Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses
Abstract
:1. Introduction
2. Results
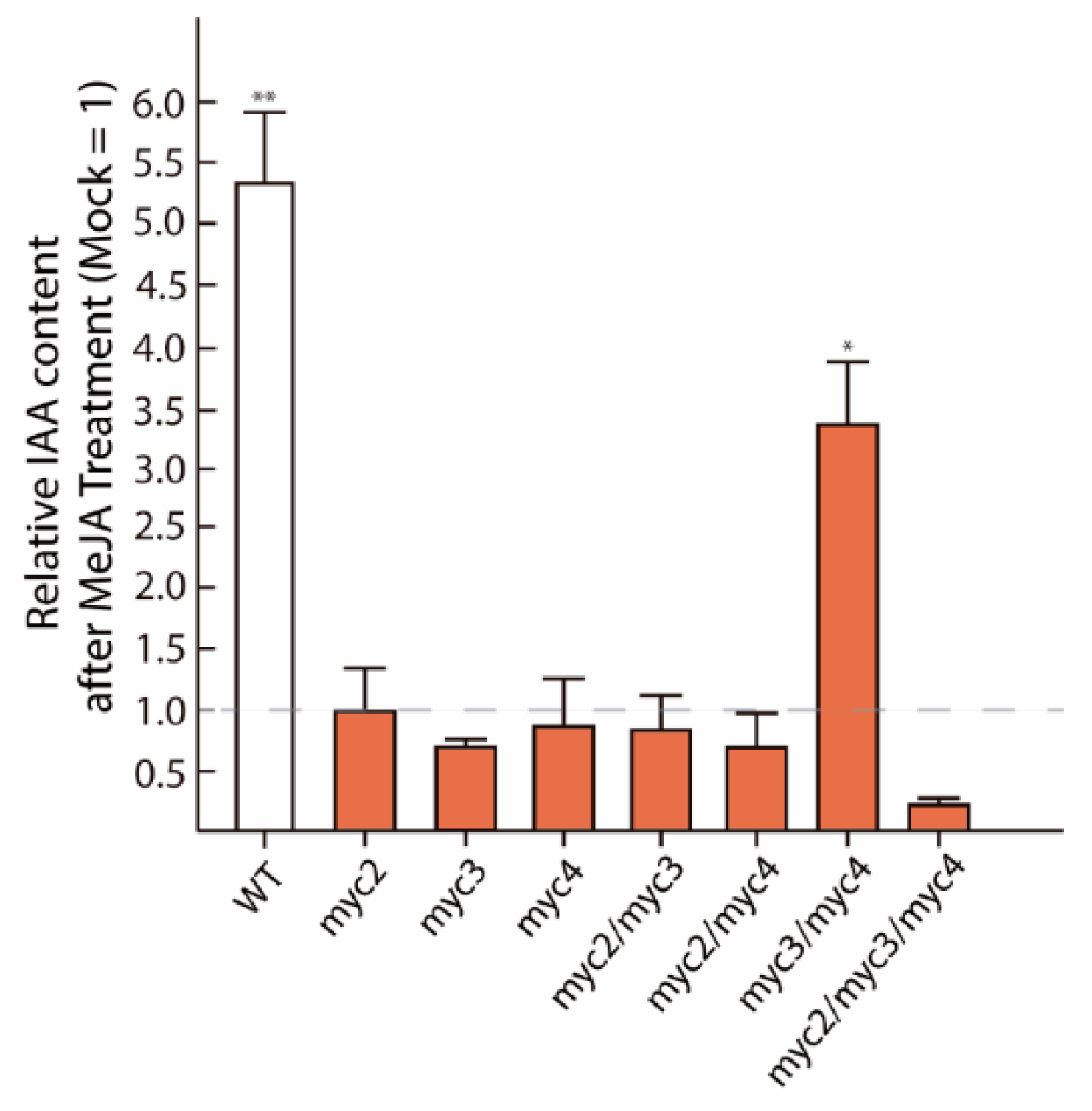
2.1. MYC2, MYC3, and MYC4 Trigger Auxin Formation after JA Treatment
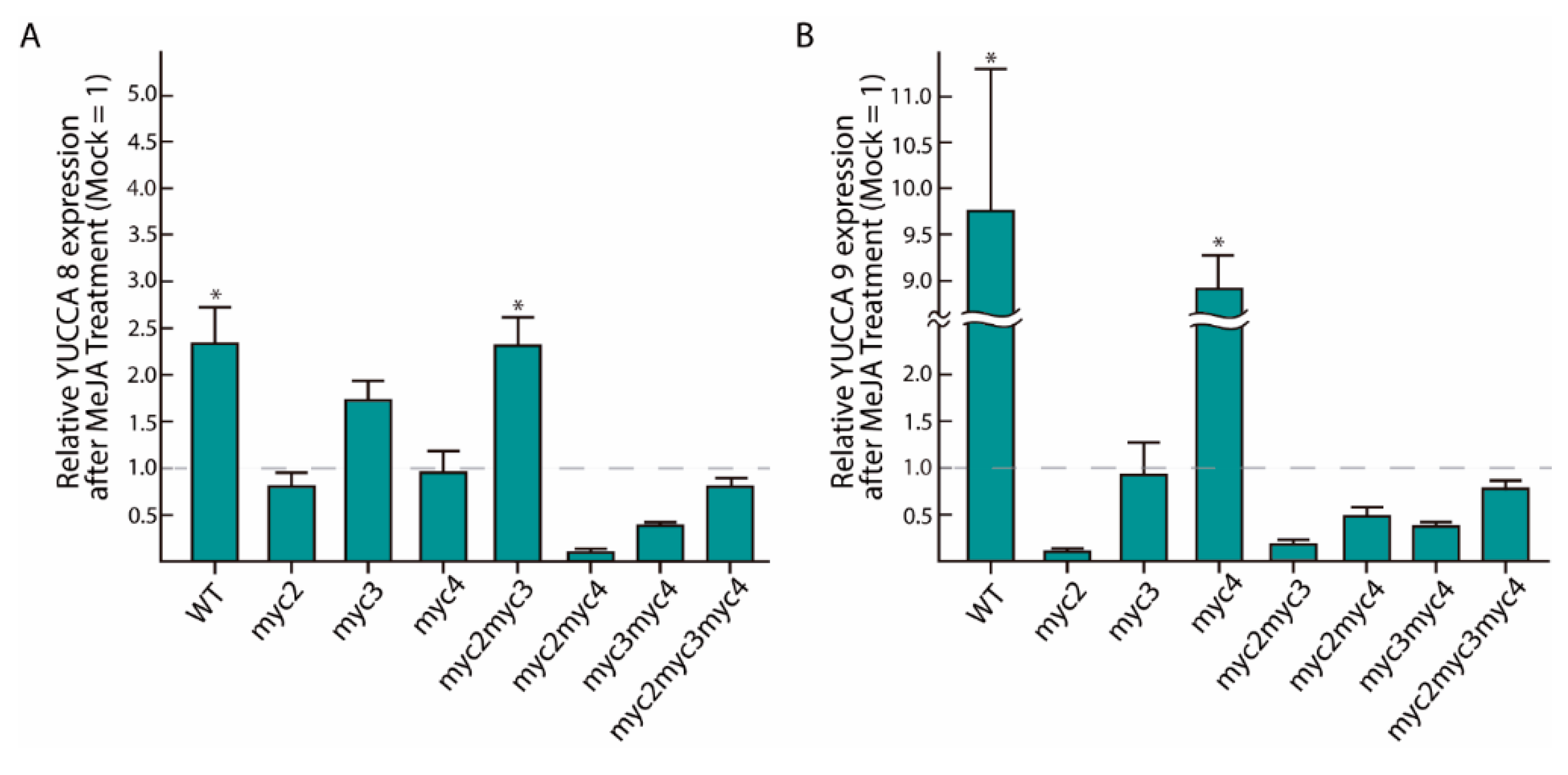
2.2. MeJA-Dependent YUC8 and YUC9 Induction Is Abolished in Myc Loss-of-Function Mutants
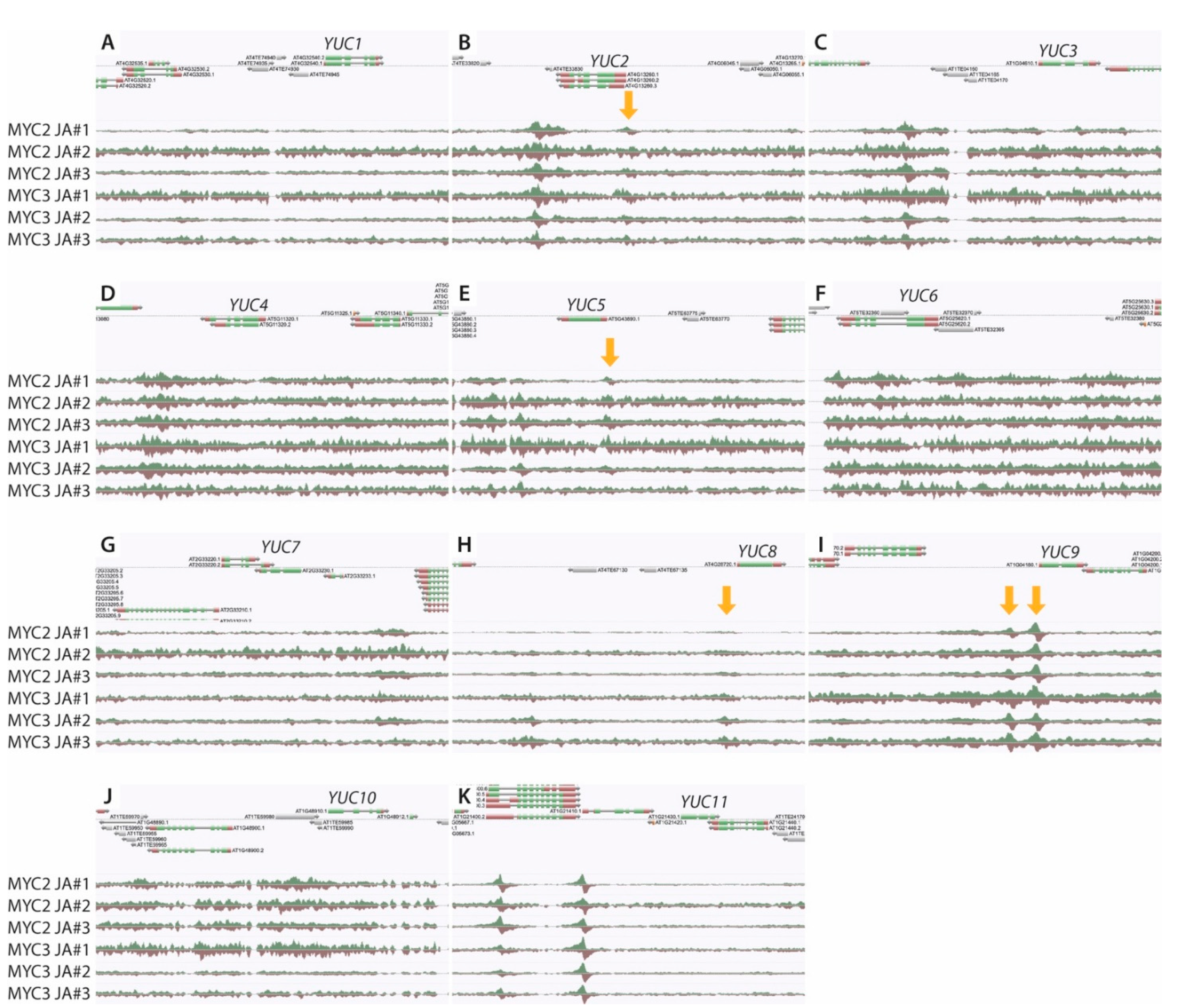
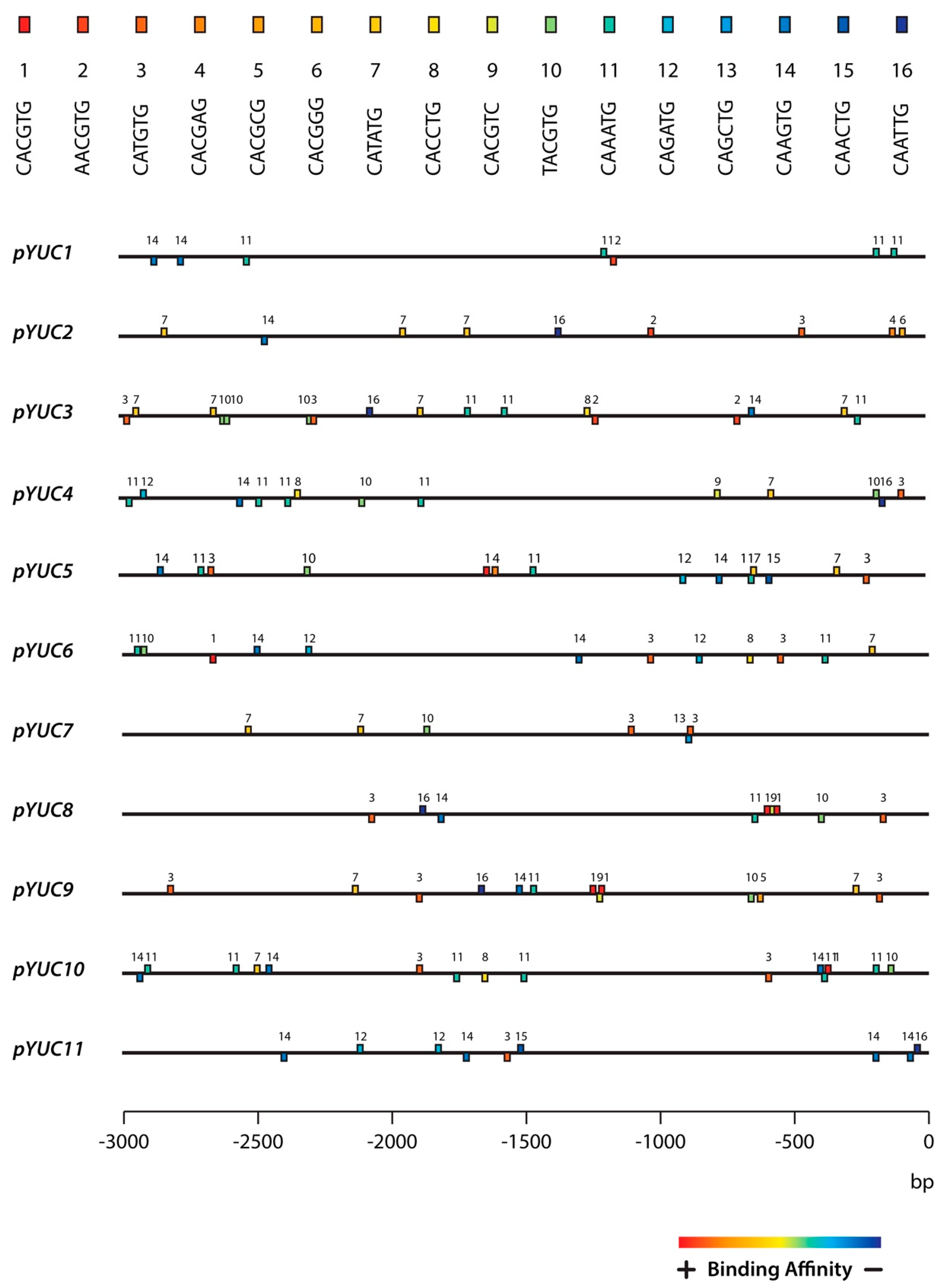
2.3. The YUC8 and YUC9 Promoters Contain a Conserved MYC2, MYC3, and MYC4 Binding Motif
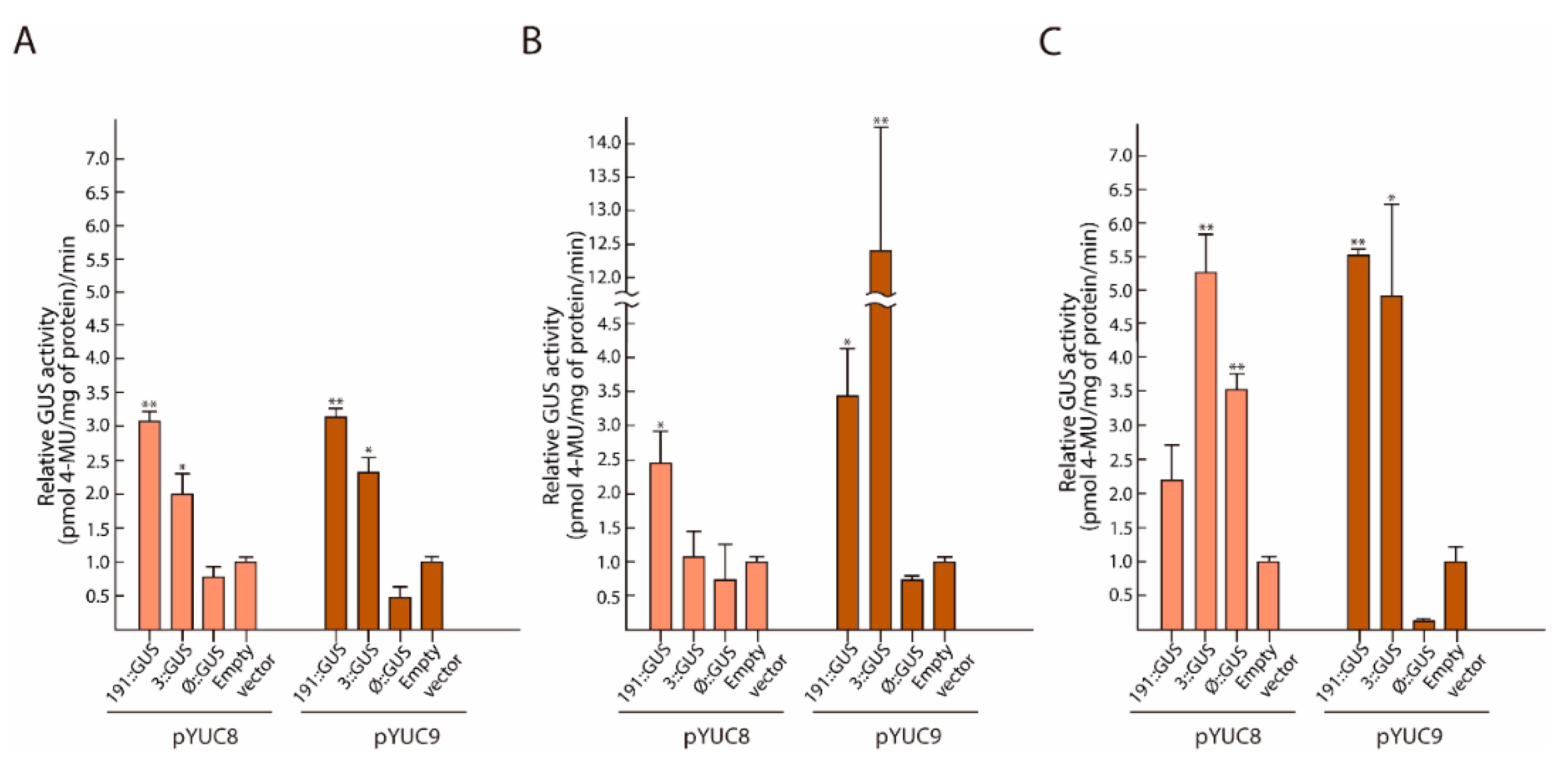
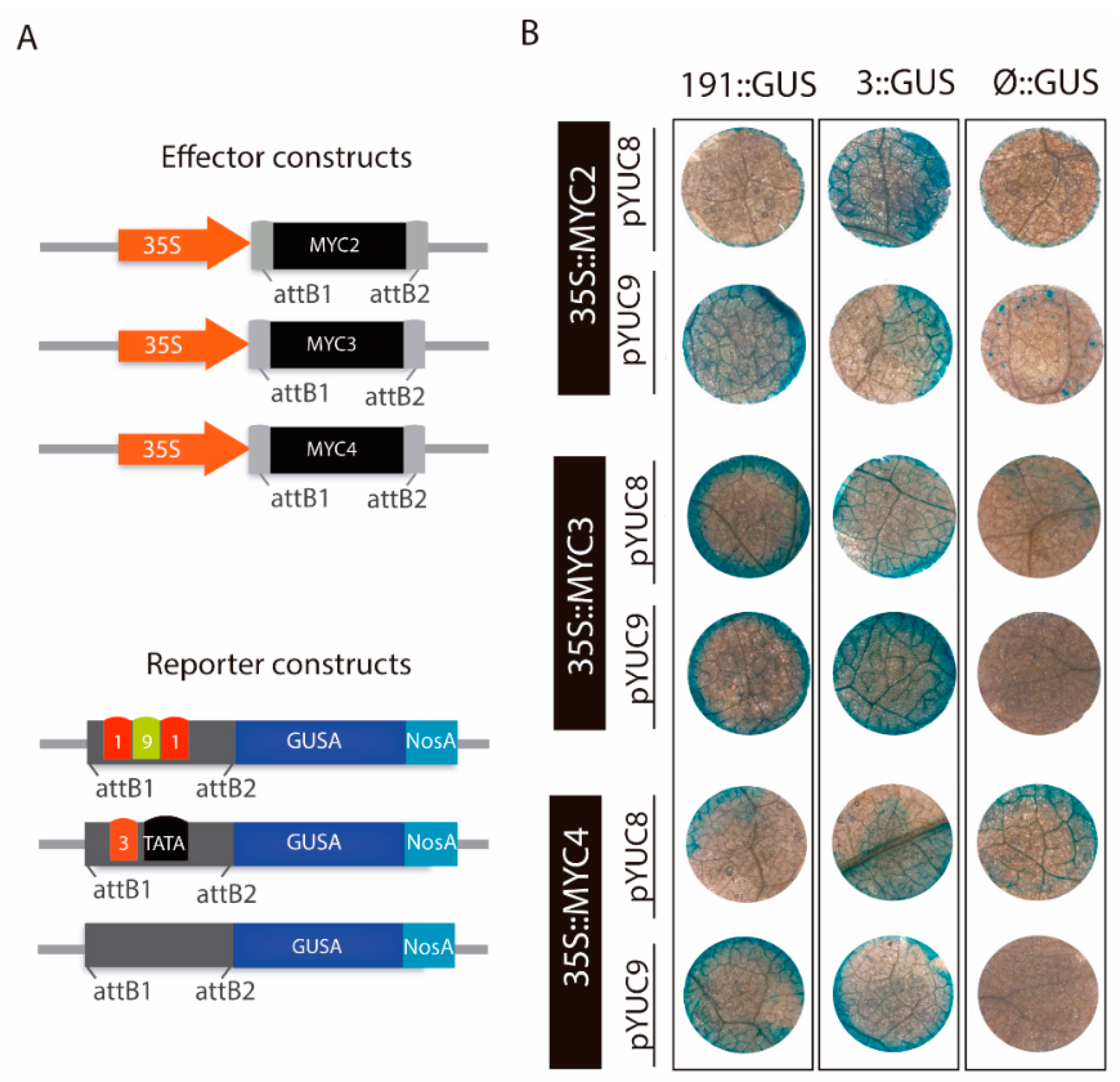
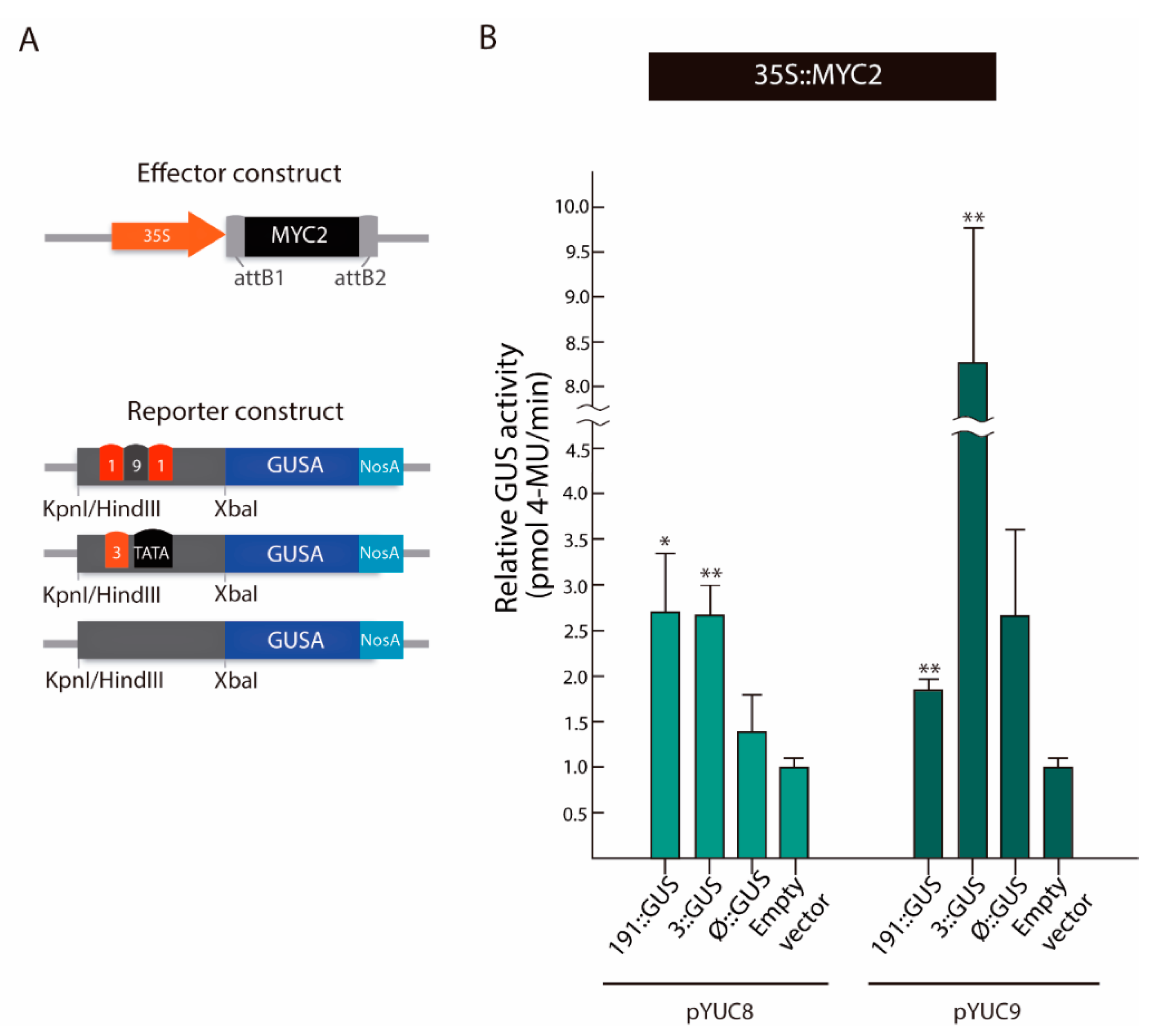
2.4. MYC2 Regulates YUC8 and YUC9 Expression through the Interaction with G-Box Elements
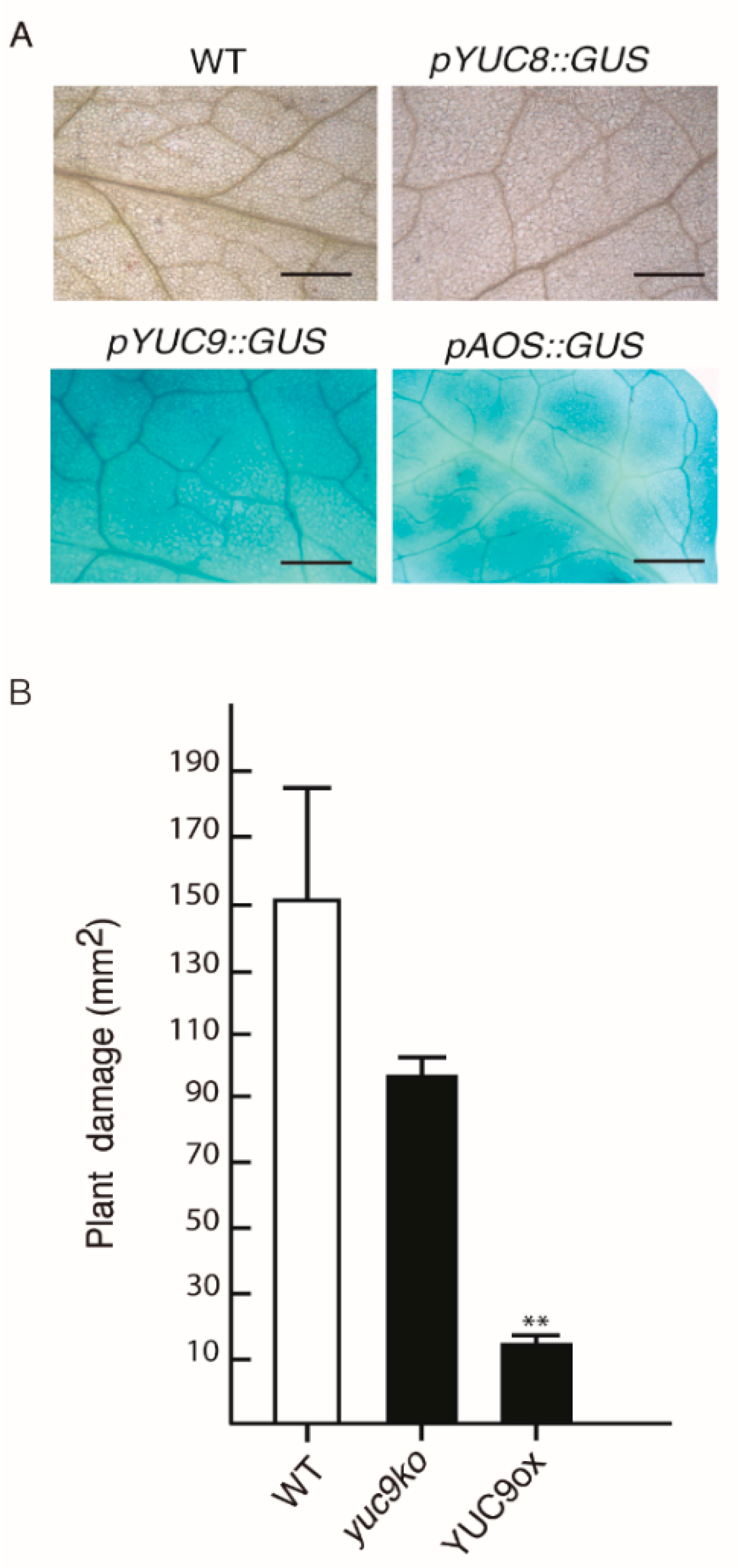
2.5. YUC9 Plays a Role in Biotic Stress Responses
3. Discussion
4. Material and Methods
4.1. Plant Material
4.2. qRT-PCR Analysis
4.3. Auxin Quantification
4.4. In Silico Analysis of YUCCA Promoter Sequences
4.5. Transient Expression Analysis in Nicotiana Benthamiana
4.6. Arabidopsis Protoplast-Based Transient Expression
4.7. Plant-Arthropod Interactions
4.8. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B

| Primer Name | Primer Sequence (5′-3′) and Restriction Sites |
|---|---|
| Promoter YUC8-(919) For | TATGGATCCAAAAGTGCAGCGTCTACCAAAA |
| Promoter YUC8-(919) Rev | TATTCTAGATTAGGTACGGAAAATGTGATT |
| Promoter YUC8-(3) For | TATGGATCCTCCGTACCTAAAAATTGGATT |
| Promoter YUC8-(3) Rev | TATTCTAGATGCTTGACGACGAAGTAATAAT |
| Promoter YUC8-(Ø) For | TATGGATCCTCGTCGTCAAGCATTATCACTGTT |
| Promoter YUC8-(Ø) Rev | TATCCATGGTCTAGATGGAAGTTGTATTGGAAATGGTTT |
| Promoter YUC9-(919) For | TATAAGCTTAACAAAATTAGGACCCGCTCT |
| Promoter YUC9-(919) Rev | TATTCTAGAGATTGAATTATATGGTAAACTCAA |
| Promoter YUC9-(3) For | TATAAGCTTACCACGAAGAAAATAACATCTC |
| Promoter YUC9-(3) Rev | TATCCATGGTCTAGAGTTAAGAGTTATAACGAGACTG |
| Promoter YUC9-(Ø) For | TATAAGCTTCAAATTATTCACATTAATAAAATAATC |
