Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland
Abstract
1. Introduction
2. Results
2.1. Phenotypic Antimicrobial Resistance Profile
2.2. Genotypic Antimicrobial Analysis in Silico
2.3. MLST, wg-SNP and wgMLST Phylogenetic Analysis
2.4. SGI1-K Structure Analysis and Plasmid Detection
3. Discussion
4. Materials and Methods
4.1. Tested S. Kentucky Isolates
4.2. Antimicrobial Susceptibility Testing of S. Kentucky Isolates
4.3. Whole-Genome Sequencing Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [PubMed]
- Khademi, F.; Vaez, H.; Ghanbari, F.; Arzanlou, M.; Mohammadshahi, J.; Sahebkar, A. Prevalence of fluoroquinolone-resistant Salmonella serotypes in Iran: A meta-analysis. Pathog. Glob. Health 2020, 114, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.N.; Fowler, R.C.; Williams, A.J.; Iwen, P.C.; Fey, P.D. Nontyphoidal Salmonella enterica Nonsusceptible to Both Levofloxacin and Ceftriaxone in Nebraska, United States 2014–2015. Foodborne Pathog. Dis. 2018, 15, 235–238. [Google Scholar] [CrossRef]
- Kariuki, S.; Gordon, M.A.; Feasey, N.; Parry, C.M. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 2015, 33 (Suppl. 3), C21–C29. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, G.; Tessema, T.S.; Beyene, G.; Aseffa, A. Molecular epidemiology of fluoroquinolone resistant Salmonella in Africa: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0192575. [Google Scholar] [CrossRef]
- Jeong, H.S.; Kim, J.A.; Shin, J.H.; Chang, C.L.; Jeong, J.; Cho, J.H.; Kim, M.N.; Kim, S.; Kim, Y.R.; Lee, C.H.; et al. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb. Drug Resist. 2011, 17, 551–557. [Google Scholar] [CrossRef]
- Casas, M.R.; Camargo, C.H.; Soares, F.B.; da Silveira, W.D.; Fernandes, S.A. Presence of plasmid-mediated quinolone resistance determinants and mutations in gyrase and topoisomerase in Salmonella enterica isolates with resistance and reduced susceptibility to ciprofloxacin. Diagn. Microbiol. Infect. Dis. 2016, 85, 85–89. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Ferrari, R.; Galiana, A.; Cremades, R.; Rodríguez, J.C.; Magnani, M.; Tognim, M.C.; Oliveira, T.C.; Royo, G. Plasmid-mediated quinolone resistance (PMQR) and mutations in the topoisomerase genes of Salmonella enterica strains from Brazil. Braz. J. Microbiol. 2013, 44, 651–656. [Google Scholar] [CrossRef][Green Version]
- Wasyl, D.; Hoszowski, A. First isolation of ESBL-producing Salmonella and emergence of multiresistant Salmonella Kentucky in turkey in Poland. Food Res. Int. 2012, 45, 958–961. [Google Scholar] [CrossRef]
