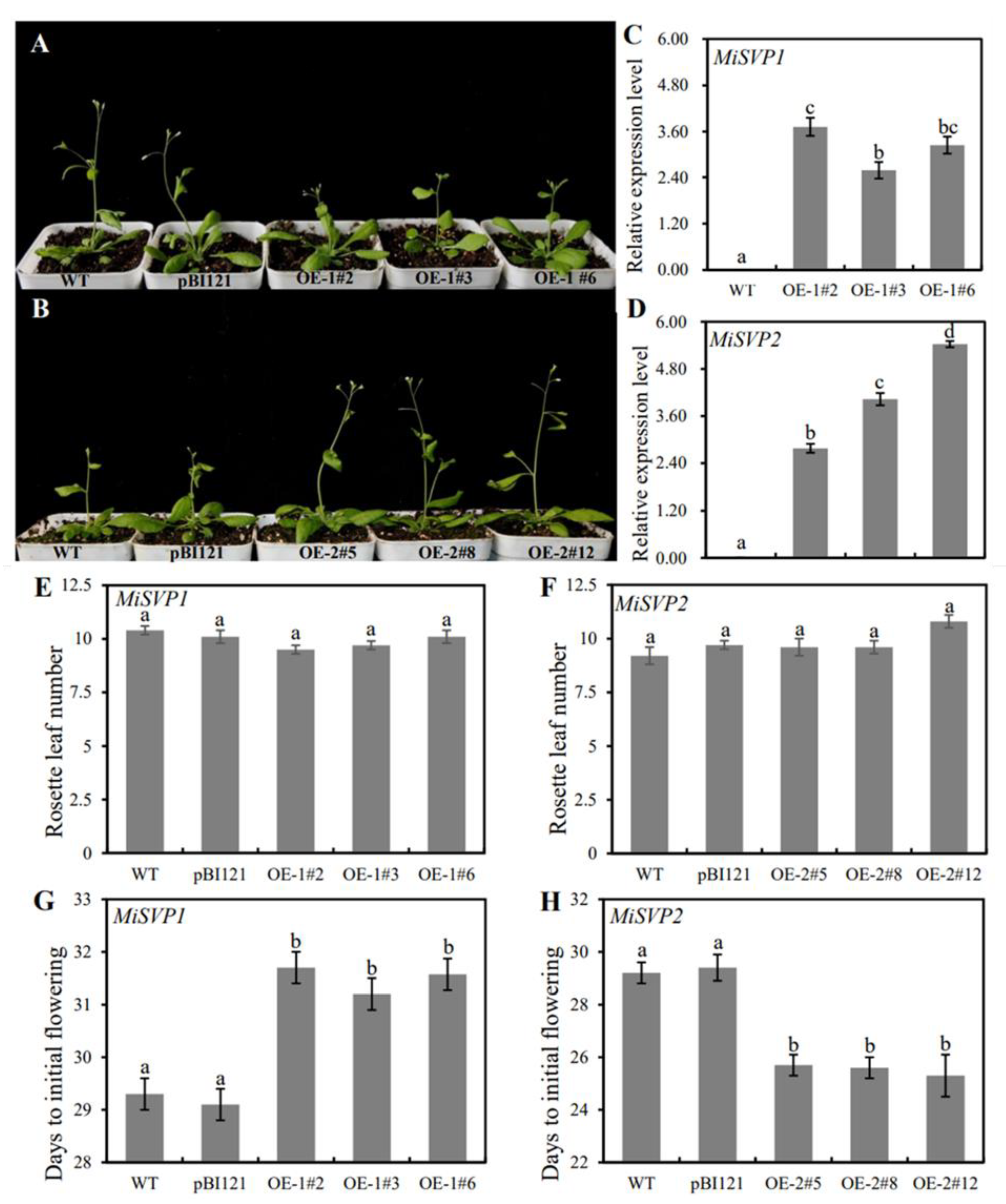
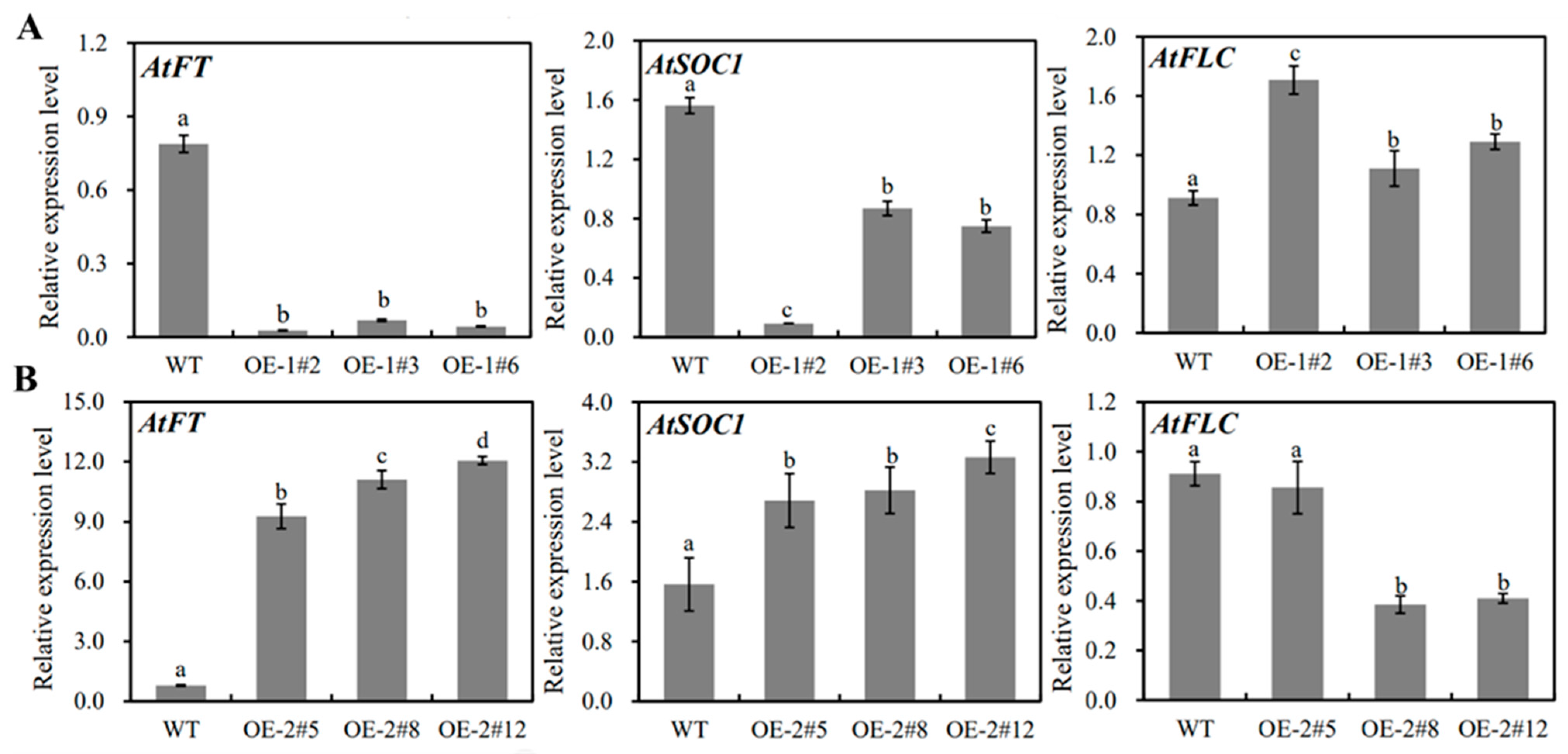
2.4. Functional Analysis of MiSVPs in Arabidopsis thaliana
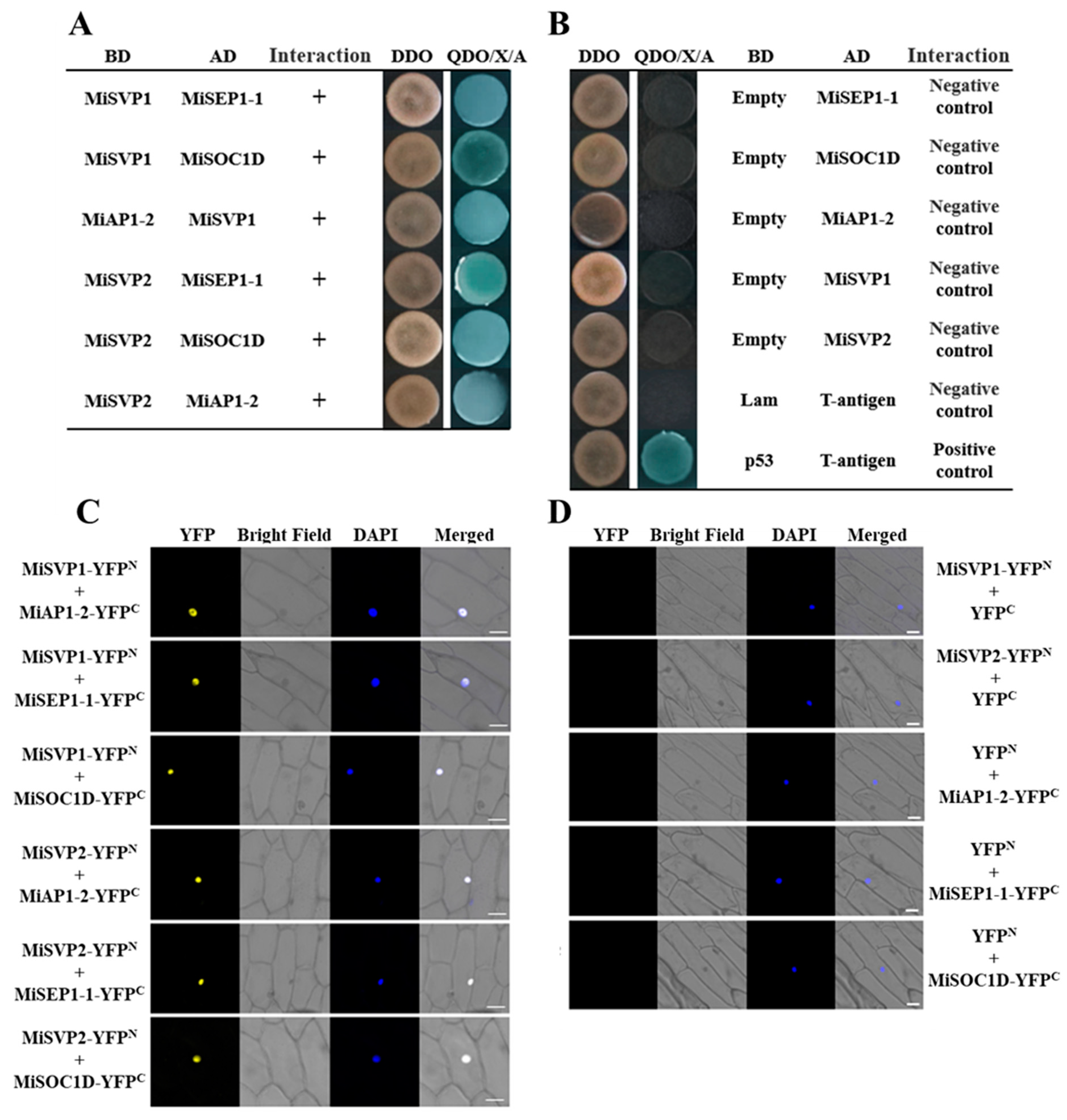
2.5. Screening and Validation of MiSVPs Interacting Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Cloning and Bioinformatics Analysis of MiSVPs
4.3. Expression Analysis of MiSVPs
4.4. Subcellular Localization
4.5. MiSVP Vector Construction and Transformation of Arabidopsis
4.6. Yeast Two-Hybrid Assay
4.7. Bimolecular Fluorescence Complementation (BiFC) Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Yang, X.F.; Qin, Q.P. Research progress on flowering pathway of woody fruit trees. Mod. Agric. Sci. Technol. 2010, 24, 111–112. [Google Scholar]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, Y.H.; Smith, R.W.; Jones, H.J.; Seaton, D.D.; Grima, R.; Halliday, K.J. Mathematical models light up plant signaling. Plant Cell 2014, 26, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Capovilla, G.; Schmid, M.; Posé, D. Control of flowering by ambient temperature. J. Exp. Bot. 2015, 66, 59–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergonzi, S.; AIbani, M.C.; Themaat, E.V.L.V.; Nordstrom, K.J.V.; Wang, R.H.; Schneeberger, K.; Perry, D.; Moerland, P.D.; George, C. Mechanisms of Age-Dependent response to winter temperature in perennial flowering of Arabisalpina. Science 2013, 340, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant. Biol. 2004, 7, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Horvath, D.P.; Sung, S.; Kim, D.; Chao, W.; Anderson, J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 2010, 73, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, H.; Saedler, P.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant. J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell. 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.; Torti, S.; Coupland, G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant. J. 2009, 60, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Ryu, H.S.; Chung, K.S.; Posé, D.; Kim, S.; Schmid, M.; Ahn, J.H. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 2013, 342, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 2006, 18, 1373–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, C.; Yu, H.X.; Fan, Y.; Zhang, X.J.; He, X.H. Research advance on the flowering mechanism of mango. Acta Hortic. 2019, 1, 17–22. [Google Scholar] [CrossRef]
- Yu, H.X.; Luo, C.; Fan, Y.; Zhang, X.J.; Huang, F.; Li, M.; He, X.H. Isolation and characterization of two APETALA1-Like genes from mango (Mangifera indica L.). Sci. Hortic. 2020, 259, 108814. [Google Scholar] [CrossRef]
- Wei, J.; Liu, D.; Liu, G.; Tang, J.; Chen, Y.Y. Molecular Cloning, Characterization, and Expression of MiSOC1: A Homolog of the Flowering Gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from Mango (Mangifera indica L). Front Plant Sci. 2016, 7, 1758. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.Y.; He, X.H.; Fan, Y.; Yu, H.X.; Wang, Y.H.; Xie, X.J.; Liu, Y.; Mo, X.; Wang, J.Y.; Luo, C. Isolation and functional characterization of three MiFTs genes from mango. Plant Physiol. Biochem. 2020, 155, 169–176. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, C.; Zhang, X.J.; Lu, X.X.; Yu, H.X.; Xie, X.J.; Fan, Z.Y.; Mo, X.; He, X.H. Overexpression of the mango MiCO gene delayed flowering time in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 2020, 143, 219–228. [Google Scholar] [CrossRef]
- Lu, S.H.; Meng, Z. Gene duplication and functional diversity of MADS-box gene family. Chin. Bull. Bot. 2007, 24, 60–70. [Google Scholar]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant. J. 2003, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.M.; Zhang, J.Z.; Mei, L.; Deng, X.X.; Hu, C.G.; Yao, J.L. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Mol. Biol. 2010, 74, 129–142. [Google Scholar] [CrossRef]