| Promoter YUC9-(Ø) Rev | TATCCATGGTCTAGATTTCTTGAGTGAGTTTTTGAATG |
| ORF MYC2 For | TATGGTACCATGACTGATTACCGGCTACAACCAACGA |
| ORF MYC2 Rev | TATGCGGCCGCTTAACCGATTTTTGAAATCAAACTTGCTCTGA |
| ORF MYC3 For | TATGGATCCATGAACGGCACAACATCATCA |
| ORF MYC3 Rev | TATGATATCTCAATAGTTTTCTCCGACTTTCGT |
| ORF MYC4 For | TATGGATCCATGTCTCCGACGAATGTTCAAGTAACCGA |
| ORF MYC4 Rev | TATGATATCTCATGGACATTCTCCAACTTTCTCCGTT |
| pENTRY-SP1 For | TATCTGATAGTGACCTGTTCGTTGCA |
| pENTRY-SP1 Rev | TATGGAGATCCGTGACGCAGTAGC |
| pBT-10-Seq Rev | TATTTGGGGTTTCTACAGGACGGACCAT |
| pENTRY-SP1 Rev | TATGGAGATCCGTGACGCAGTAGC |
| YUC8-qPCR For | CGTCTCAAGCTTCACCTTCC |
| YUC8-qPCR Rev | AGCCACTGGTCTCATCGAAC |
| YUC9-qPCR For | TTCTCGCCACCGGTTATCGTAG |
| YUC9-qPCR Rev | AGCGATGTTAACGGCGTCTACTG |
| APT1-qPCR For | TCGTGCTGTTCCTTGCAACCG |
| APT1-qPCR Rev | GCGGAGGAGAAGAGGCGGAGT |
| UBI10-qPCR For | TTGGAGGATGGCAGAACTCTTGCT |
| UBI10-qPCR Rev | AGTTTTCCCAGTCAACGTCTTAACGAAA |
References
- Kögl, F.; Erxleben, H.; Haagen-Smit, A.J. Über die Isolierung der Auxine a und b aus pflanzlichen Materialien. 9. Mitteilung über pflanzliche Wachstumsstoffe. Hoppe-Seyler’s Z. Für Physiol. Chem. 1934, 225, 215–229. [Google Scholar] [CrossRef]
- Thimann, K.V.; Koepfli, J.B. Identity of the Growth-Promoting and Root-Forming Substances of Plants. Nature 1935, 135, 101–102. [Google Scholar] [CrossRef]
- Went, F.W. Auxin, the plant growth-hormone. Bot. Rev. 1935, 1, 162–182. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abel, S.; Theologis, A. Odyssey of auxin. Cold Spring Harb. Perspect. Biol. 2010, 2, a004572. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef] [PubMed]
- McGuire, R.; Agrawal, A.A. Trade-offs between the shade-avoidance response and plant resistance to herbivores? Tests with mutant Cucumis sativus. Funct. Ecol. 2005, 19, 1025–1031. [Google Scholar] [CrossRef]
- Walters, D.; Heil, M. Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 2007, 71, 3–17. [Google Scholar] [CrossRef]