- Weill, F.X.; Bertrand, S.; Guesnier, F.; Baucheron, S.; Cloeckaert, A.; Grimont, P.A. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 2006, 12, 1611–1612. [Google Scholar] [CrossRef] [PubMed]
- Le Hello, S.; Hendriksen, R.S.; Doublet, B.; Fisher, I.; Nielsen, E.M.; Whichard, J.M.; Bouchrif, B.; Fashae, K.; Granier, S.A.; Jourdan-Da Silva, N.; et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 2011, 204, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Le Hello, S.; Bekhit, A.; Granier, S.A.; Barua, H.; Beutlich, J.; Zając, M.; Münch, S.; Sintchenko, V.; Bouchrif, B.; Fashae, K.; et al. The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front. Microbiol. 2013, 4, 395. [Google Scholar] [CrossRef]
- Mahindroo, J.; Thanh, D.P.; Nguyen, T.N.T.; Mohan, B.; Thakur, S.; Baker, S.; Taneja, N. Endemic fluoroquinolone-resistant Salmonella enterica serovar Kentucky ST198 in northern India. Microb. Genom. 2019, 5, e000275. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Boyd, D.A.; Finley, R.; Fakharuddin, K.; Langner, S.; Allen, V.; Ang, L.; Bekal, S.; El Bailey, S.; Haldane, D.; et al. Ciprofloxacin-resistant Salmonella enterica serovar Kentucky in Canada. Emerg. Infect. Dis. 2013, 19, 999–1001. [Google Scholar] [CrossRef]
- Park, A.K.; Shin, E.; Kim, S.; Park, J.; Jeong, H.J.; Chun, J.H.; Hwang, K.J.; Kim, J. Traveller-associated high-level ciprofloxacin-resistant Salmonella enterica Serovar Kentucky in the Republic of Korea. J. Glob. Antimicrob. Resist. 2020, 22, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.H.; Paul, N.C.; Guard, J. Complete Genome Sequence of a Ciprofloxacin-Resistant Salmonella enterica subsp. enterica Serovar Kentucky Sequence Type 198 Strain, PU131, Isolated from a Human Patient in Washington State. Genome Announc. 2018, 6, e00125-18. [Google Scholar] [CrossRef]
- Mancini, S.; Marchesi, M.; Imkamp, F.; Wagner, K.; Keller, P.M.; Quiblier, C.; Bodendoerfer, E.; Courvalin, P.; Bottger, E.C. Population-based inference of aminoglycoside resistance mechanisms in Escherichia coli. EBioMedicine 2019, 46, 184–192. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union One health 2018 Zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar]
- Le Hello, S.; Weill, F.X.; Guibert, V.; Praud, K.; Cloeckaert, A.; Doublet, B. Early strains of multidrug-resistant Salmonella enterica serovar Kentucky sequence type 198 from Southeast Asia harbor Salmonella genomic island 1-J variants with a novel insertion sequence. Antimicrob. Agents Chemother. 2012, 56, 5096–5102. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, 6007. [Google Scholar] [CrossRef]
