Heterologous Complementation of SPO11-1 and -2 Depends on the Splicing Pattern
Abstract
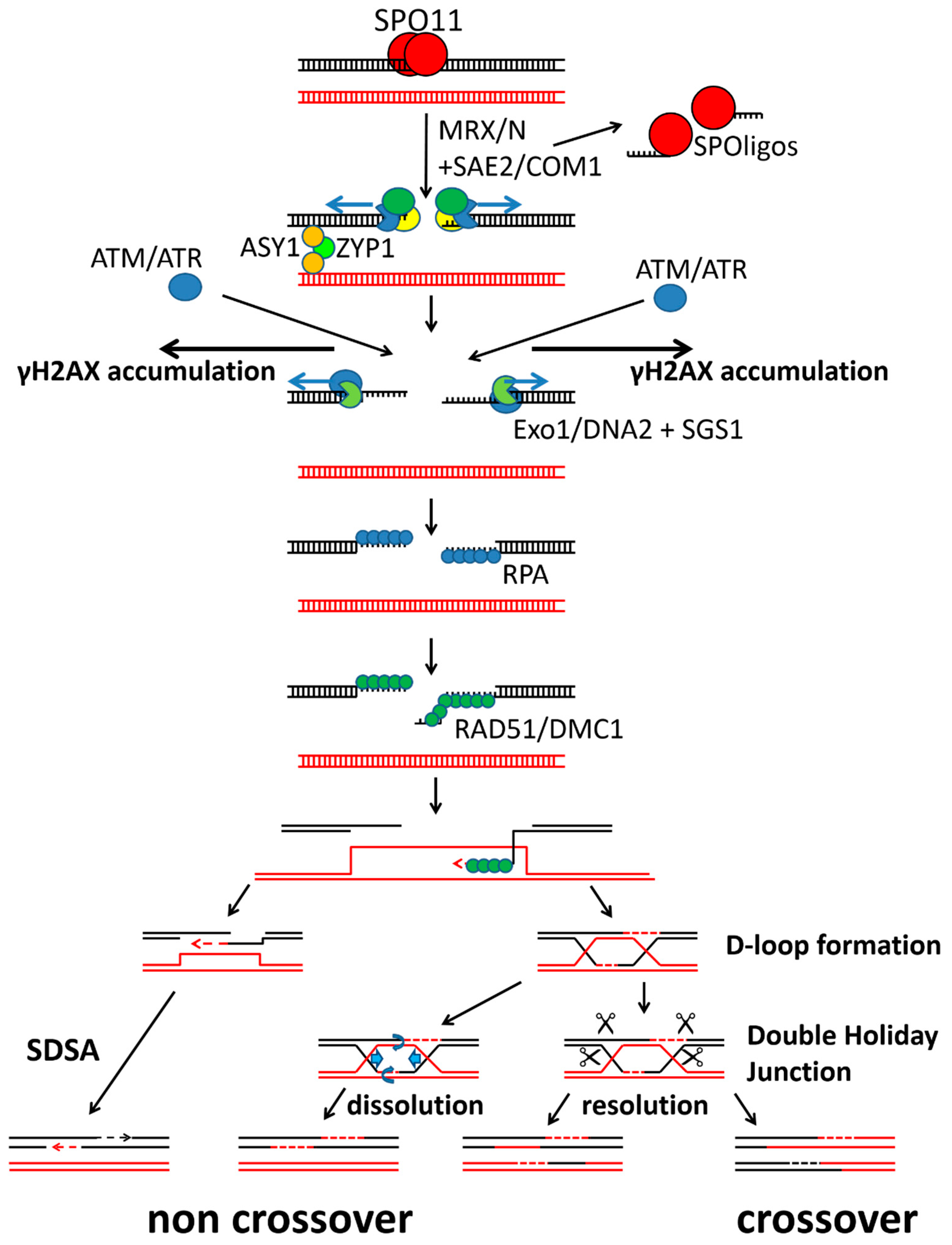
1. Introduction
2. Results
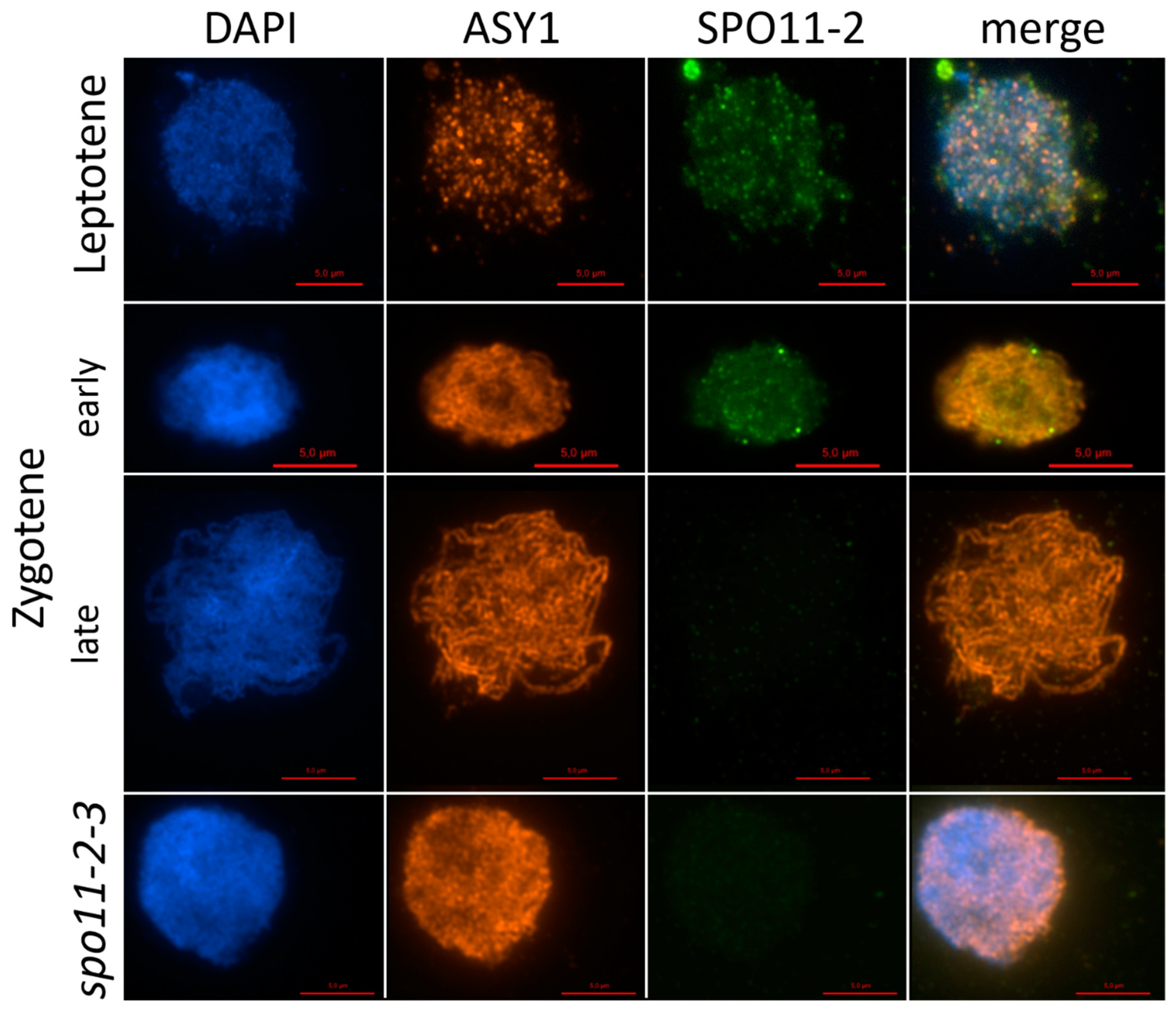
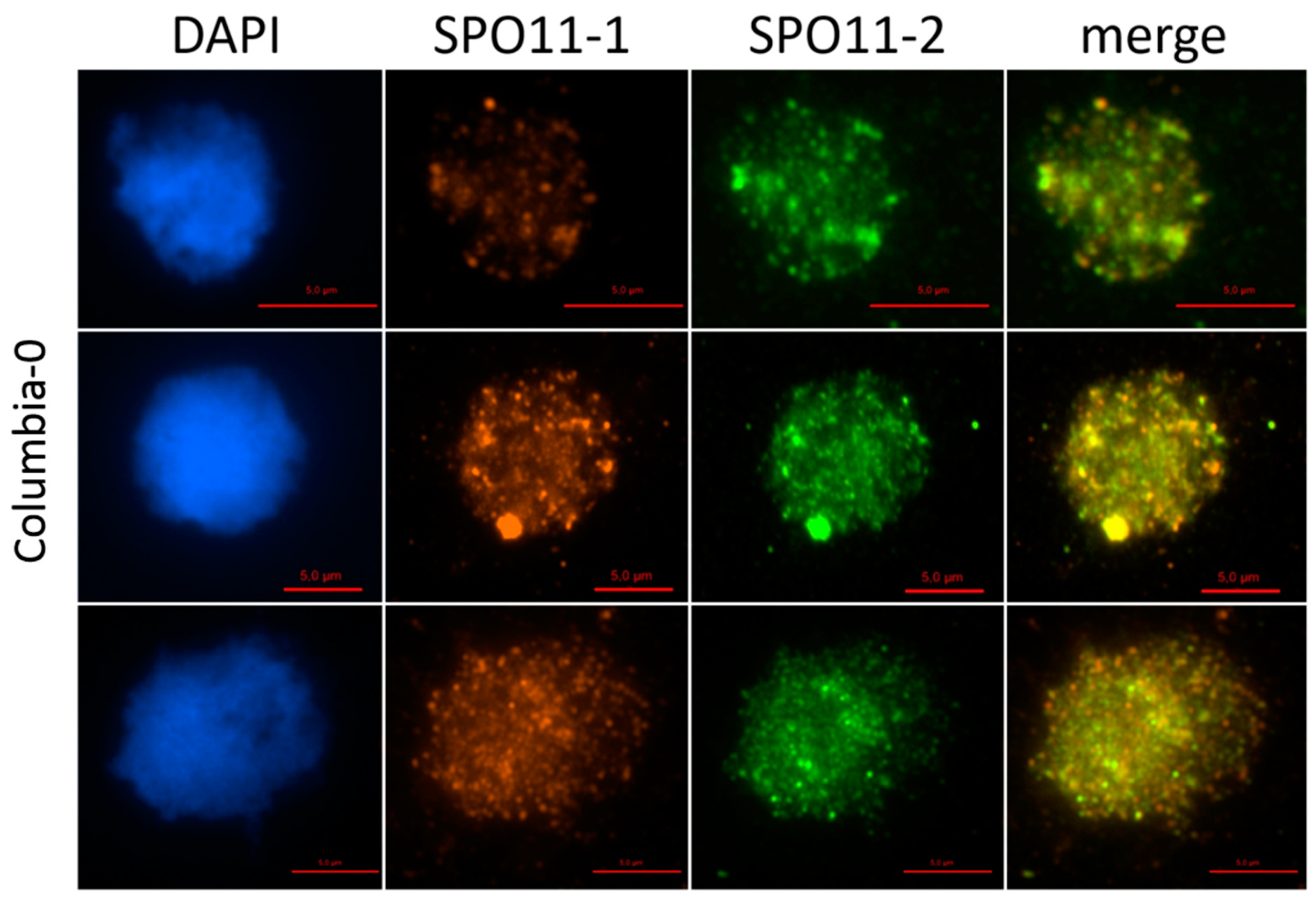
2.1. AthSPO11 is Located on the Chromosome during Leptotene and Early Zygotene
2.2. The Function of AthSPO11 Is Sequence Specific
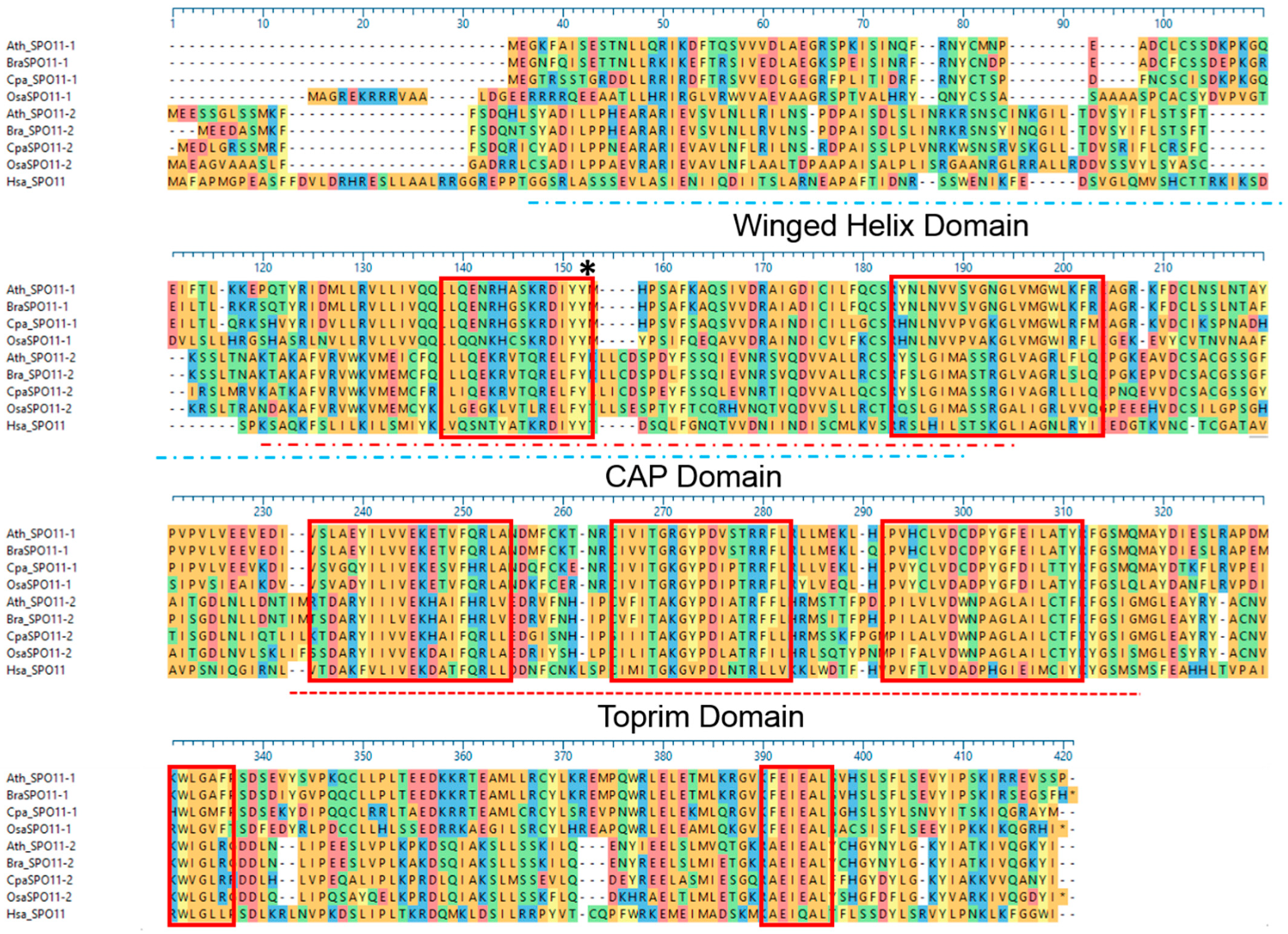
2.3. The Function of SPO11 Is, to a Certain Extent, Species Specific
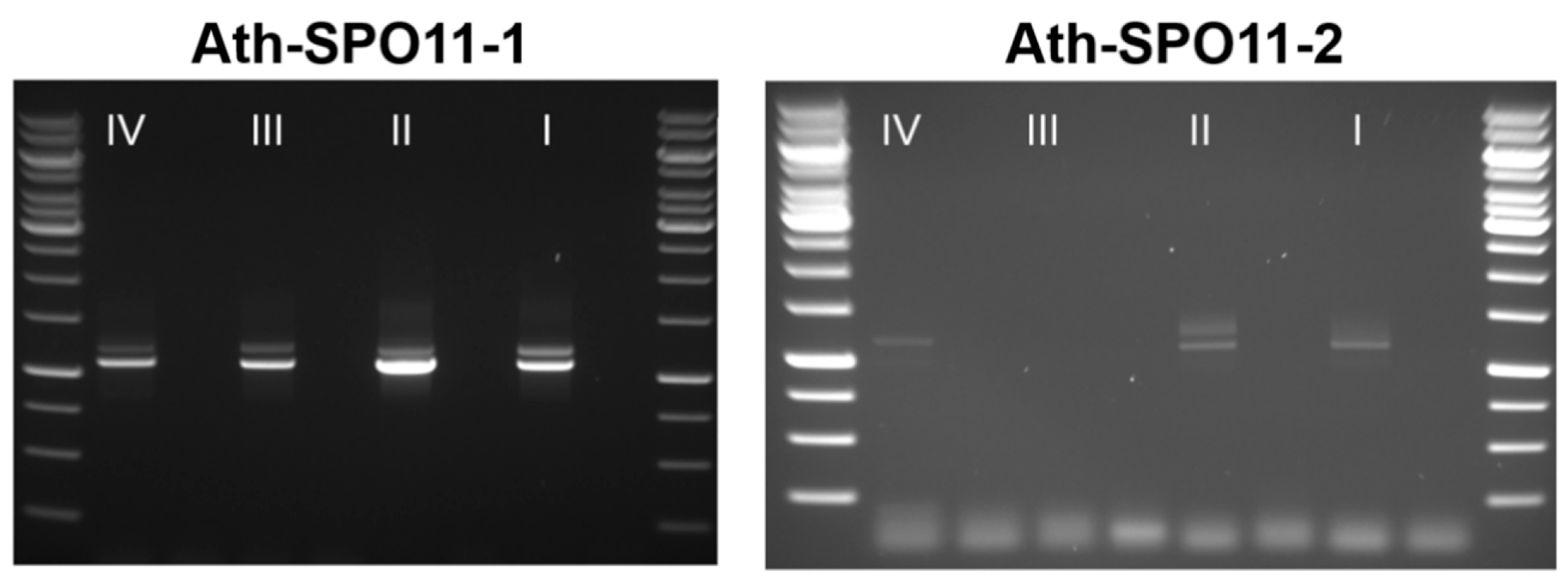
2.4. The Splicing Landscape of SPO11 Homologs Changes when Transformed in A. thaliana
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Molecular Characterization of the Mutant Lines
4.3. Protein Prediction
4.4. Plasmid Construction and Plant Transformation
4.5. RNA Extraction and RT-PCR
4.6. Molecular Characterization of SPO11 Splice Variants
4.7. Complementation Experiments
4.8. Preparation of Pollen Mother Cells
4.9. Statistical Analyses
4.10. Protein Alignments
4.11. Antibody Production
4.12. Immunolocalization Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The Molecular Biology of Meiosis in Plants. Ann. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Roeder, G.S. Meiotic chromosomes: It takes two to tango. Genes Dev. 1997, 11, 2600–2621. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998, 32, 619–697. [Google Scholar] [CrossRef] [PubMed]
