Deciphering Structural Alterations Associated with Activity Reductions of Genetic Polymorphisms in Cytochrome P450 2A6 Using Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results and Discussion
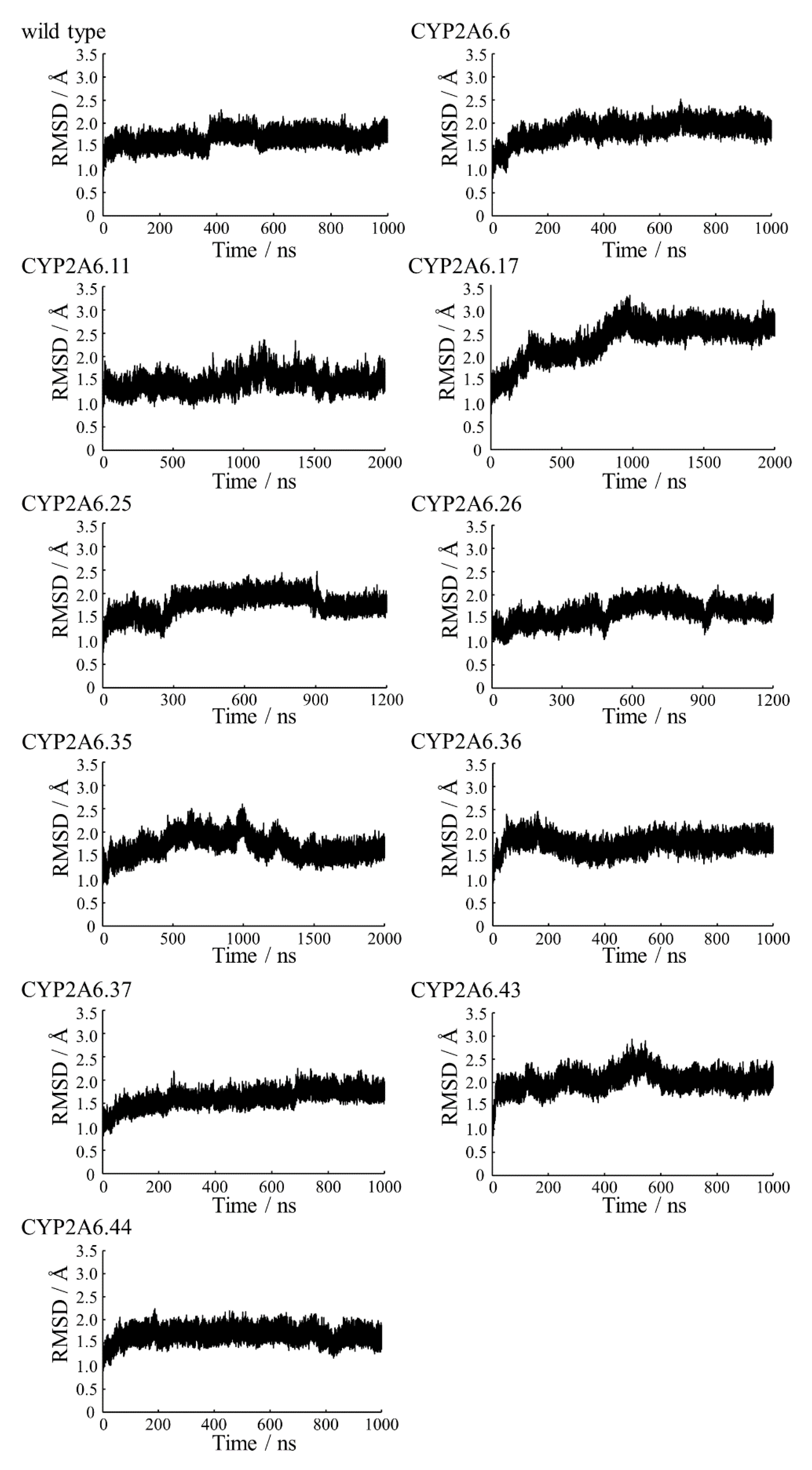
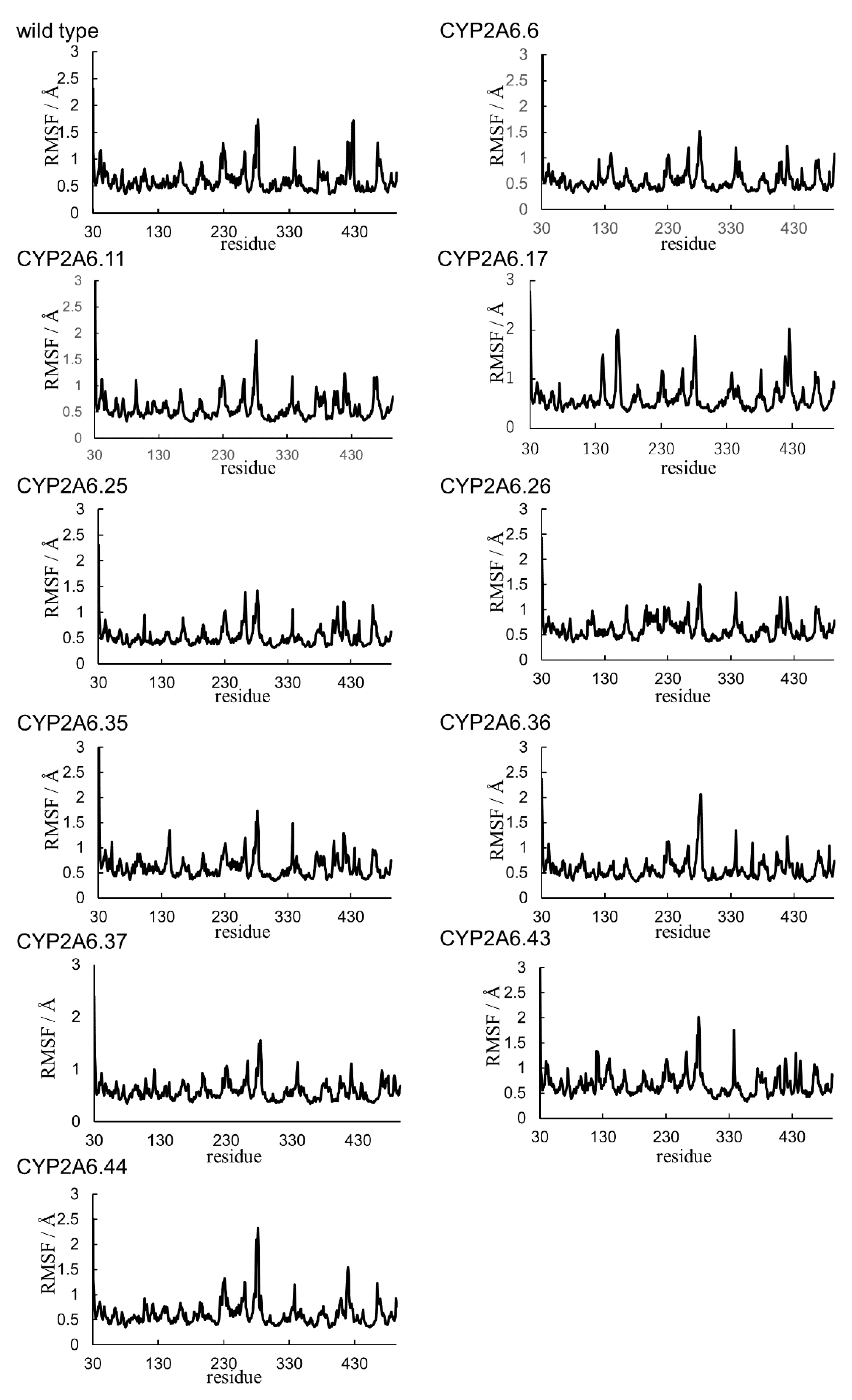
2.1. CYP2A6.6 Affected the Interaction with the Redox Partner
2.2. CYP2A6.11 Affected Secondary Structure Formation and Interaction with Heme
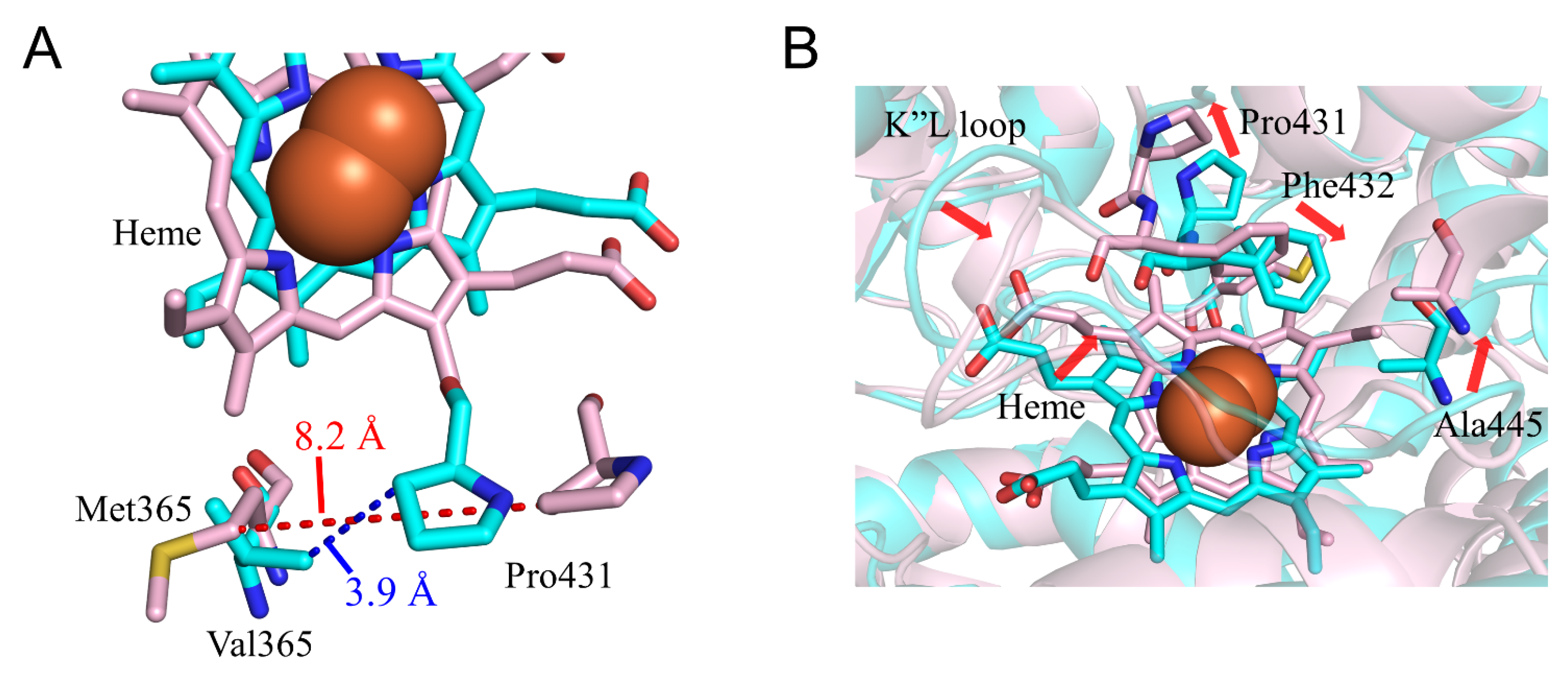
2.3. CYP2A6.17 Affected Secondary Structure Formation and Interaction with Heme and Redox Partoner
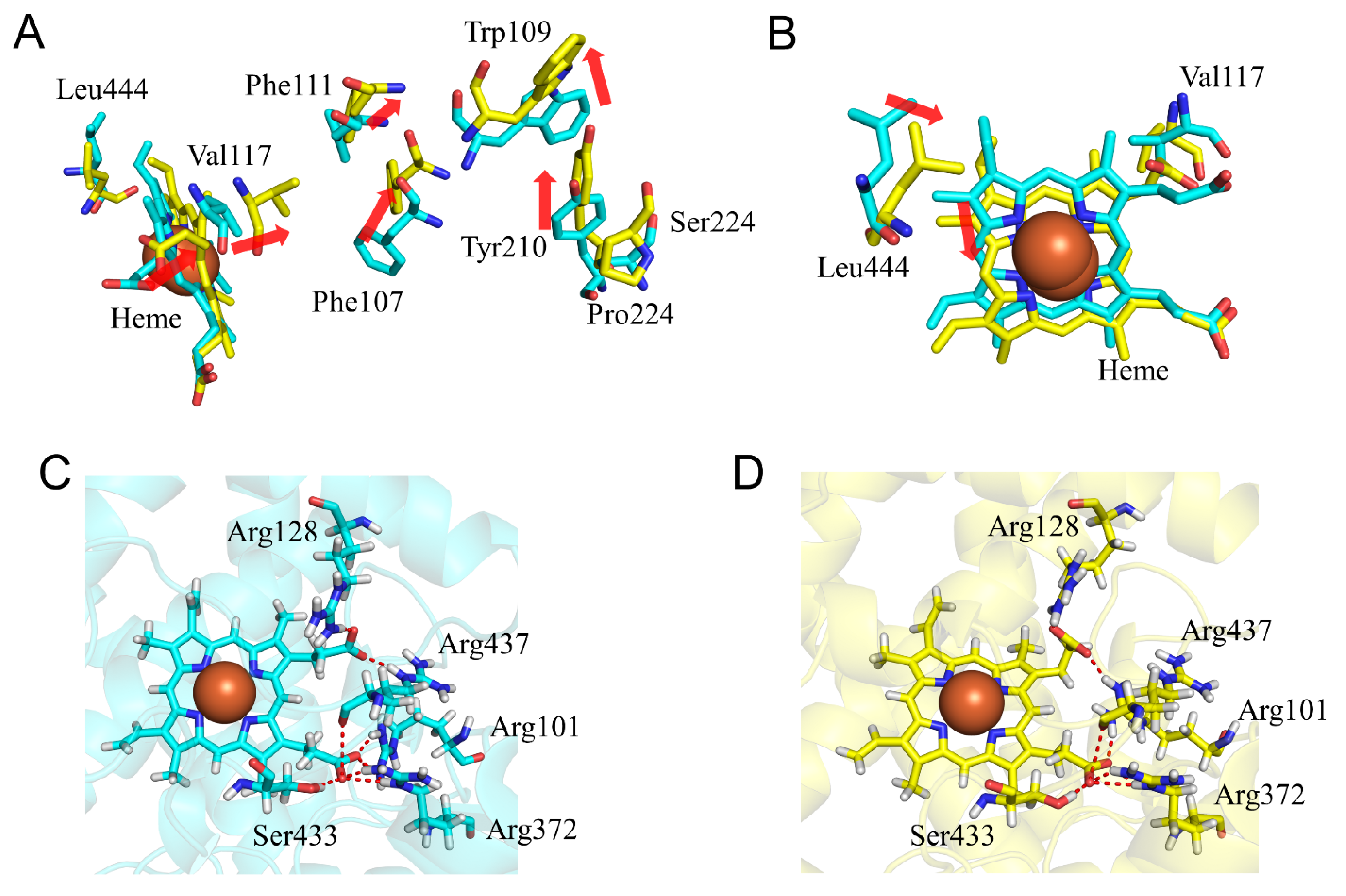
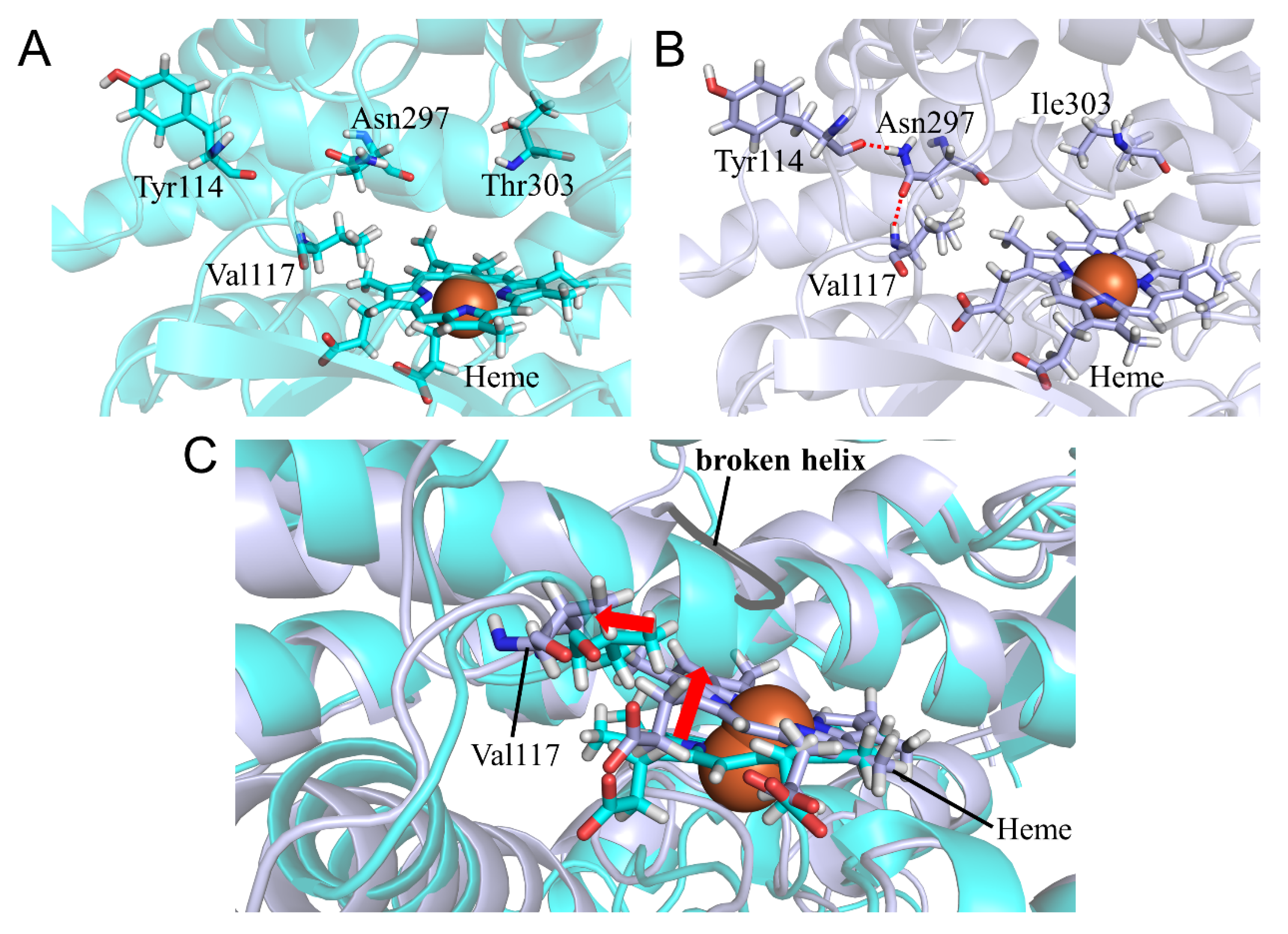
2.4. CYP2A6.25 and CYP2A6.26 Affected Secondary Structure Formation and Interaction with Heme and Substrates
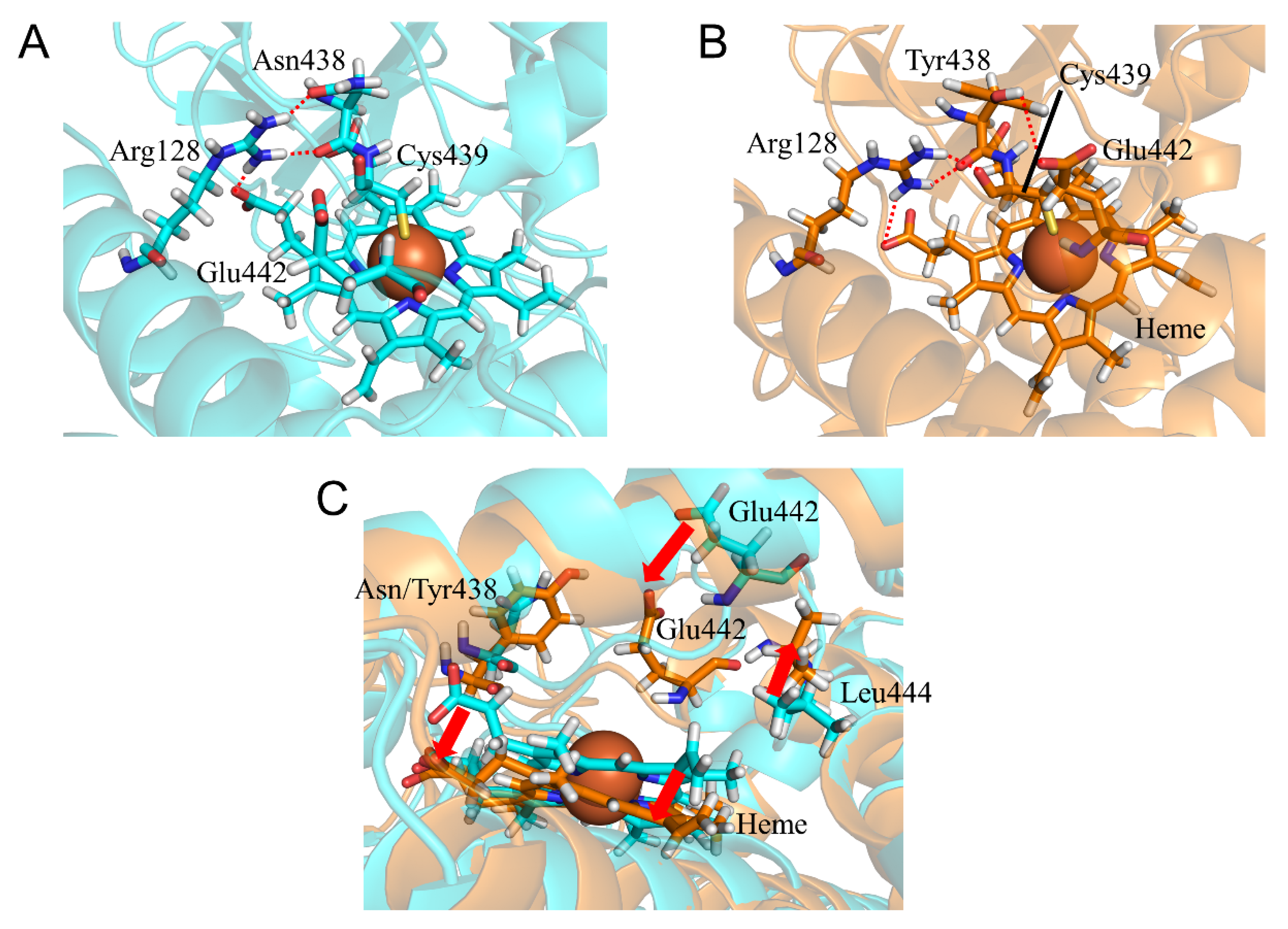
2.5. CYP2A6.35, CYP2A6.36, and CYP2A6.37 Affected Interaction with Heme and Substrates
2.6. CYP2A6.43 Affected the Interaction with Substrates
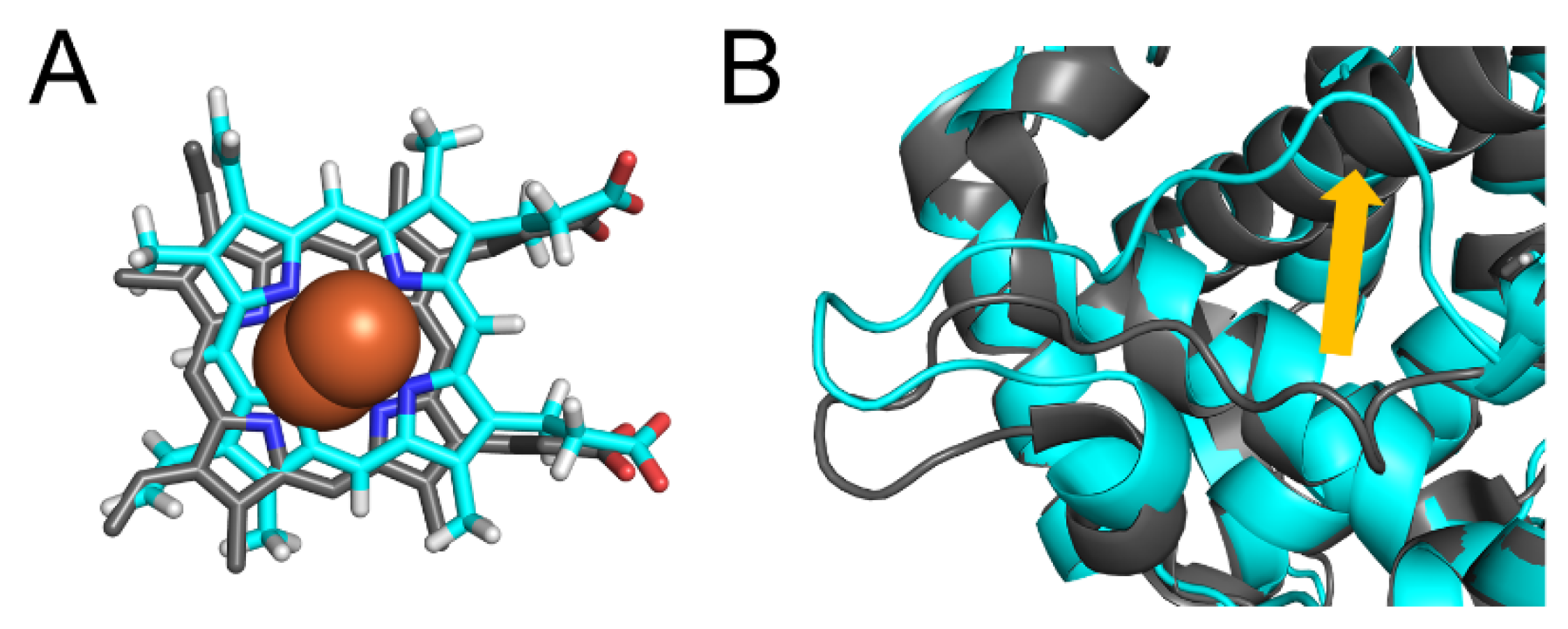
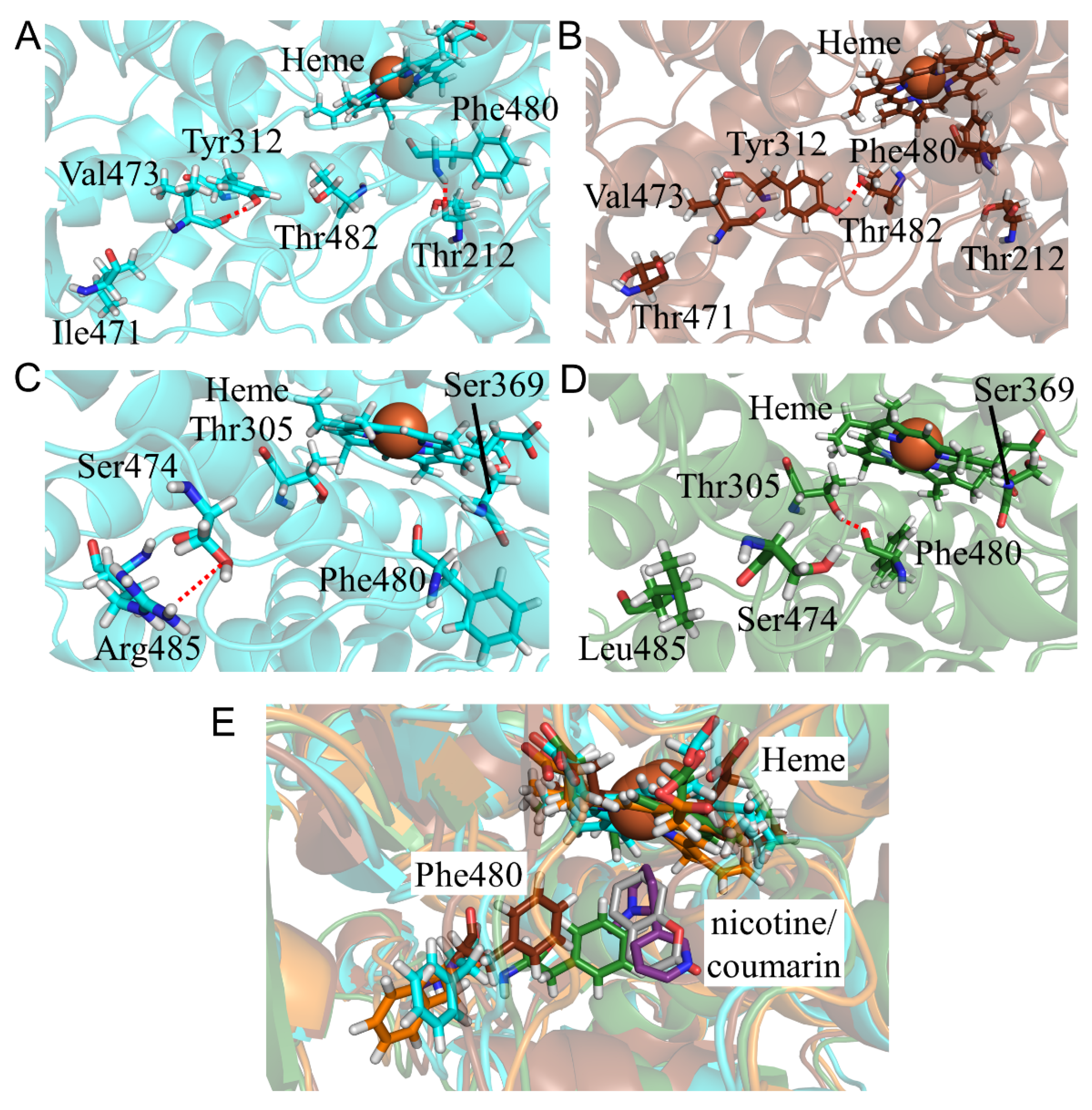
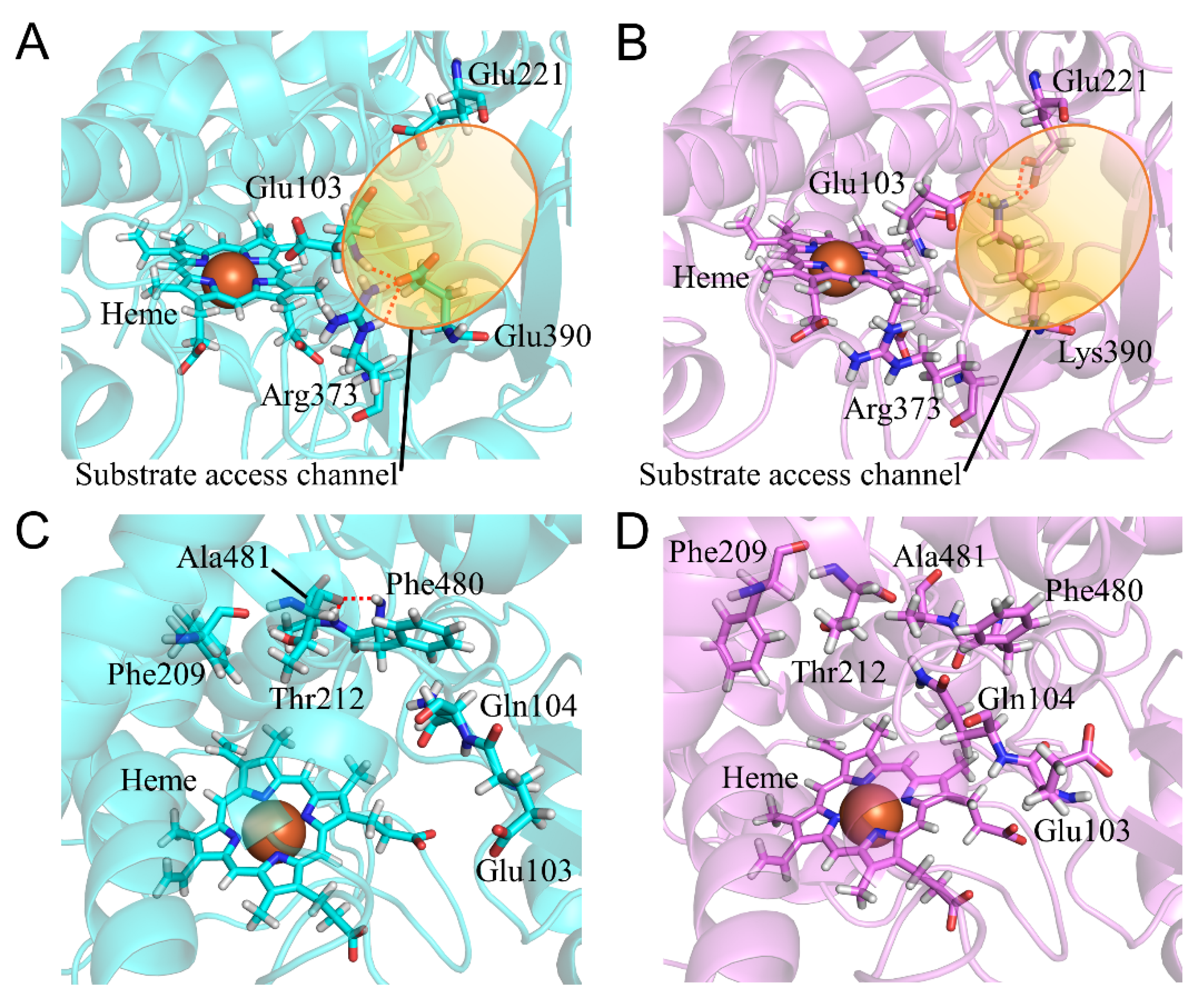
2.7. CYP2A6.44 Changed Substrate Access Channel
3. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guengerich, F.P. Human Cytochrome P450 Enzymes. In Cytochrome P450 Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano, P.R., Ed.; Springer: Cham, Switzerland, 2015; pp. 523–786. [Google Scholar]
- Rendic, S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002, 34, 83–448. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Hyland, R.; Jones, B.C.; Smith, D.A.; Hurst, S.; Goosen, T.C.; Peterkin, V.; Koup, J.R.; Ball, S.E. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004, 32, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Wienkers, L.C.; Heath, T.G. Predicting in vivo drug interactions from in vitro drug discovery data. Nat. Rev. Drug Discov. 2005, 4, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Schlichting, I.; Berendzen, J.; Chu, K.; Stock, A.M.; Maves, S.A.; Benson, D.E.; Sweet, R.M.; Ringe, D.; Petsko, G.A.; Sligar, S.G. The catalytic pathway of cytochrome p450cam at atomic resolution. Science 2000, 287, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Krest, C.M.; Onderko, E.L.; Yosca, T.H.; Calixto, J.C.; Karp, R.F.; Livada, J.; Rittle, J.; Green, M.T. Reactive intermediates in cytochrome p450 catalysis. J. Biol. Chem. 2013, 288, 17074–17081. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef]
- Shimizu, T.; Tateishi, T.; Hatano, M.; Fujii-Kuriyama, Y. Probing the role of lysines and arginines in the catalytic function of cytochrome P450d by site-directed mutagenesis. Interaction with NADPH-cytochrome P450 reductase. J. Biol. Chem. 1991, 266, 3372–3375. [Google Scholar] [CrossRef]
- Chang, Y.T.; Stiffelman, O.B.; Vakser, I.A.; Loew, G.H.; Bridges, A.; Waskell, L. Construction of a 3D model of cytochrome P450 2B4. Protein Eng. 1997, 10, 119–129. [Google Scholar] [CrossRef]