- Zając, M.; Wasyl, D.; Hoszowski, A.; Le Hello, S.; Szulowski, K. Genetic lineages of Salmonella enterica serovar Kentucky spreading in pet reptiles. Vet. Microbiol. 2013, 166, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Mąka, Ł.; Maćkiw, E.; Stasiak, M.; Wołkowicz, T.; Kowalska, J.; Postupolski, J.; Popowska, M. Ciprofloxacin and nalidixic acid resistance of Salmonella spp. isolated from retail food in Poland. Int. J. Food Microbiol. 2018, 276, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wasyl, D.; Hoszowski, A.; Zając, M. Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet. Microbiol. 2014, 171, 307–314. [Google Scholar] [CrossRef]
- Levings, R.S.; Partridge, S.R.; Djordjevic, S.P.; Hall, R.M. SGI1-K, a Variant of the SGI1 Genomic Island Carrying a Mercury Resistance Region, in Salmonella enterica Serovar Kentucky. Antimicrob. Agents Chemother. 2006, 51, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Holt, K.E.; Hall, R.M. The complete sequence of Salmonella genomic island SGI1-K. J. Antimicrob. Chemother. 2015, 70, 305–306. [Google Scholar] [CrossRef]
- Chen, H.; Song, J.; Zeng, X.; Chen, D.; Chen, R.; Chen, Q.; Zhoum, K. National Prevalence of Salmonella enterica Serotype Kentucky ST198 with High-Level Resistance to Ciprofloxacin and Extended-Spectrum Cephalosporins in China, 2013 to 2017. Systems 2021, 6, e00935-20. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars; WHO Collaborating Centre for Research on Salmonella, Institute Pasteur: Paris, France, 2007. [Google Scholar]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Deatherage Kaiser, B.L.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella Serotype Determination Utilizing High-throughput Genome Sequencing Data. J. Clin. Micobiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total Genome Sequenced Bacteria. J. Clin. Micobiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Abudahab, K.; Goater, R.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.; Holden, M.; Yeats, C.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]



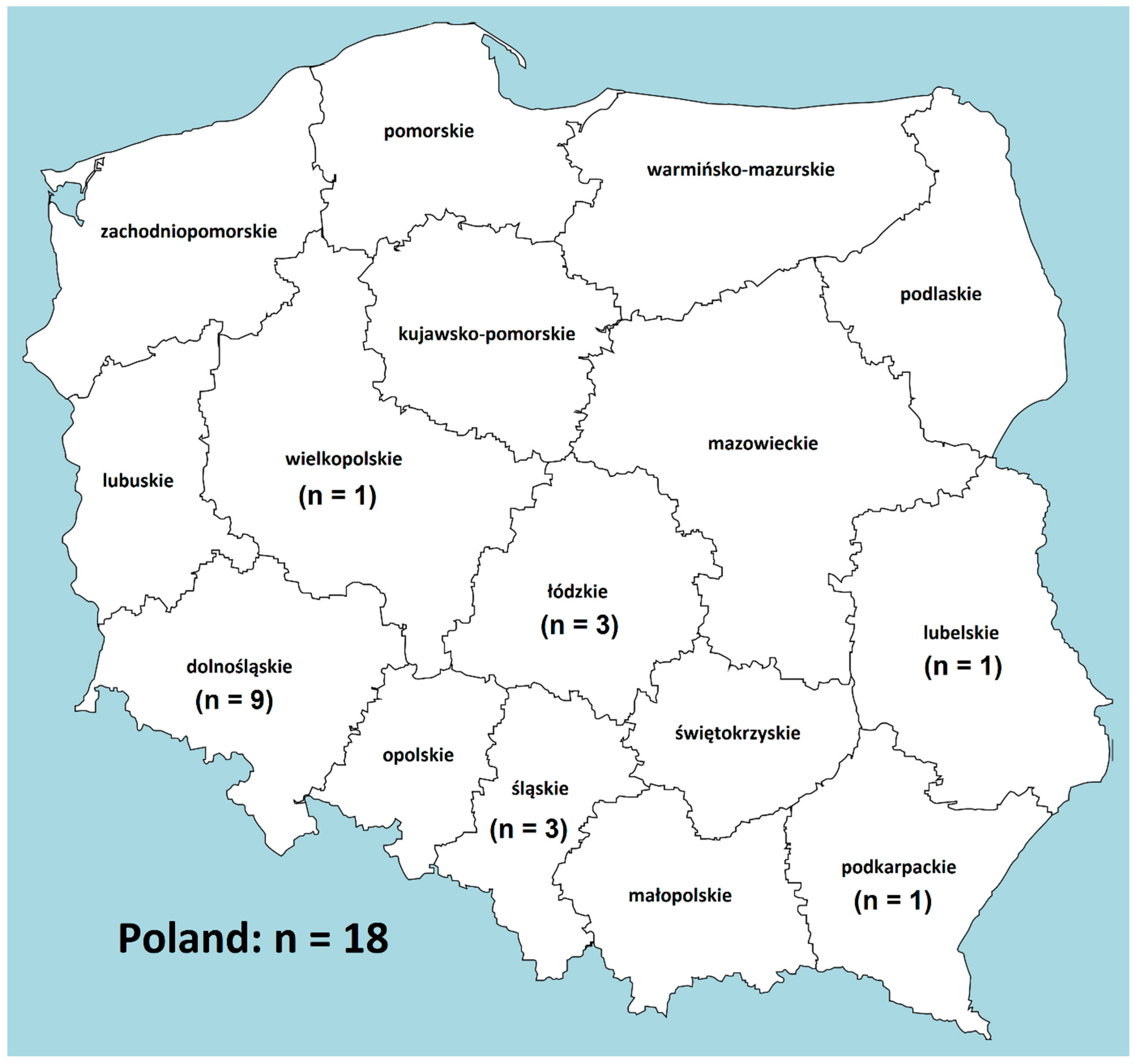
| No. | Isolate ID | Date of Isolation | Sex Female/Male | Age | Patient Status | Province | MIC (mg/L) 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | FOX | CTX | CAZ | GEN | AMK | NAL | CIP | TET | CHL | SXT | |||||||
| 1 | 3/19 | 08.03.2019 | M | 54 | patient | podkarpackie | >256 | 2 | 0.12 | 0.38 | 64 | 1.5 | 1024 | 16 | 32 | 2 | 0.047 |
| 2 | 169/18 | ND | M | ND | patient | śląskie | >256 | 2 | 0.06 | 0.38 | 0.38 | 1.5 | >1024 | 12 | 1.5 | 3 | 0.064 |