- Paques, F.; Haber, J.E. Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999, 63, 349–404. [Google Scholar] [CrossRef] [PubMed]
- Hartung, F.; Suer, S.; Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 18836–18841. [Google Scholar] [CrossRef]
- Knoll, A.; Puchta, H. The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 2011, 62, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- De Massy, B. Initiation of Meiotic Recombination: How and Where? Conservation and Specificities among Eukaryotes. Annu. Rev. Genet. 2013, 47, 563–599. [Google Scholar] [CrossRef]
- Bergerat, A.; de Massy, B.; Gadelle, D.; Varoutas, P.-C.; Nicolas, A.; Forterre, P. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 1997, 386, 414–417. [Google Scholar] [CrossRef]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-Specific DNA Double-Strand Breaks Are Catalyzed by Spo11, a Member of a Widely Conserved Protein Family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef]
- Grelon, M.; Vezon, D.; Gendrot, G.; Pelletier, G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001, 20, 589–600. [Google Scholar] [CrossRef]
- Malik, S.-B.; Ramesh, M.A.; Hulstrand, A.M.; Logsdon, J.M., Jr. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol. Biol. Evol. 2007, 24, 2827–2841. [Google Scholar] [CrossRef]
- Alani, E.; Padmore, R.; Kleckner, N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 1990, 61, 419–436. [Google Scholar] [CrossRef]
- Cao, L.; Alani, E.; Kleckner, N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 1990, 61, 1089–1101. [Google Scholar] [CrossRef]
- Liu, J.; Wu, T.C.; Lichten, M. The location and structure of double-strand DNA breaks induced during yeast meiosis: Evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995, 14, 4599–4608. [Google Scholar] [CrossRef]
- Nairz, K.; Klein, F. mre11S—A yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997, 11, 2272–2290. [Google Scholar] [CrossRef]
- Prinz, S.; Amon, A.; Klein, F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 1997, 146, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Buhler, C.; Lebbink, J.H.G.; Bocs, C.; Ladenstein, R.; Forterre, P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 2001, 276, 37215–37222. [Google Scholar] [CrossRef]
- Amiard, S.; Charbonnel, C.; Allain, E.; Depeiges, A.; White, C.I.; Gallego, M.E. Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 2010, 22, 3020–3033. [Google Scholar] [CrossRef]