- Kempel, A.; Schadler, M.; Chrobock, T.; Fischer, M.; van Kleunen, M. Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc. Natl. Acad. Sci. USA 2011, 108, 5685–5689. [Google Scholar] [CrossRef] [Green Version]
- Leone, M.; Keller, M.M.; Cerrudo, I.; Ballare, C.L. To grow or defend? Low red : Far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 2014, 204, 355–367. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Kurashige, N.S.; Agrawal, A.A. Phenotypic plasticity to light competition and herbivory in Chenopodium album (Chenopodiaceae). Am. J. Bot. 2005, 92, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Griebel, T.; Zeier, J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008, 147, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Mutka, A.M.; Fawley, S.; Tsao, T.; Kunkel, B.N. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 2013, 74, 746–754. [Google Scholar] [CrossRef]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Poudel, A.N.; Jewell, J.B.; Kitaoka, N.; Staswick, P.; Matsuura, H.; Koo, A.J. Hormone crosstalk in wound stress response: Wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2107–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, C.; Lakshmanan, V.; Bais, H.P. Interplant Aboveground Signaling Prompts Upregulation of Auxin Promoter and Malate Transporter as Part of Defensive Response in the Neighboring Plants. Front. Plant Sci. 2017, 8, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttcher, C.; Pollmann, S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009, 276, 4693–4704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, A.J.; Howe, G.A. The wound hormone jasmonate. Phytochemistry 2009, 70, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Forner, S.; Strnad, M.; Hause, B. Jasmonates in flower and seed development. Biochimie 2013, 95, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.-M.; Pollmann, S. How Auxin May Contribute to the Regulation of Plant Defense Responses against Herbivory. Austin J. Plant Biol. 2018, 4, 1–5. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [Green Version]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, N.; Zhang, Y.; Li, Y. Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 2020, 11, 1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Rice, J.H.; Chen, N.; Baum, T.J.; Hewezi, T. Synchronization of Developmental Processes and Defense Signaling by Growth Regulating Transcription Factors. PLoS ONE 2014, 9, e98477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lei, Y.; Lu, C.; Wang, L.; Wu, J. MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 2020, 62, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Huang, H.; Wang, J.; Liu, B.; Qi, T.; Xie, D. MYC5 is Involved in Jasmonate-Regulated Plant Growth, Leaf Senescence and Defense Responses. Plant Cell Physiol. 2017, 58, 1752–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Moerkercke, A.; Duncan, O.; Zander, M.; Šimura, J.; Broda, M.; Vanden Bossche, R.; Lewsey, M.G.; Lama, S.; Singh, K.B.; Ljung, K.; et al. A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. USA 2019, 116, 23345–23356. [Google Scholar] [CrossRef]
- Kiran, K.; Ansari, S.A.; Srivastava, R.; Lodhi, N.; Chaturvedi, C.P.; Sawant, S.V.; Tuli, R. The TATA-box sequence in the basal promoter contributes to determining light-dependent gene expression in plants. Plant Physiol. 2006, 142, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Saldierna Guzmán, J.P.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A.; et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. S1), S153–S164. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-oxylipin crosstalk: Relationship of antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.C.; Goossens, A. Jasmonate signalling: A copycat of auxin signalling? Plant Cell Environ. 2013, 36, 2071–2084. [Google Scholar] [CrossRef]