- Bridges, A.; Gruenke, L.; Chang, Y.T.; Vakser, I.A.; Loew, G.; Waskell, L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 1998, 273, 17036–17049. [Google Scholar] [CrossRef]
- Hamdane, D.; Xia, C.; Im, S.C.; Zhang, H.; Kim, J.J.; Waskell, L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009, 284, 11374–11384. [Google Scholar] [CrossRef]
- Pelkonen, O.; Rautio, A.; Raunio, H.; Pasanen, M. CYP2A6: A human coumarin 7-hydroxylase. Toxicology 2000, 144, 139–147. [Google Scholar] [CrossRef]
- Di, Y.M.; Chow, V.D.; Yang, L.P.; Zhou, S.F. Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr. Drug Metab. 2009, 10, 754–780. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhu, A.S.; Yang, C.Q.; Xu, C.Y.; Yang, D.Q.; Lou, Z.H.; Zhang, G.J. Cytochrome P450 2A6 is associated with macrophage polarization and is a potential biomarker for hepatocellular carcinoma. FEBS Open Bio. 2021, 11, 670–683. [Google Scholar] [CrossRef]
- Yano, J.K.; Hsu, M.H.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat. Struct. Mol. Biol. 2005, 12, 822–823. [Google Scholar] [CrossRef] [PubMed]
- Raunio, H.; Rautio, A.; Gullstén, H.; Pelkonen, O. Polymorphisms of CYP2A6 and its practical consequences. Br. J. Clin. Pharmacol. 2001, 52, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Hendrychová, T.; Anzenbacherová, E.; Hudeček, J.; Skopalík, J.; Lange, R.; Hildebrandt, P.; Otyepka, M.; Anzenbacher, P. Flexibility of human cytochrome P450 enzymes: Molecular dynamics and spectroscopy reveal important function-related variations. Biochim. Biophys. Acta 2011, 1814, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Guengerich, F.P. Ethnic-related differences in coumarin 7-hydroxylation activities catalyzed by cytochrome P4502A6 in liver microsomes of Japanese and Caucasian populations. Xenobiotica 1996, 26, 395–403. [Google Scholar] [CrossRef]
- Hassanzadeh Khayyat, M.; Vahdati Mashhadian, N.; Eghbal, S.; Jalali, N. Inter-individual variability of coumarin 7-hydroxylation (CYP2A6 activity) in an Iranian population. Iran. J. Basic Med. Sci. 2013, 16, 610–614. [Google Scholar]
- Fujieda, M.; Yamazaki, H.; Saito, T.; Kiyotani, K.; Gyamfi, M.A.; Sakurai, M.; Dosaka-Akita, H.; Sawamura, Y.; Yokota, J.; Kunitoh, H.; et al. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis 2004, 25, 2451–2458. [Google Scholar] [CrossRef]
- Hosono, H.; Kumondai, M.; Maekawa, M.; Yamaguchi, H.; Mano, N.; Oda, A.; Hirasawa, N.; Hiratsuka, M. Functional characterization of 34 CYP2A6 allelic variants by assessment of nicotine C-Oxidation and coumarin 7-hydroxylation activities. Drug Metab. Dispos. 2017, 45, 279–285. [Google Scholar] [CrossRef] [PubMed]
- López-Flores, L.A.; Pérez-Rubio, G.; Falfán-Valencia, R. Distribution of polymorphic variants of CYP2A6 and their involvement in nicotine addiction. EXCLI J. 2017, 16, 174–196. [Google Scholar] [PubMed]