| 3 | 85/18 | 28.04.2018 | F | 12 | patient | dolnośląskie | >256 | 2 | 0.06 | 0.38 | 0.38 | 2 | 1024 | 12 | 1 | 2 | 1.5 |
| 4 | 86/18 | 16.03.2018 | F | 35 | patient | dolnośląskie | >256 | 2 | 0.06 | 0.38 | 0.38 | 2 | 1024 | 12 | 32 | 3 | 0.047 |
| 5 | 87/18 | 28.04.2018 | M | 75 | patient | dolnośląskie | >256 | 3 | 0.12 | 0.25 | 0.38 | 2 | >1024 | 12 | 1 | 2 | 0.75 |
| 6 | 325/18 | 09.06.2018 | F | ND | patient | dolnośląskie | >256 | 1.5 | 0.06 | 0.38 | 0.25 | 2 | >1024 | 12 | 1 | 3 | 0.064 |
| 7 | 329/18 | 15.05.2018 | M | ND | patient | dolnośląskie | >256 | 3 | 0.03 | 0.125 | 0.38 | 2 | 1024 | 12 | 1 | 3 | 0.047 |
| 8 | 438/18 | ND | M | ND | patient | wielkopolskie | >256 | 1.5 | 0.06 | 0.38 | 12 | 1.5 | 1024 | 12 | 32 | 2 | 0.19 |
| 9 | 188/18 | 05.06.2018 | M | 42 | patient | łódzkie | >256 | 1.5 | 0.06 | 0.38 | 8 | 2 | 1024 | 12 | 32 | 3 | 0.19 |
| 10 | 88/18 | 28.04.2018 | M | 65 | patient | dolnośląskie | >256 | 1.5 | 0.06 | 0.19 | 0.25 | 1.5 | >1024 | 8 | 1 | 3 | 0.047 |
| 11 | 172/18 | 24.05.2018 | F | ND | patient | dolnośląskie | >256 | 2 | 0.06 | 0.25 | 0.38 | 2 | 1024 | 8 | 1 | 2 | 0.064 |
| 12 | 384/19 | July 2019 | M | ND | patient | śląskie | >256 | 2 | 0.06 | 0.25 | 16 | 3 | 1024 | 8 | 32 | 3 | 0.19 |
| 13 | 457/19 | 12.09.2019 | F | 1 | patient | łódzkie | >256 | 1.5 | 0.06 | 0.25 | 0.38 | 2 | 1024 | 8 | 0.75 | 2 | 0.047 |
| 14 | 93/18 | 27.01.2018 | F | ND | carrier | dolnośląskie | >256 | 2 | 0.06 | 0.38 | 0.38 | 2 | 1024 | 8 | 1 | 2 | 0.047 |
| 15 | 368/18 | ND | F | ND | patient | śląskie | >256 | 1.5 | 0.06 | 0.38 | 12 | 1.5 | >1024 | 6 | 24 | 3 | 0.094 |
| 16 | 145/19 | 04.06.2019 | M | ND | ND | lubelskie | >256 | 2 | 0.06 | 0.25 | 12 | 2 | >1024 | 6 | 32 | 2 | 0.19 |
| 17 | 383/18 | 18.08.2018 | F | ND | patient | dolnośląskie | 0.5 | 2 | 0.06 | 0.19 | 0.25 | 1.5 | 4 | 0.016 | 1 | 2 | 0.047 |
| 18 | 412/18 | 01.10.2018 | M | 86 | patient | łódzkie | 0.38 | 1.5 | 0.03 | 0.19 | 0.25 | 1.5 | 4 | 0.016 | 1 | 2 | 0.047 |
| No. | Isolate ID | MLST Type | Plasmids | Resistance Phenotype | Resistance Genotype by WGS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactams | Aminoglycosides | Sulfonamides | Trimethoprim | Tetracyclines | Fluoroquinolones | ||||||||
| QRDR Amino Acid Change in | PMQR | ||||||||||||
| GyrA | ParC Ser80 | ||||||||||||
| Ser83 | Asp87 | ||||||||||||
| 1 | 3/19 | 198 | Col156 IncR | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaa, aac(6′)-Iid aac(3)-IId aph(3′’)-Ib aph(6)-Id | tet(A) | Phe | Tyr | Ile | qnrS1 | ||
| 2 | 169/18 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 3 | 85/18 | 198 | IncI1-I | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa aadA1 | dfrA1 | Phe | Tyr | Ile | |||
