Effects of Cardiolipin on the Conformational Dynamics of Membrane-Anchored Bcl-xL
Abstract
:1. Introduction
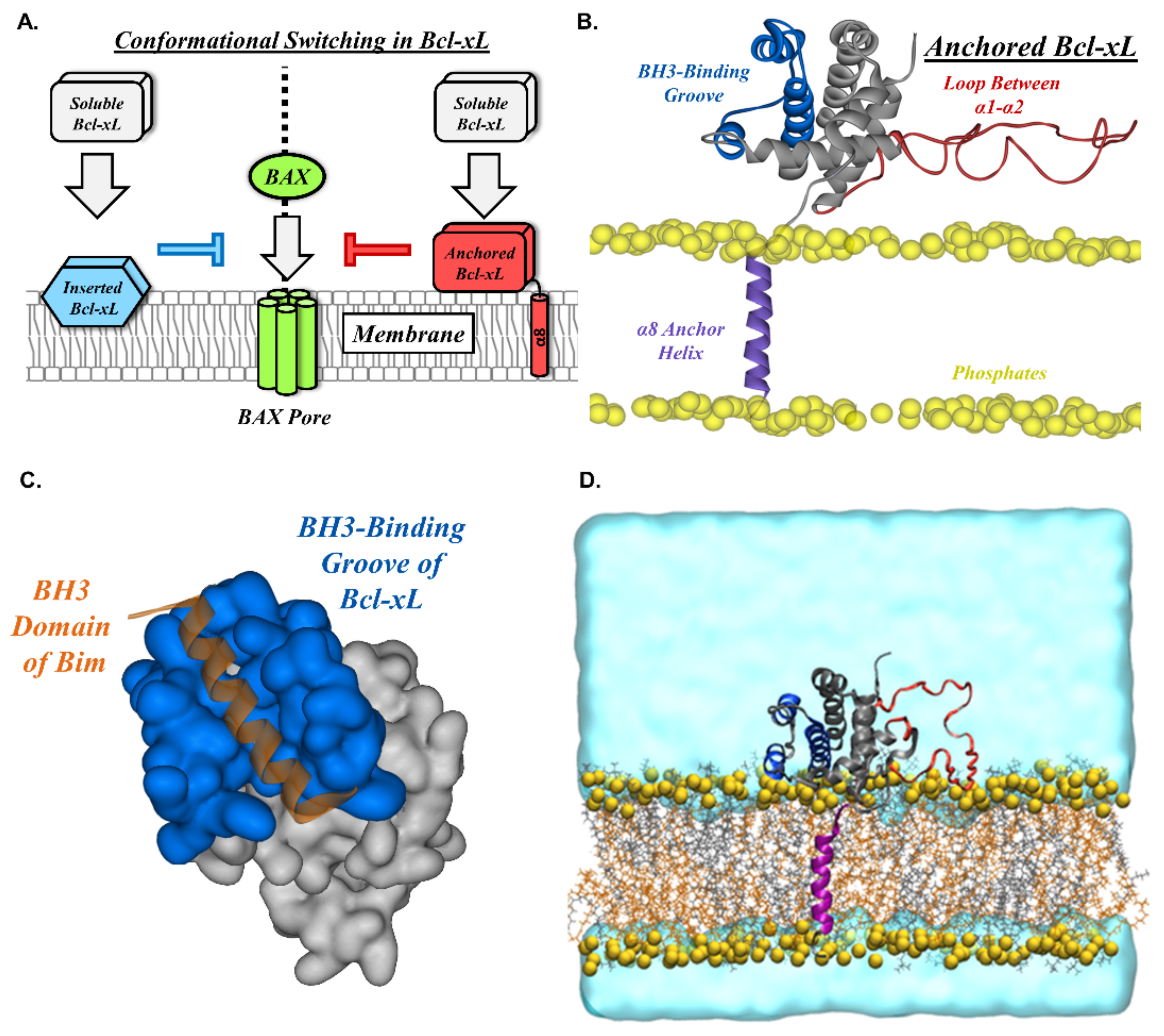
2. Results
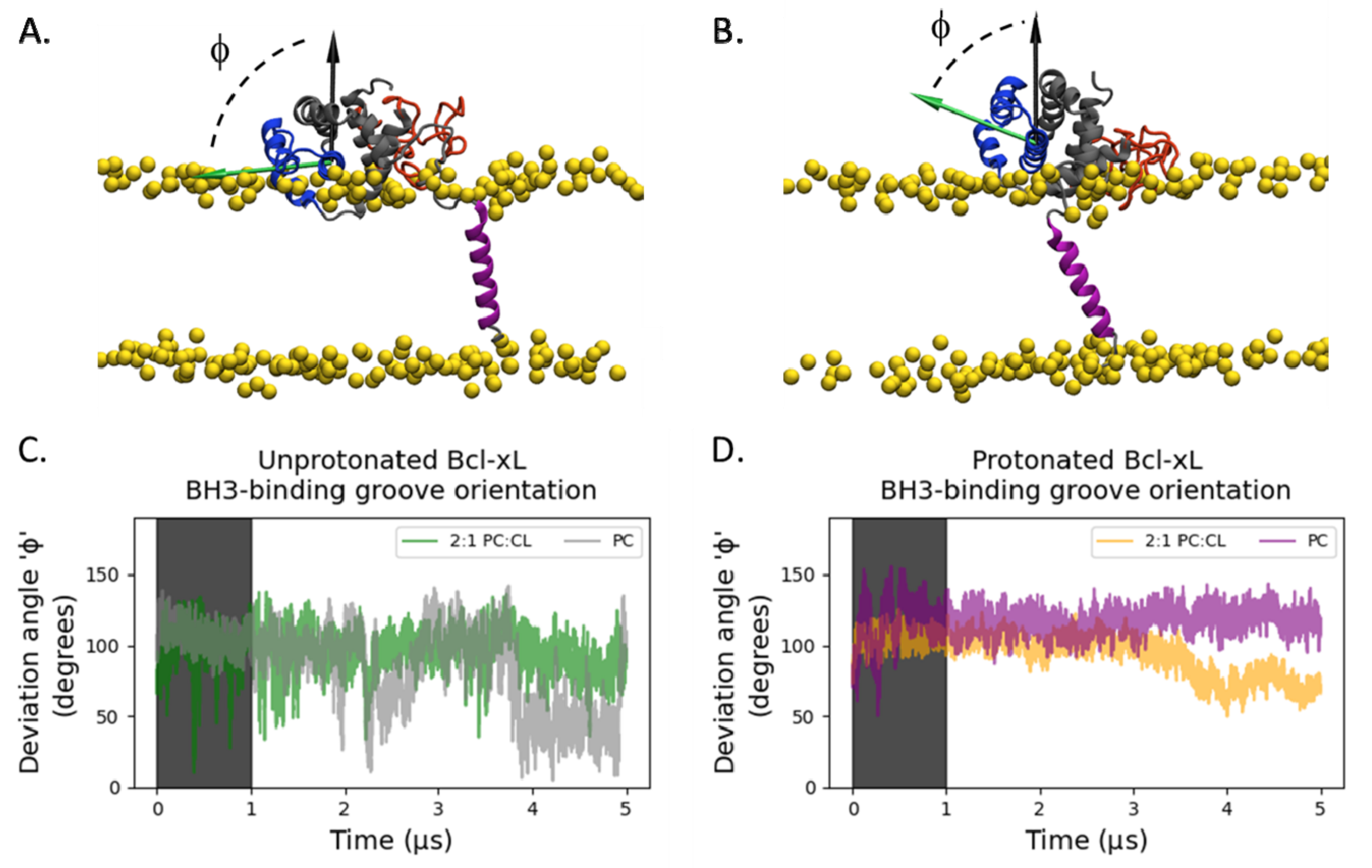
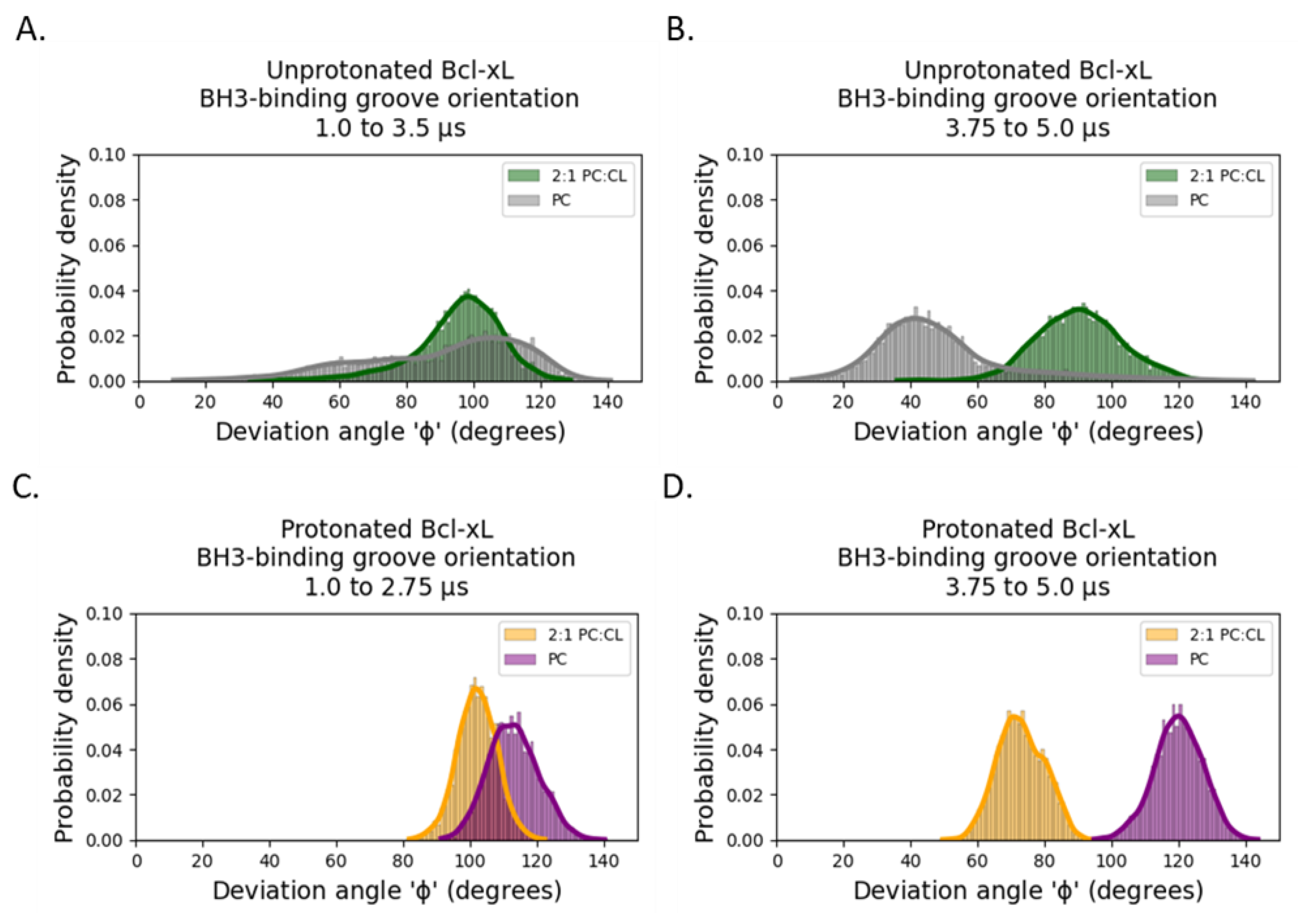
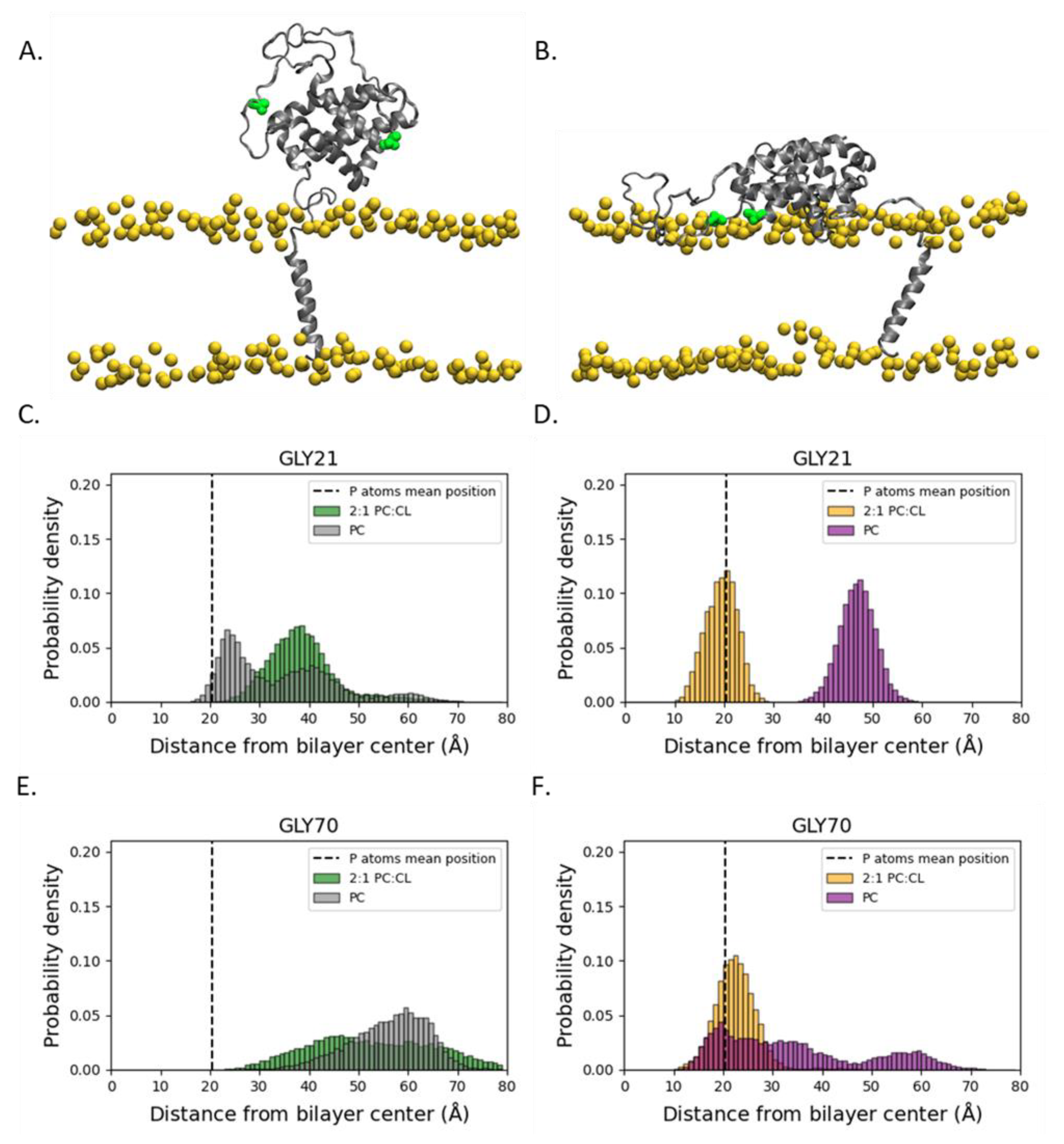
2.1. Orientation of the Soluble Head and Accessibility of the BH3-Binding Groove
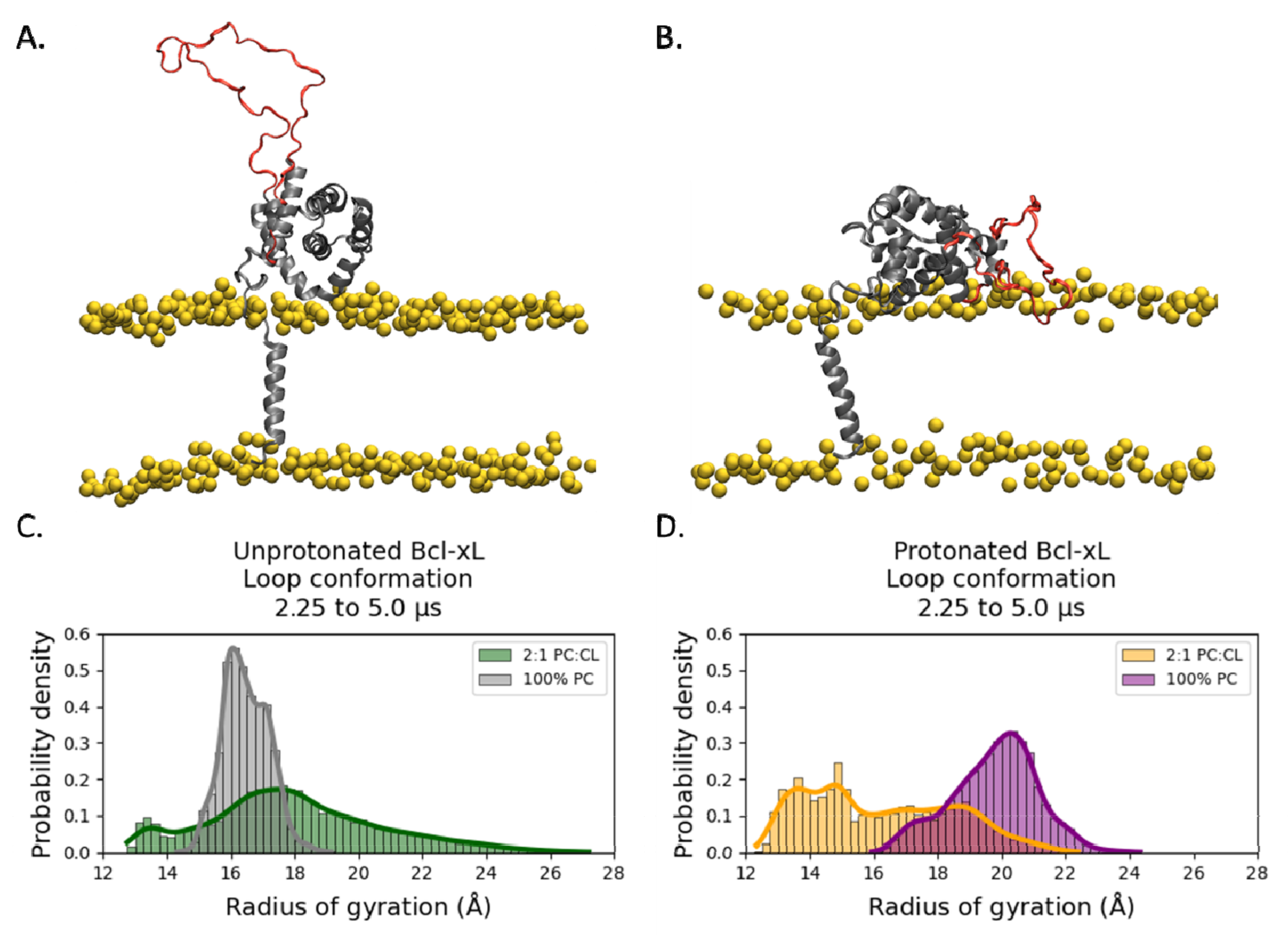
2.2. Conformational Dynamics of the α1-α2 Loop
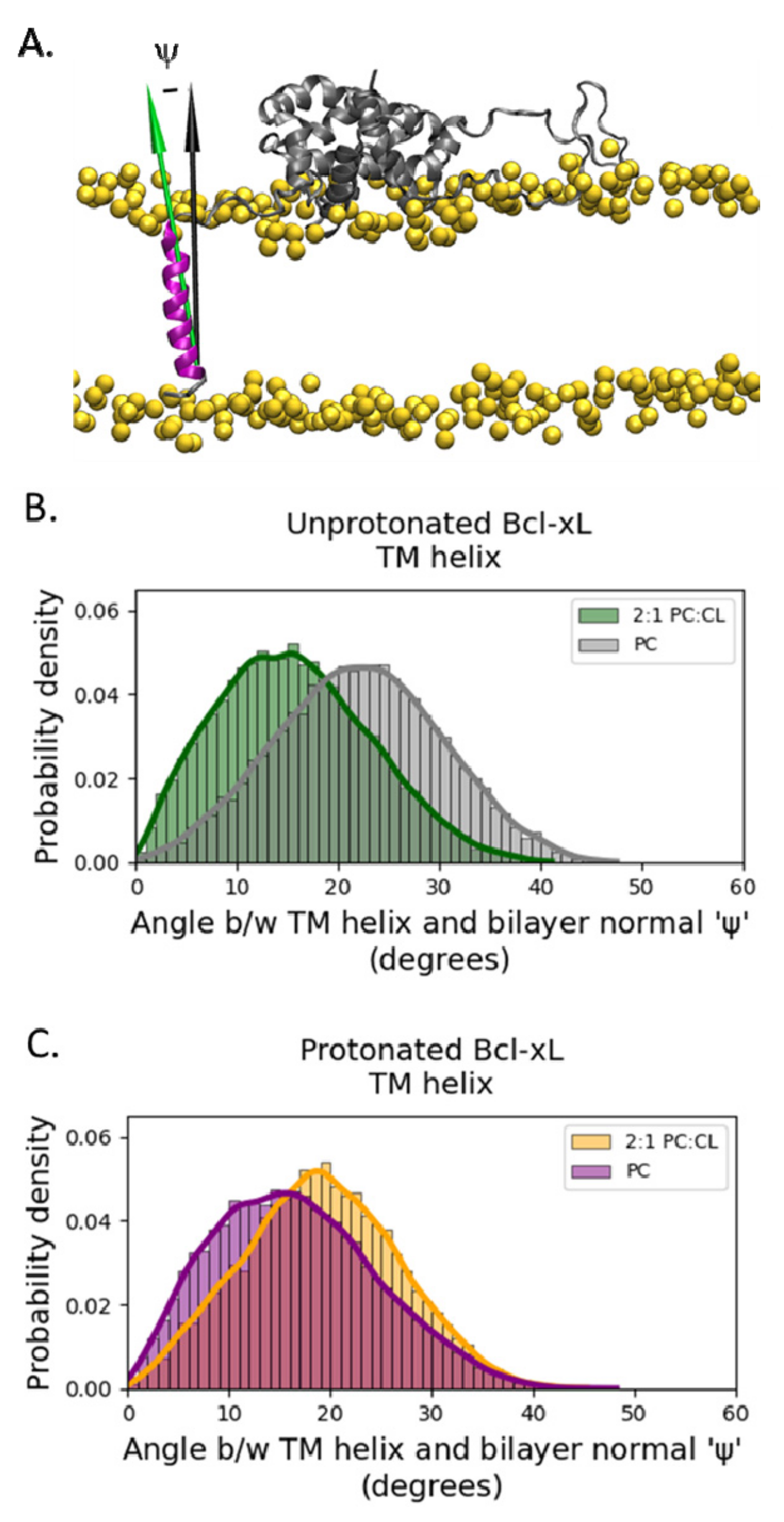
2.3. Transmembrane Helix Orientation
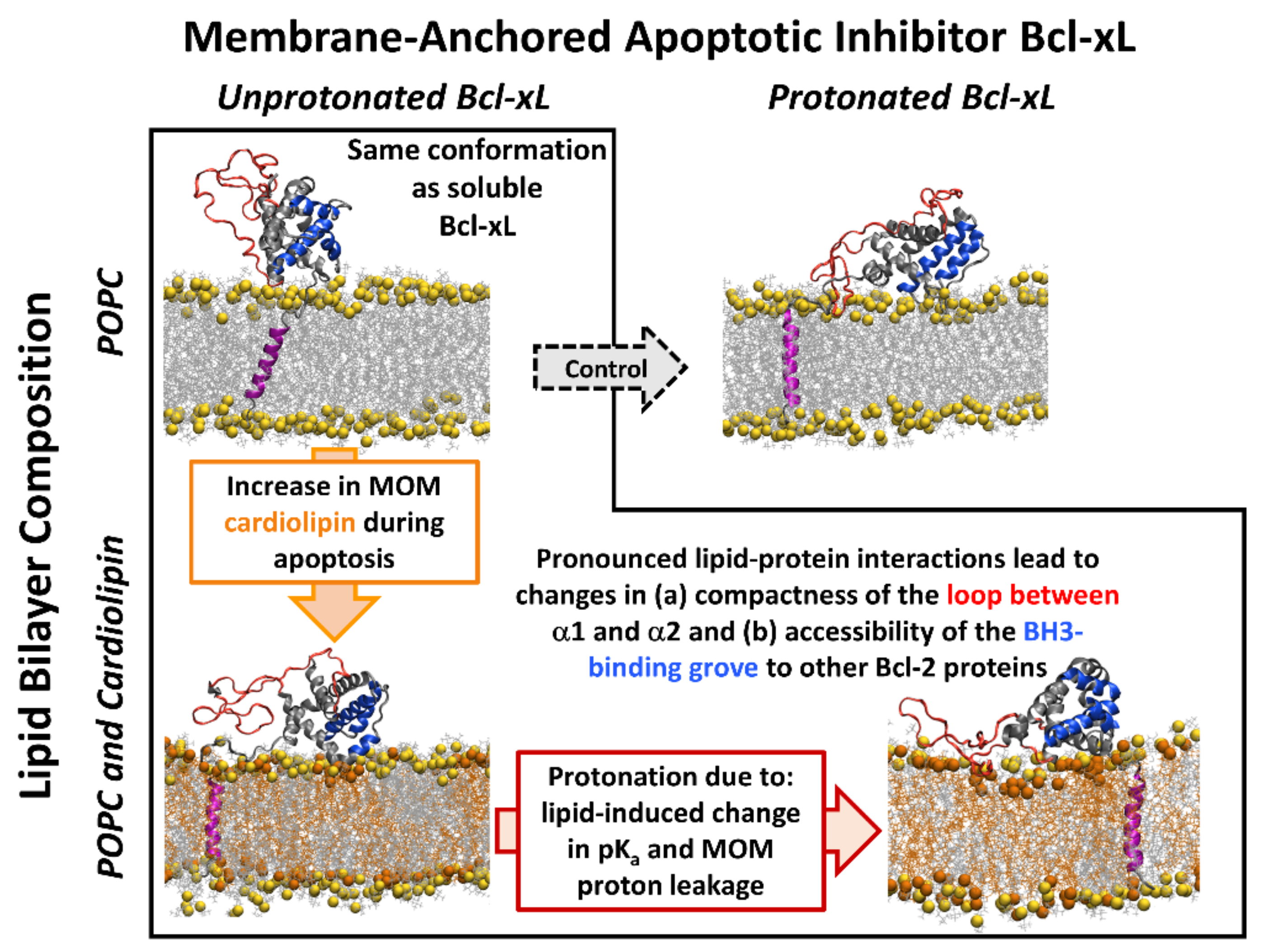
3. Discussion
4. Materials and Methods
4.1. Simulation Systems
4.2. Molecular Dynamics Simulations
4.3. Microsecond-Timescale Simulations
4.4. Trajectory Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gillies, L.A.; Kuwana, T. Apoptosis regulation at the mitochondrial outer membrane. J. Cell. Biochem. 2014, 115, 632–640. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Cossman, J.; Jaffe, E.; Croce, C.M. Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985, 228, 1440–1443. [Google Scholar] [CrossRef]