- Shabek, N.; Zheng, N. Plant ubiquitin ligases as signaling hubs. Nat. Struct. Mol. Biol. 2014, 21, 293–296. [Google Scholar] [CrossRef]
- Tiryaki, I.; Staswick, P.E. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002, 130, 887–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, C.; Pan, J.; Peng, W.; Genschik, P.; Hobbie, L.; Hellmann, H.; Estelle, M.; Gao, B.; Peng, J.; Sun, C.; et al. Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 2005, 42, 514–524. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [Green Version]
- Gutjahr, C.; Riemann, M.; Müller, A.; Düchting, P.; Weiler, E.W.; Nick, P. Cholodny-Went revisited: A role for jasmonate in gravitropism of rice coleoptiles. Planta 2005, 222, 575–585. [Google Scholar] [CrossRef]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inze, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.T.; Xu, P.; Zhao, P.X.; Liu, R.; Yu, L.H.; Xiang, C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef]
- Santamaría, M.E.; Martínez, M.; Arnaiz, A.; Ortego, F.; Grbic, V.; Díaz, I. MATI, a Novel Protein Involved in the Regulation of Herbivore-Associated Signaling Pathways. Front. Plant Sci. 2017, 8, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubigsteltig, I.; Laudert, D.; Weiler, E.W. Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 1999, 208, 463–471. [Google Scholar] [CrossRef]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Santamaría, M.E.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 2014, 164, 384–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, R.A.; Robert, C.A.; Arce, C.C.; Ferrieri, A.P.; Xu, S.; Jimenez-Aleman, G.H.; Baldwin, I.T.; Erb, M. Auxin Is Rapidly Induced by Herbivore Attack and Regulates a Subset of Systemic, Jasmonate-Dependent Defenses. Plant Physiol. 2016, 172, 521–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensoussan, N.; Santamaria, M.E.; Zhurov, V.; Díaz, I.; Grbic, M.; Grbic, V. Plant-Herbivore Interaction: Dissection of the Cellular Pattern of Tetranychus urticae Feeding on the Host Plant. Front. Plant Sci. 2016, 7, 1105. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.N.; Yun, J.; Likhacheva, A.V.; Alonso, J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 2007, 19, 2169–2185. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.F.; Kende, H. Auxin-induced ethylene biosynthesis in subapical stem sections of etiolated seedlings of Pisum sativum L. Planta 1979, 146, 649–656. [Google Scholar] [CrossRef]
- Yu, Y.B.; Adams, D.O.; Yang, S.F. Regulation of Auxin-induced Ethylene Production in Mung Bean Hypocotyls: Role of 1-Aminocyclopropane-1-Carboxylic Acid. Plant Physiol. 1979, 63, 589–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.F.; Hoffman, N.E. Ethylene Biosynthesis and its Regulation in Higher Plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Abel, S.; Nguyen, M.D.; Chow, W.; Theologis, A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J. Biol. Chem. 1995, 270, 19093–19099. [Google Scholar] [CrossRef] [Green Version]
- Tsuchisaka, A.; Yu, G.; Jin, H.; Alonso, J.M.; Ecker, J.R.; Zhang, X.; Gao, S.; Theologis, A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 2009, 183, 979–1003. [Google Scholar] [CrossRef] [Green Version]
- Hentrich, M.; Sánchez-Parra, B.; Pérez-Alonso, M.M.; Carrasco-Loba, V.; Carrillo, L.; Vicente-Carbajosa, J.; Medina, J.; Pollmann, S. YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav. 2013, 8, e26363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Box, M.S.; Coustham, V.; Dean, C.; Mylne, J.S. Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 2011, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Pollmann, S. Mass Spectrometric Monitoring of Plant Hormone Cross Talk During Biotic Stress Responses in Potato (Solanum tuberosum L.). Methods Mol. Biol. 2021, 2354, 143–154. [Google Scholar] [CrossRef]