- Miles, J.S.; McLaren, A.W.; Forrester, L.M.; Glancey, M.J.; Lang, M.A.; Wolf, C.R. Identification of the human liver cytochrome P-450 responsible for coumarin 7-hydroxylase activity. Biochem. J. 1990, 267, 365–371. [Google Scholar] [CrossRef]
- Daigo, S.; Takahashi, Y.; Fujieda, M.; Ariyoshi, N.; Yamazaki, H.; Koizumi, W.; Tanabe, S.; Saigenji, K.; Nagayama, S.; Ikeda, K.; et al. A novel mutant allele of the CYP2A6 gene (CYP2A6*11) found in a cancer patient who showed poor metabolic phenotype towards tegafur. Pharm. Genom. 2002, 12, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Mwenifumbo, J.C.; Al Koudsi, N.; Ho, M.K.; Zhou, Q.; Hoffmann, E.B.; Sellers, E.M.; Tyndale, R.F. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum. Mutat. 2008, 29, 679–688. [Google Scholar] [CrossRef]
- Al Koudsi, N.; Ahluwalia, J.S.; Lin, S.K.; Sellers, E.M.; Tyndale, R.F. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharm. J. 2009, 9, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Piliguian, M.; Zhu, A.Z.; Zhou, Q.; Benowitz, N.L.; Ahluwalia, J.S.; Sanderson Cox, L.; Tyndale, R.F. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharm. Genom. 2014, 24, 118–128. [Google Scholar] [CrossRef][Green Version]
- Bouard, C.; Terreux, R.; Tissier, A.; Jacqueroud, L.; Vigneron, A.; Ansieau, S.; Puisieux, A.; Payen, L. Destabilization of the TWIST1/E12 complex dimerization following the R154P point-mutation of TWIST1: An in silico approach. BMC Struct. Biol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Narang, S.S.; Shuaib, S.; Goyal, D.; Goyal, B. Assessing the effect of D59P mutation in the DE loop region in amyloid aggregation propensity of β2-microglobulin: A molecular dynamics simulation study. J. Cell. Biochem. 2018, 119, 782–792. [Google Scholar] [CrossRef]
- Kato, K.; Nakayoshi, T.; Sato, M.; Kurimoto, E.; Oda, A. Molecular dynamics simulations for three-dimensional structures of orotate phosphoribosyltransferases constructed from a simplified amino acid set. ACS Omega 2020, 5, 13069–13076. [Google Scholar] [CrossRef]
- Kobayashi, K.; Takahashi, O.; Hiratsuka, M.; Yamaotsu, N.; Hirono, S.; Watanabe, Y.; Oda, A. Evaluation of influence of single nucleotide polymorphisms in cytochrome P450 2B6 on substrate recognition using computational docking and molecular dynamics simulation. PLoS ONE 2014, 9, e96789. [Google Scholar]
- Fukuyoshi, S.; Kometani, M.; Watanabe, Y.; Hiratsuka, M.; Yamaotsu, N.; Hirono, S.; Manabe, N.; Takahashi, O.; Oda, A. Molecular dynamics simulations to investigate the influences of amino acid mutations on protein three-dimensional structures of cytochrome P450 2D6.1, 2, 10, 14A, 51, and 62. PLoS ONE 2016, 11, e0152946. [Google Scholar] [CrossRef]
- Watanabe, Y.; Fukuyoshi, S.; Hiratsuka, M.; Yamaotsu, N.; Hirono, S.; Takahashi, O.; Oda, A. Prediction of three-dimensional structures and structural flexibilities of wild-type and mutant cytochrome P450 1A2 using molecular dynamics simulations. J. Mol. Graph. Model. 2016, 68, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Skopalík, J.; Anzenbacher, P.; Otyepka, M. Flexibility of human cytochromes P450: Molecular dynamics reveals differences between CYPs 3A4, 2C9, and 2A6, which correlate with their substrate preferences. J. Phys. Chem. B 2008, 112, 8165–8173. [Google Scholar] [CrossRef]