- Neale, M.J.; Pan, J.; Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 2005, 436, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008, 455, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.; Kohli, J.; Ludin, K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009, 5, e1000722. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Phelps, S.E.L.; Gray, S.; Neale, M.J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Edlinger, B.; Schlogelhofer, P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 2011, 62, 1545–1563. [Google Scholar] [CrossRef] [PubMed]
- Lambing, C.; Franklin, F.C.H.; Wang, C.-J.R. Understanding and manipulating meiotic recombination in plants. Plant Physiol. 2017, 173, 1530–1542. [Google Scholar] [CrossRef]
- Bellani, M.A.; Boateng, K.A.; McLeod, D.; Camerini-Otero, R.D. The expression profile of the major mouse SPO11 isoforms indicates that SPO11β introduces double strand breaks and suggests that SPO11α has an additional role in prophase in both spermatocytes and oocytes. Mol. Cell. Boil. 2010, 30, 4391–4403. [Google Scholar] [CrossRef]
- Kauppi, L.; Barchi, M.; Baudat, F.; Romanienko, P.J.; Keeney, S.; Jasin, M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 2011, 331, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Cole, F.; Keeney, S.; Jasin, M. Evolutionary conservation of meiotic DSB proteins: More than just Spo11. Genes Dev. 2010, 24, 1201–1207. [Google Scholar] [CrossRef]