| 4 | 86/18 | 198 | Col8282 | AMP, NA, CIP, TET | blaTEM-1B | aac(6′)-Iaa | tet(A) | Phe | Gly | Ile | |||
| 5 | 87/18 | 198 | IncI1-I | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa aadA1 | dfrA1 | Phe | Tyr | Ile | |||
| 6 | 325/18 | 198 | IncI1-I | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 7 | 329/18 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 8 | 438/18 | 198 | None detected | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaa aac(3)-Id | sul1 | tet(A) | Phe | Tyr | Ile | ||
| 9 | 188/18 | 198 | None detected | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaa aac(3)-Id | sul1 | tet(A) | Phe | Tyr | Ile | ||
| 10 | 88/18 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 11 | 172/18 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 12 | 384/19 | 198 | None detected | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaa aac(3)-Id | sul1 | tet(A) | Phe | Tyr | Ile | ||
| 13 | 457/19 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 14 | 93/18 | 198 | None detected | AMP, NA, CIP | blaTEM-1B | aac(6′)-Iaa | Phe | Tyr | Ile | ||||
| 15 | 368/18 | 198 | None detected | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaaaac(3)-Id | sul1 | tet(A) | Phe | Tyr | Ile | ||
| 16 | 145/19 | 198 | None detected | AMP, NA, CIP, TET, GEN | blaTEM-1B | aac(6′)-Iaaaac(3)-Id | sul1 | tet(A) | Phe | Tyr | Ile | ||
| 17 | 383/18 | 314 | None detected | - | aac(6′)-Iaa | WT | WT | WT | |||||
| 18 | 412/18 | 696 | None detected | - | aac(6′)-Iaa | WT | WT | WT | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wołkowicz, T.; Zacharczuk, K.; Gierczyński, R.; Nowakowska, M.; Piekarska, K. Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland. Int. J. Mol. Sci. 2021, 22, 9381. https://doi.org/10.3390/ijms22179381
Wołkowicz T, Zacharczuk K, Gierczyński R, Nowakowska M, Piekarska K. Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland. International Journal of Molecular Sciences. 2021; 22(17):9381. https://doi.org/10.3390/ijms22179381
Chicago/Turabian StyleWołkowicz, Tomasz, Katarzyna Zacharczuk, Rafał Gierczyński, Magdalena Nowakowska, and Katarzyna Piekarska. 2021. "Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland" International Journal of Molecular Sciences 22, no. 17: 9381. https://doi.org/10.3390/ijms22179381
APA StyleWołkowicz, T., Zacharczuk, K., Gierczyński, R., Nowakowska, M., & Piekarska, K. (2021). Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland. International Journal of Molecular Sciences, 22(17), 9381. https://doi.org/10.3390/ijms22179381