- Hendrychova, T.; Berka, K.; Navratilova, V.; Anzenbacher, P.; Otyepka, M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr. Drug. Metab. 2012, 13, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Vanhoof, G.; Goossens, F.; de Meester, I.; Hendriks, D.; Scharpe, S. Proline motif in peptides and their biological processing. FASEB J. 1995, 9, 9736–9744. [Google Scholar] [CrossRef]
- Kato, K.; Furuhashi, T.; Kato, K.; Oda, A.; Kurimoto, E. The assembly mechanism of coiled-coil domains of the yeast cargo receptors Emp46p/47p and the mutational alteration of pH-dependency of complex formation. J. Biochem. 2018, 163, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Dou, T.; Wang, Z.; Wu, J.; Yang, L.; Zeng, S.; Deng, M.; Lü, M.; Liang, S. Inhibition of human cytochrome P450 2A6 by 7-hydroxycoumarin analogues: Analysis of the structure-activity relationship and isoform selectivity. Eur. J. Pharm. Sci. 2019, 136, 104944. [Google Scholar] [CrossRef] [PubMed]
- DeVore, N.M.; Scott, E.E. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes. J. Biol. Chem. 2012, 287, 26576–26585. [Google Scholar] [CrossRef]
- Cojocaru, V.; Winn, P.J.; Wade, R.C. The ins and outs of cytochrome P450s. Biochim. Biophys. Acta 2007, 1770, 390–401. [Google Scholar] [CrossRef]
- Schoch, G.A.; Yano, J.K.; Sansen, S.; Dansette, P.M.; Stout, C.D.; Johnson, E.F. Determinants of cytochrome P450 2C8 substrate binding: Structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J. Biol. Chem. 2008, 283, 17227–17237. [Google Scholar] [CrossRef]
- Fishelovitch, D.; Shaik, S.; Wolfson, H.J.; Nussinov, R. Theoretical characterization of substrate access/exit channels in the human cytochrome P450 3A4 enzyme: Involvement of phenylalanine residues in the gating mechanism. J. Phys. Chem. B 2009, 113, 13018–13025. [Google Scholar] [CrossRef]
- Joung, I.S.; Cheatham, T.E., III. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER16; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Yamaotsu, N.; Hirono, S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J. Comput. Chem. 2005, 26, 818–826. [Google Scholar] [CrossRef] [PubMed]


| Nicotine | Coumarin | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Amino Acid Changes | Km/μM | Vmax/pmol min−1 | CLint/ nL·min−1 | Km/μM | Vmax/pmol min−1 | CLint/ μL·min−1 |
| CYP2A6.1 | – | 33.3 ± 5.44 | 5.91 ± 1.26 | 176 ± 11.2 (100%) | 1.94 ± 0.06 | 5.89 ± 0.34 | 3.04 ± 0.08 (100%) |
| CYP2A6.6 | R128Q | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.11 | S224P | 142 ± 31.3 | 4.31 ± 0.34 | 31.6 ± 5.88 * (18%) | 3.30 ± 0.37 | 2.57 ± 0.03 | 0.79 ± 0.009 *** (26%) |