- Strasser, A.; Cory, S.; Adams, J.M. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011, 30, 3667–3683. [Google Scholar] [CrossRef]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Billen, L.P.; Kokoski, C.L.; Lovell, J.F.; Leber, B.; Andrews, D.W. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol. 2008, 6, e147. [Google Scholar] [CrossRef]
- Ding, J.; Mooers, B.H.M.; Zhang, Z.; Kale, J.; Falcone, D.; McNichol, J.; Huang, B.; Zhang, X.C.; Xing, C.; Andrews, D.W.; et al. After Embedding in membranes antiapoptotic bcl-xl protein binds both bcl-2 homology region 3 and helix 1 of proapoptotic bax protein to inhibit apoptotic mitochondrial permeabilization. J. Biol. Chem. 2014, 289, 11873–11896. [Google Scholar] [CrossRef] [Green Version]
- Subburaj, Y.; Cosentino, K.; Axmann, M.; Pedrueza-Villalmanzo, E.; Hermann, E.; Bleicken, S.; Spatz, J.; García-Sáez, A.J. Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat. Commun. 2015, 6, 8042. [Google Scholar] [CrossRef] [Green Version]
- Moldoveanu, T.; Follis, A.V.; Kriwacki, R.W.; Green, D.R. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014, 39, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Leber, B.; Lin, J.; Andrews, D.W. Embedded together: The life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 2007, 12, 897–911. [Google Scholar] [CrossRef] [Green Version]