- Chen, Y.A.; Wen, Y.C.; Chang, W.C. AtPAN: An integrated system for reconstructing transcriptional regulatory networks in Arabidopsis thaliana. BMC Genom. 2012, 13, 85. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Lehmann, T.; Sánchez-Parra, B.; Björk, R.G.; Karim, S.; Amirjani, M.R.; Aronsson, H.; Wilkinson, M.D.; et al. Endogenous indole-3-acetamide levels contribute to the crosstalk between auxin and abscisic acid, and trigger plant stress responses in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehlert, A.; Weltmeier, F.; Wang, X.; Mayer, C.S.; Smeekens, S.; Vicente-Carbajosa, J.; Droge-Laser, W. Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 2006, 46, 890–900. [Google Scholar] [CrossRef]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schutze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Droge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006, 25, 3133–3143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Lukasik, E.; Gawehns, F.; Takken, F.L. The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 2012, 835, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: ß-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Kirby, J.; Kavanagh, T.A. NAN fusions: A synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. Plant J. 2002, 32, 391–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sprenger-Haussels, M.; Weisshaar, B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 2000, 22, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Koncz, C. PEG-mediated protoplast transformation with naked DNA. Methods Mol. Biol. 1998, 82, 267–276. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, R.; Oñate-Sánchez, L.; Weltmeier, F.; Ehlert, A.; Díaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Dröge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, S.R. Quantitation of GUS Activity by Fluorometry. In GUS Protocols; Gallagher, S.R., Ed.; Academic Press: San Diego, CA, USA, 1992; pp. 47–59. [Google Scholar] [CrossRef]