| CYP2A6.17 | V365M | 25.9 ± 6.16 | 1.70 ± 0.21 | 68.3 ± 12.2 * (39%) | 1.72 ± 0.03 | 2.76 ± 0.08 | 1.60 ± 0.002 ** (53%) |
| CYP2A6.25 | F118L | 59.9 ± 5.94 | 1.67 ± 0.09 | 28.0 ± 2.48 * (16%) | N.D. | N.D. | N.D. |
| CYP2A6.26 | F118L R128L S131A | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.35 | N438Y | 75.7 ± 14.9 | 3.98 ± 0.51 | 53.5 ± 5.31 * (30%) | 2.50 ± 0.28 | 4.63 ± 0.35 | 1.86 ± 0.08 *** (61%) |
| CYP2A6.36 | N438Y I471T | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.37 | N438Y I471T R485L | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| CYP2A6.43 | T303I | 1049 ± 683 | 4.76 ± 2.00 | 5.33 ± 1.28 * (3%) | 8.55 ± 1.47 | 1.34 ± 0.09 | 0.16 ± 0.02 *** (5%) |
| CYP2A6.44 | E390K N418D E419D | 1097 ± 284 | 2.39 ± 0.43 | 2.22 ± 0.16 * (1%) | N.D. | N.D. | N.D. |
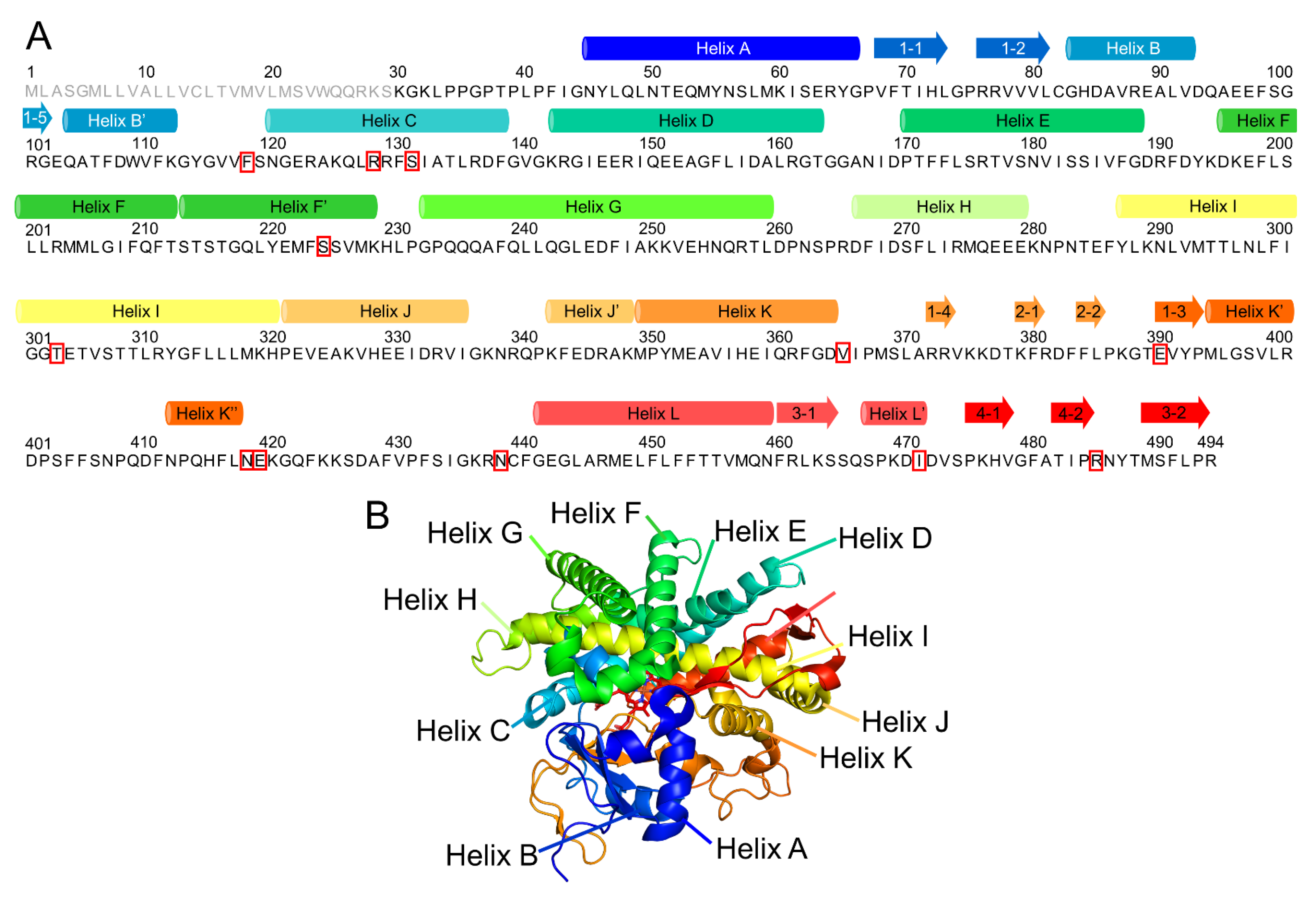
| Wild Type | 2A6.6 | 2A6.11 | 2A6.17 | 2A6.25 | |
|---|---|---|---|---|---|
| HA | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 |
| β1-1 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 |
| β1-2 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 |
| HB | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 |
| β1-5 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 |
| HB’ | 105–111 | 105–108 | 105–111 | 105–108 | 105–111 |
| HC | 121–138 | 122–135 | 122–138 | 125–134 | 121–138 |
| HD | 143–163 | 145–162 | 143–162 | 145–160 | 142–162 |
| HE | 171–186 | 170–186 | 170–186 | 170–186 | 170–186 |
| HF | 196–212 | 196–211 | 196–212 | 196–212 | 196–213 |
| HF’ | 215–227 | 215–227 | 215–222, 224–227 | 215–227 | 215–227 |
| HG | 233–256 | 233–257 | 233–257 | 233–256 | 233–256 |
| HH | 267–277 | 267–277 | 267–277 | 267–277 | 267–279 |
| HI | 288–319 | 288–318 | 288–319 | 288–319 | 288–296, 302–319 |
| HJ | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 |
| HJ’ | 343–347 | 343–347 | 343–348 | 343–348 | 343–347 |
| HK | 350–362 | 350–362 | 350–362 | 350–362 | 350–362 |
| β1-4 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 |
| β2-1 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 |
| β2-2 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 |
| β1-3 | 391–393 | 391–393 | 390–393 | 390–393 | 390–393 |
| HK’ | 395–399 | 395–398 | 395–398 | 395–398 | 395–398 |
| HK’’ | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 |
| HL | 447–459 | 442–459 | 441–443, 447–459 | 435–437, 441–443, 448–457 | 442–459 |
| β3-1 | 460–463 | 460–463 | 460–463 | 460–462 | 460–464 |
| HL’ | 468–470 | 468–470 | 468–470 | 468–470 | 468–470 |
| β4-1 | 477–478 | 477–478 | 477–478 | 477–478 | 477–478 |
| β4-2 | 482–483 | 482–483 | 482–483 | 482–483 | 482–484 |
| β3-2 | 489–493 | 489–493 | 489–493 | 491–493 | 489–493 |
| 2A6.26 | 2A6.35 | 2A6.36 | 2A6.37 | 2A6.43 | 2A6.44 | |
|---|---|---|---|---|---|---|
| HA | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 | 54–65 |
| β1-1 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 | 68–73 |