- Bogner, C.; Leber, B.; Andrews, D.W. Apoptosis: Embedded in membranes. Curr. Opin. Cell Biol. 2010, 22, 845–851. [Google Scholar] [CrossRef]
- Chi, X.; Kale, J.; Leber, B.; Andrews, D.W. Regulating cell death at, on, and in membranes. Biochim. Biophys. Acta—Mol. Cell Res. 2014, 1843, 2100–2113. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Marassi, F.M. Reconstitution and Characterization of BCL-2 Family Proteins in Lipid Bilayer Nanodiscs. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 1877, pp. 233–246. [Google Scholar]
- Yao, Y.; Fujimoto, L.M.; Hirshman, N.; Bobkov, A.A.; Antignani, A.; Youle, R.J.; Marassi, F.M. Conformation of BCL-XL upon membrane integration. J. Mol. Biol. 2015, 427, 2262–2270. [Google Scholar] [CrossRef] [Green Version]
- Ryzhov, P.; Tian, Y.; Yao, Y.; Bobkov, A.A.; Im, W.; Marassi, F.M. Conformational States of the Cytoprotective Protein Bcl-xL. Biophys. J. 2020, 119, 1324–1334. [Google Scholar] [CrossRef]
- Vasquez-Montes, V.; Kyrychenko, A.; Vargas-Uribe, M.; Rodnin, M.V.; Ladokhin, A.S. Conformational Switching in Bcl-xL: Enabling Non-Canonic Inhibition of Apoptosis Involves Multiple Intermediates and Lipid Interactions. Cells 2020, 9, 539. [Google Scholar] [CrossRef] [Green Version]