- Romanienko, P.J.; Camerini-Otero, R.D. Cloning, characterization, and localization of mouse and human SPO11. Genomics 1999, 61, 156–169. [Google Scholar] [CrossRef]
- Metzler-Guillemain, C.; de Massy, B. Identification and characterization of an SPO11 homolog in the mouse. Chromosoma 2000, 109, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Hartung, F.; Puchta, H. Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 2000, 28, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Sprink, T.; Hartung, F. The splicing fate of plant SPO11 genes. Front. Plant Sci. 2014, 5, 214. [Google Scholar] [CrossRef]
- Habu, T.; Taki, T.; West, A.; Nishimune, Y.; Morita, T. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 1996, 24, 470–477. [Google Scholar] [CrossRef]
- Nakagawa, T.; Ogawa, H. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells 1997, 2, 65–79. [Google Scholar] [CrossRef]
- Terzi, L.C.; Simpson, G.G. Regulation of flowering time by RNA processing. Nuclear pre-mRNA Process. Plants 2008, 326, 201–218. [Google Scholar]
- Stacey, N.J.; Kuromori, T.; Azumi, Y.; Roberts, G.; Breuer, C.; Wada, T.; Maxwell, A.; Roberts, K.; Sugimoto-Shirasu, K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006, 48, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Hartung, F.; Angelis, K.J.; Meister, A.; Schubert, I.; Melzer, M.; Puchta, H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 2002, 12, 1787–1791. [Google Scholar] [CrossRef]
- Simkova, K.; Moreau, F.; Pawlak, P.; Vriet, C.; Baruah, A.; Alexandre, C.; Hennig, L.; Apel, K.; Laloi, C. Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 16360–16365. [Google Scholar] [CrossRef]
- An, X.J.; Deng, Z.Y.; Wang, T. OsSpo11-4, a rice homologue of the archaeal TopVIA protein, mediates double-strand DNA cleavage and interacts with OsTopVIB. PLoS ONE 2011, 6, e20327. [Google Scholar] [CrossRef]
- Shingu, Y.; Tokai, T.; Agawa, Y.; Toyota, K.; Ahamed, S.; Kawagishi-Kobayashi, M.; Komatsu, A.; Mikawa, T.; Yamamoto, M.-T.; Wakasa, K.; et al. The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay. BMC Mol. Biol. 2012, 13, 1. [Google Scholar] [CrossRef]
- Vrielynck, N.; Chambon, A.; Vezon, D.; Pereira, L.; Chelysheva, L.; de Muyt, A.; Mézard, C.; Mayer, C.; Grelon, M. A DNA topoisomerase VI–like complex initiates meiotic recombination. Science 2016, 351, 939–943. [Google Scholar] [CrossRef]