- Li, Y.S.; Zhou, Y.Z.; Yang, W.R.; Cheng, T.G.; Wang, J.; Zhang, Q.X. Isolation and functional characterization of SVP-like genes in Prunus mume. Sci. Hortic. 2017, 215, 91–101. [Google Scholar] [CrossRef]
- Zhao, Q. Isolation, Expression and Function Analysis of SVP-Like Genes in Strawberry; Shenyang Agricultural University: Shenyang, China, 2016. [Google Scholar]
- Wu, R.M.; Walton, E.F.; Richardson, A.C.; Wood, M.; Hellens, R.P.; Varkonyi-Gasic, E. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 2012, 63, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Wang, T.C.; Warren, B.A.W.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 2017, 68, 1071–1082. [Google Scholar] [CrossRef]
- Jaudal, M.; Monash, J.; Zhang, L.; Wen, J.; Mysore, K.S.; Macknight, R.; Putterill, J. Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 2014, 65, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Gao, Y.; Fan, M.; Yuan, L.; Wu, Z.; Zhang, Q. Overexpression of Chrysanthemum morifolium SVP gene delays blossoming and regulates inflorescence architecture in transgenic Arabidopsis. Can J. Plant Sci. 2017, 97, 1130–1139. [Google Scholar]
- Tang, X.L.; Liang, M.X.; Han, J.J.; Cheng, J.S.; Zhang, H.X.; Liu, X.H. Ectopic expression of LoSVP, a MADS-domain transcription factor from lily, leads to delayed flowering in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 289–298. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Zhang, Z.; Lin, S.; Lin, S.; Yang, X. The role of EjSVPs in flower initiation in Eriobotrya japonica. Int. J. Mol. Sci. 2019, 20, 5933. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Tomes, S.; Karunairetnam, S.; Tustin, S.D.; Hellens, R.P.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. SVP-like MADS box genes control dormancy and budbreak in apple. Front Plant Sci. 2017, 8, 477. [Google Scholar] [CrossRef]
- Wang, S.X.; Zuo, X.Y.; Xing, L.B.; Fan, S.; Zhuang, D.; Han, M.Y.; Zhuang, L.S. Cloning, expression and promoter activity analysis of apple flower suppressor protein SVP gene. Acta Hortic. Sin. 2019, 46, 1445–1457. [Google Scholar]