- Vasquez-Montes, V.; Vargas-Uribe, M.; Pandey, N.K.; Rodnin, M.V.; Langen, R.; Ladokhin, A.S. Lipid-modulation of membrane insertion and refolding of the apoptotic inhibitor Bcl-xL. Biochim. Biophys. Acta-Proteins Proteom. 2019, 1867, 691–700. [Google Scholar] [CrossRef]
- Ugarte-Uribe, B.; García-Sáez, A.J. Apoptotic foci at mitochondria: In and around bax pores. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Popgeorgiev, N.; Jabbour, L.; Gillet, G. Subcellular localization and dynamics of the Bcl-2 family of proteins. Front. Cell Dev. Biol. 2018, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Martinou, J.C.; Youle, R.J. Mitochondria in Apoptosis: Bcl-2 Family Members and Mitochondrial Dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [Green Version]
- Cristea, I.M.; Degli Esposti, M. Membrane lipids and cell death: An overview. Chem. Phys. Lipids 2004, 129, 133–160. [Google Scholar] [CrossRef]
- Crimi, M.; Esposti, M.D. Apoptosis-induced changes in mitochondrial lipids. Biochim. Biophys. Acta-Mol. Cell Res. 2011, 1813, 551–557. [Google Scholar] [CrossRef]
- Wang, C.; Youle, R.J. The role of mitochondria in apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef] [Green Version]