| β1-2 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 | 76–81 |
| HB | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 | 84–91 |
| β1-5 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 | 100–101 |
| HB’ | 105–110 | 108–111 | 105–110 | 105–111 | 105–110 | 107–111 |
| HC | 124–138 | 121–139 | 121–138 | 122–137 | 122–138 | 121–137 |
| HD | 145–162 | 145–162 | 143–161 | 143–161 | 145–161 | 143–161 |
| HE | 171–186 | 171–186 | 171–186 | 171–186 | 171–186 | 171–186 |
| HF | 196–212 | 196–212 | 196–211 | 196–213 | 196–213 | 196–212 |
| HF’ | 215–227 | 215–227 | 215–227 | 215–227 | 215–227 | 215–227 |
| HG | 233–256 | 233–257 | 233–256 | 233–256 | 233–257 | 233–256 |
| HH | 267–278 | 267–277 | 267–278 | 267–278 | 267–280 | 267–277 |
| HI | 288–319 | 288–319 | 288–319 | 288–319 | 288–297, 305–319 | 288–319 |
| HJ | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 | 321–334 |
| HJ’ | 343–347 | 343–348 | 343–347 | 343–347 | 343–346 | 343–348 |
| HK | 350–363 | 350–362 | 350–362 | 350–362 | 350–362 | 350–362 |
| β1-4 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 | 372–373 |
| β2-1 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 | 378–380 |
| β2-2 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 | 383–385 |
| β1-3 | 390–393 | 390–393 | 390–393 | 390–393 | 390–393 | 390–393 |
| HK’ | 395–398 | 395–398 | 395–399 | 395–398 | 395–398 | 395–398 |
| HK’’ | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 | 413–416 |
| HL | 442–459 | 442–459 | 446–457 | 447–459 | 449–457 | 442–459 |
| β3-1 | 460–463 | 460–464 | 460–464 | 460–463 | 460–463 | 460–463 |
| HL’ | 468–470 | 468–470 | 468–470 | 470–472 | 468–470 | 468–470 |
| β4-1 | 477–478 | – | 477–478 | 477–478 | 477–478 | 477–478 |
| β4-2 | 482–483 | – | 482–483 | 482–483 | 482–483 | 482–483 |
| β3-2 | 489–493 | 489–493 | 489–493 | 489–493 | 489–493 | 489–493 |
| Donor | Donor H | Acceptor | Wild Type | 2A6.6 |
|---|---|---|---|---|
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Asn438 O | 99.92 | 0 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 Oδ | 88.72 | 0 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 O | 33.52 | 0 |
| Phe429 N | Phe429 H | Ser425 O | 0 | 77.88 |
| Ala428 N | Ala428 H | Ser425 Oγ | 42.66 | 61.68 |
| His84 Nε2 | His84 Hε2 | Asp427 Oδ2 | 0 | 46.76 |
| His84 Nε2 | His84 Hε2 | Asp427 Oδ1 | 0 | 38.64 |
| Val430 N | Val430 H | Ala428 O | 0 | 52.86 |
| Donor | Donor H | Acceptor | Wild Type | 2A6.11 |
|---|---|---|---|---|
| Arg101 Nη1 | Arg101 Hη1 | Heme O1A | 100 | 0 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O1A | 99.98 | 0.06 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O2A | 0 | 98.88 |
| Arg101 Nη1 | Arg101 Hη1 | Heme O2D | 0 | 97.48 |
| Arg101 Nη2 | Arg101 Hη2 | Heme O2D | 0 | 88.66 |
| Arg101 Nε | Arg101 Hε | Heme O2A | 0 | 65.4 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 81.86 |
| Arg372 Nη2 | Arg372 Hη2 | Heme O2A | 99.1 | 48.12 |
| Arg372 Nε | Arg372 Hε | Heme O1A | 98.88 | 74.96 |