- Sun, L.M.; Zhang, J.Z.; Hu, C.G. Characterization and expression analysis of PtAGL24, a SHORT VEGETATIVE PHASE/AGAMOUS-LIKE 24 (SVP/AGL24)-type MADS-Box gene from Trifoliate Orange (Poncirus trifoliata L. Raf.). Front Plant Sci. 2016, 7, 823. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zhu, G.; Wei, Y.; Gao, J.; Liang, G.; Peng, L.; Lu, C.; Jin, J. Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genom. 2019, 20, 53. [Google Scholar] [CrossRef] [Green Version]
- Haghighi, R.; Tabatabaei, B.E.S.T.; Maibody, S.A.M.M.; Talebi, M.; Molina, R.V.; Nebauer, S.G.; Renau-Morata, B. A flowering inhibitor of the temperature-dependent pathway in Crocus sativus L. Mol. Biol. Rep. 2020, 47, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, F.X.; Hong, Y.C.; Yao, J.J.; Ren, Z.Z.; Shi, H.Z.; Zhu, J.K. The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol. Plant. 2018, 11, 1184–1197. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Park, S.H.; Lee, J.S.; Ahn, J.H. A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. BBA-Bioenerg. 2007, 1769, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, S.C.; An, G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2010, 54, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.M.; Wang, T.C.; Tony, M.G.; Charlotte, V.; Andrew, C.A.; Roger, P.H.; Erika, V.G. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 2014, 65, 4985–4995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.M.; Wang, T.C.; Andrew, C.A.; Richard, C.M.; Erika, V.G. Overexpression of both AcSVP1 and AcSVP4 delays budbreak in kiwifruit A. chinensis var. deliciosa, but only AcSVP1 delays flowering in model plants. Environ. Exp. Bot. 2018, 153, 262–270. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, S.; Li, Y.; Li, M.; Souer, E. Leaf-like sepals induced by ectopic expression of a SHORT VEGETATIVE PHASE (SVP)-Like MADS-box gene from the basal eudicot Epimedium sagittatum. Front Plant Sci. 2016, 28, 1461. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.Y.; Yu, Q.; Zhang, Y.; Yao, C.C.; Han, Y.J. StMADS11 subfamily gene PfMADS16 from Polypogon fugax regulates early flowering and seed development. Front Plant Sci. 2020, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.N. Cloning and Functional Analysis of SVP-Like and SOC1-Like Genes in Phyllostachys Violascens; Beijing Forestry University: Beijing, China, 2016. [Google Scholar]
- Wu, D.S. Study on the Function of Carva Cathayensis SVP Gene; Zhejiang Agriculture & Forestry University: Hangzhou, China,, 2019. [Google Scholar]
- Liu, W.Z.; Xie, Y.B.; Ma, J.Y.; Luo, X.T.; Nie, P.; Zuo, Z.X.; Lahrmann, U.; Zhao, Q.; Zheng, Y.Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, C.; He, X.H.; Chen, H.; Hu, Y.; Ou, S.J. Molecular cloning and expression analysis of four actin genes (MiACT) from mango. Biol. Plantarum. 2013, 57, 238–244. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplifed method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, X.; Luo, C.; Yu, H.; Chen, J.; Liu, Y.; Xie, X.; Fan, Z.; He, X. Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango. Int. J. Mol. Sci. 2021, 22, 9802. https://doi.org/10.3390/ijms22189802
Mo X, Luo C, Yu H, Chen J, Liu Y, Xie X, Fan Z, He X. Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango. International Journal of Molecular Sciences. 2021; 22(18):9802. https://doi.org/10.3390/ijms22189802
Chicago/Turabian StyleMo, Xiao, Cong Luo, Haixia Yu, Jinwen Chen, Yuan Liu, Xiaojie Xie, Zhiyi Fan, and Xinhua He. 2021. "Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango" International Journal of Molecular Sciences 22, no. 18: 9802. https://doi.org/10.3390/ijms22189802
APA StyleMo, X., Luo, C., Yu, H., Chen, J., Liu, Y., Xie, X., Fan, Z., & He, X. (2021). Isolation and Functional Characterization of Two SHORT VEGETATIVE PHASE Homologous Genes from Mango. International Journal of Molecular Sciences, 22(18), 9802. https://doi.org/10.3390/ijms22189802