- Cazaux, M.; Navarro, M.; Bruinsma, K.A.; Zhurov, V.; Negrave, T.; Van Leeuwen, T.; Grbic, V.; Grbic, M. Application of two-spotted spider mite Tetranychus urticae for plant-pest interaction studies. J. Vis. Exp. 2014, 89, 51738. [Google Scholar] [CrossRef] [Green Version]
- Koch, E.; Slusarenko, A. Arabidopsis Is Susceptible to Infection by a Downy Mildew Fungus. Plant Cell 1990, 2, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular Localization of H2O2 in Plants. H2O2 Accumulation in Papillae and Hypersensitive Response during the Barley-Powdery Mildew Interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Miki, T.; Ae Park, J.; Nagao, K.; Murayama, N.; Horiuchi, T. Control of Segregation of Chromosomal DNA by Sex Factor F in Escherichia Coli. Mutants of DNA Gyrase Subunit A Suppress LetD (CcdB) Product Growth Inhibition. J. Mol. Biol. 1992, 225, 39–52. [Google Scholar] [CrossRef]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—A Flexible Tool for Reliable High-Throughput Primer Design for Quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Alonso, M.-M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. https://doi.org/10.3390/ijms22189768
Pérez-Alonso M-M, Sánchez-Parra B, Ortiz-García P, Santamaría ME, Díaz I, Pollmann S. Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. International Journal of Molecular Sciences. 2021; 22(18):9768. https://doi.org/10.3390/ijms22189768
Chicago/Turabian StylePérez-Alonso, Marta-Marina, Beatriz Sánchez-Parra, Paloma Ortiz-García, Maria Estrella Santamaría, Isabel Díaz, and Stephan Pollmann. 2021. "Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses" International Journal of Molecular Sciences 22, no. 18: 9768. https://doi.org/10.3390/ijms22189768
APA StylePérez-Alonso, M.-M., Sánchez-Parra, B., Ortiz-García, P., Santamaría, M. E., Díaz, I., & Pollmann, S. (2021). Jasmonic Acid-Dependent MYC Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. International Journal of Molecular Sciences, 22(18), 9768. https://doi.org/10.3390/ijms22189768