- Cosentino, K.; García-Sáez, A.J. Mitochondrial alterations in apoptosis. Chem. Phys. Lipids 2014, 181, 62–75. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Grimsley, G.R.; Scholtz, J.M. Protein Ionizable Groups: pK Values and Their Contribution to Protein Stability and Solubility. J. Biol. Chem. 2009, 284, 13285–13289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pahari, S.; Sun, L.; Alexov, E. PKAD: A database of experimentally measured pKa values of ionizable groups in proteins: A database of experimentally measured pKa values of ionizable groups in proteins. Databases 2019, 2019, baz024. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, V.H.; Vila-Viçosa, D.; Reis, P.B.P.S.; MacHuqueiro, M. PKa Values of Titrable Amino Acids at the Water/Membrane Interface. J. Chem. Theory Comput. 2016, 12, 930–934. [Google Scholar] [CrossRef]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef]
- Muchmore, S.W.; Sattler, M.; Liang, H.; Meadows, R.P.; Harlan, J.E.; Yoon, H.S.; Nettesheim, D.; Chang, B.S.; Thompson, C.B.; Wong, S.L.; et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 1996, 381, 335–341. [Google Scholar] [CrossRef]
- Raltchev, K.; Pipercevic, J.; Hagn, F. Production and Structural Analysis of Membrane-Anchored Proteins in Phospholipid Nanodiscs. Chem.—A Eur. J. 2018, 24, 5493–5499. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Jo, S.; Cheng, X.; Islam, S.M.; Huang, L.; Rui, H.; Zhu, A.; Lee, H.S.; Qi, Y.; Han, W.; Vanommeslaeghe, K.; et al. CHARMM-GUI PDB manipulator for advanced modeling and simulations of proteins containing nonstandard residues. In Advances in Protein Chemistry and Structural Biology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 96, pp. 235–265. [Google Scholar]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2007, 2, e880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.-J.; Kern, N.R.; Kim, S.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2018, 15, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y.; Cheng, X.; Lee, J.; Vermaas, J.V.; Pogorelov, T.V.; Tajkhorshid, E.; Park, S.; Klauda, J.B.; Im, W. CHARMM-GUI HMMM Builder for Membrane Simulations with the Highly Mobile Membrane-Mimetic Model. Biophys. J. 2015, 109, 2012–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- GrubmüLler, H.; Heller, H.; Windemuth, A.; Schulten, K. Generalized verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991, 6, 121–142. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Shaw, D.E.; Grossman, J.P.; Bank, J.A.; Batson, B.; Butts, J.A.; Chao, J.C.; Deneroff, M.M.; Dror, R.O.; Even, A.; Fenton, C.H.; et al. Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, New Orleans, LA, USA, 16–21 November 2014; pp. 41–53. [Google Scholar]
- Tuckerman, M.; Berne, B.J.; Martyna, G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992, 97, 1990–2001. [Google Scholar] [CrossRef] [Green Version]
- Shan, Y.; Klepeis, J.L.; Eastwood, M.P.; Dror, R.O.; Shaw, D.E. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005, 122, 54101. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé-Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Lippert, R.A.; Predescu, C.; Ierardi, D.J.; Mackenzie, K.M.; Eastwood, M.P.; Dror, R.O.; Shaw, D.E. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 2013, 139, 164106. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
| Simulated System | Protonation State | Membrane Composition |
|---|---|---|
| Bcl-xL with 50 mM NaCl | Protonated Bcl-xL | POPC |
| Bcl-xL with 50 mM NaCl | Unprotonated Bcl-xL | POPC |
| Bcl-xL with neutralizing NaCl | Protonated Bcl-xL | 2:1 POPC:TOCL |
| Bcl-xL with neutralizing NaCl | Unprotonated Bcl-xL | 2:1 POPC:TOCL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyagi, V.; Vasquez-Montes, V.; Freites, J.A.; Kyrychenko, A.; Tobias, D.J.; Ladokhin, A.S. Effects of Cardiolipin on the Conformational Dynamics of Membrane-Anchored Bcl-xL. Int. J. Mol. Sci. 2021, 22, 9388. https://doi.org/10.3390/ijms22179388
Tyagi V, Vasquez-Montes V, Freites JA, Kyrychenko A, Tobias DJ, Ladokhin AS. Effects of Cardiolipin on the Conformational Dynamics of Membrane-Anchored Bcl-xL. International Journal of Molecular Sciences. 2021; 22(17):9388. https://doi.org/10.3390/ijms22179388
Chicago/Turabian StyleTyagi, Vivek, Victor Vasquez-Montes, J. Alfredo Freites, Alexander Kyrychenko, Douglas J. Tobias, and Alexey S. Ladokhin. 2021. "Effects of Cardiolipin on the Conformational Dynamics of Membrane-Anchored Bcl-xL" International Journal of Molecular Sciences 22, no. 17: 9388. https://doi.org/10.3390/ijms22179388
APA StyleTyagi, V., Vasquez-Montes, V., Freites, J. A., Kyrychenko, A., Tobias, D. J., & Ladokhin, A. S. (2021). Effects of Cardiolipin on the Conformational Dynamics of Membrane-Anchored Bcl-xL. International Journal of Molecular Sciences, 22(17), 9388. https://doi.org/10.3390/ijms22179388