| Arg372 Nη2 | Arg372 Hη2 | Heme O1A | 57.9 | 0 |
| Arg372 Nε | Arg372 Hε | Heme O2A | 51.34 | 94.66 |
| Ser433 Nγ | Ser433 Hγ | Heme O2A | 100 | 99.9 |
| Arg437 Nε | Arg437 Hε | Heme O2D | 99.32 | 0.1 |
| Arg437 Nη1 | Arg437 Hη1 | Heme O2D | 65.76 | 0.08 |
| Arg437 Nε | Arg437 Hε | Heme O1D | 4.5 | 99.78 |
| Donor | Donor H | Acceptor | Wild Type | 2A6.25 | 2A6.26 |
|---|---|---|---|---|---|
| Val116 N | Val116 H | Asn297 Oδ1 | 0.02 | 96.38 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O2D | 99.94 | 99.84 | 0 |
| Donor H | Donor | Acceptor | Wild Type | 2A6.35 | 2A6.36 | 2A6.37 |
|---|---|---|---|---|---|---|
| Arg128 Nη1 | Arg128 Hη1 | Asn/Tyr438 O | 99.92 | 99.98 | 83.84 | 99.78 |
| Arg128 Nη2 | Arg128 Hη2 | Asn/Tyr438 O | 88.28 | 77.68 | 0.20 | 97.34 |
| Arg128 Nη2 | Arg128 Hη2 | Asn438 Oδ/Tyr438 Oη | 33.52 | 0 | 7.22 | 0.02 |
| Arg128 Nη2 | Arg128 Hη2 | Cys439 O | 13.86 | 71.68 | 99.44 | 37.70 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O1D | 99.94 | 0 | 0 | 0 |
| Arg128 Nη1 | Arg128 Hη1 | Heme O2D | 0 | 76.96 | 100 | 83.90 |
| Tyr312 Nη | Tyr312 Hη | Val473 O | 58.62 | 0 | 0 | 0 |
| Tyr312 Nη | Tyr312 Hη | Thr482 Oγ | 12.90 | 0 | 54.78 | 0 |
| Phe480 N | Phe480 H | Thr212 O | 73.92 | 0 | 5.10 | 17.24 |
| Ser369 N | Ser369 H | Phe480 O | 39.4 | 0 | 28.08 | 0 |
| Arg485 Nη2 | Arg485 Hη2 | Ser474 Oγ | 69.98 | 74.78 | 62.9 | – |
| Arg485 Nε | Arg485 Hε | Ser474 Oγ | 47.80 | 20.36 | 45.84 | – |
| Arg485 Nε | Arg485 Hε | Ser474 O | 32.84 | 83.22 | 44.02 | – |
| Thr305 Oγ | Thr305 Hγ | Phe480 O | 0 | 0 | 0 | 82.26 |
| Donor | Donor H | Acceptor | Wild Type | 2A6.43 |
|---|---|---|---|---|
| Val117 N | Val117 H | Asn297 Oδ1 | 0 | 98.28 |
| Asn297 Nδ2 | Asn297 Hδ2 | Tyr114 O | 0 | 91.92 |
| Donor | Donor H | Acceptor | Wild Type | 2A6.44 |
|---|---|---|---|---|
| Glu103 N | Glu103 H | Glu390 Oε1 | 96.67 | – |
| Arg373 Nε | Arg373 Hε | Glu390 Oε1 | 99.40 | – |
| Arg373 Nη2 | Arg373 Hη2 | Glu390 Oε1 | 95.10 | – |
| Arg373 Nη2 | Arg373 Hη2 | Glu390 Oε2 | 73.22 | – |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 Oε1 | – | 14.18 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 Oε1 | – | 19.30 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 Oε1 | – | 35.42 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 Oε2 | – | 17.50 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 Oε2 | – | 8.72 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 Oε2 | – | 6.76 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu103 O | – | 19.96 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu103 O | – | 32.06 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu103 O | – | 22.28 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu221 Oε1 | – | 38.20 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu221 Oε1 | – | 28.04 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu221 Oε1 | – | 25.60 |
| Lys390 Nζ1 | Lys390 Hζ1 | Glu221 Oε2 | – | 36.88 |
| Lys390 Nζ2 | Lys390 Hζ2 | Glu221 Oε2 | – | 27.58 |
| Lys390 Nζ3 | Lys390 Hζ3 | Glu221 Oε2 | – | 25.52 |
| Phe480 N | Phe480 H | Thr212 O | 73.92 | 0 |
| Ala481 N | Ala481 H | Thr212 O | 78.72 | 0 |
| Protein | Amino Acid Changes | Predicted Structural Effects Involved in the Enzymatic Activity |
|---|---|---|
| CYP2A6.6 | R128Q | conformational change involved in interaction with the redox partner |
| CYP2A6.11 | S224P | heme deviation, change of secondary structures |
| CYP2A6.17 | V365M | heme deviation, change of secondary structures, conformational change involved in interaction with the redox partner |
| CYP2A6.25 | F118L | structural change of substrate-interacting residues, heme deviation, change of secondary structures |
| CYP2A6.26 | F118L R128L S131A | structural change of substrate-interacting residues, heme deviation, change of secondary structures |
| CYP2A6.35 | N438Y | heme deviation, Cys439 conformational change |
| CYP2A6.36 | N438Y I471T | structural change of substrate-interacting residues, heme deviation, Cys439 conformational change |
| CYP2A6.37 | N438Y I471T R485L | structural change of substrate-interacting residues, heme deviation, Cys439 conformational change |
| CYP2A6.43 | T303I | structural change of substrate-interacting residues |
| CYP2A6.44 | E390K N418D E419D | structural change of substrate access channel, structural change of substrate-interacting residues |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, K.; Nakayoshi, T.; Nokura, R.; Hosono, H.; Hiratsuka, M.; Ishikawa, Y.; Kurimoto, E.; Oda, A. Deciphering Structural Alterations Associated with Activity Reductions of Genetic Polymorphisms in Cytochrome P450 2A6 Using Molecular Dynamics Simulations. Int. J. Mol. Sci. 2021, 22, 10119. https://doi.org/10.3390/ijms221810119
Kato K, Nakayoshi T, Nokura R, Hosono H, Hiratsuka M, Ishikawa Y, Kurimoto E, Oda A. Deciphering Structural Alterations Associated with Activity Reductions of Genetic Polymorphisms in Cytochrome P450 2A6 Using Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2021; 22(18):10119. https://doi.org/10.3390/ijms221810119
Chicago/Turabian StyleKato, Koichi, Tomoki Nakayoshi, Rika Nokura, Hiroki Hosono, Masahiro Hiratsuka, Yoshinobu Ishikawa, Eiji Kurimoto, and Akifumi Oda. 2021. "Deciphering Structural Alterations Associated with Activity Reductions of Genetic Polymorphisms in Cytochrome P450 2A6 Using Molecular Dynamics Simulations" International Journal of Molecular Sciences 22, no. 18: 10119. https://doi.org/10.3390/ijms221810119
APA StyleKato, K., Nakayoshi, T., Nokura, R., Hosono, H., Hiratsuka, M., Ishikawa, Y., Kurimoto, E., & Oda, A. (2021). Deciphering Structural Alterations Associated with Activity Reductions of Genetic Polymorphisms in Cytochrome P450 2A6 Using Molecular Dynamics Simulations. International Journal of Molecular Sciences, 22(18), 10119. https://doi.org/10.3390/ijms221810119