- Robert, T.; Nore, A.; Brun, C.; Maffre, C.; Crimi, B.; Guichard, V.; Bourbon, H.-M.; de Massy, B. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 2016, 351, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Wang, J.; Jiang, L.; Wang, M.; Wolfe, S.; Pawlowski, W.P.; Wang, Y.; He, Y. The number of meiotic double-strand breaks influences crossover distribution in Arabidopsis. Plant Cell Online 2018, 30, 2628–2638. [Google Scholar] [CrossRef]
- Nichols, M.D.; DeAngelis, K.; Keck, J.L.; Berger, J.M. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999, 18, 6177–6188. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Moran, E.; Santos, J.-L.; Jones, G.H.; Franklin, F.C.H. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007, 21, 2220–2233. [Google Scholar] [CrossRef]
- de Muyt, A.; Vezon, D.; Gendrot, G.; Gallois, J.-L.; Stevens, R.; Grelon, M. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 2007, 26, 4126–4137. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yin, Z.; Zeng, Y.; Zhang, Q.; Chen, L.; He, Y.; Lu, P.; Ye, D.; Zhang, X. MTOPVIB interacts with AtPRD1 and plays important roles in formation of meiotic DNA double-strand breaks in Arabidopsis. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Da Ines, O.; Michard, R.; Fayos, I.; Bastianelli, G.; Nicolas, A.; Guiderdoni, E.; White, C.; Sourdille, P. Bread Wheat TaSPO11-1 exhibits evolutionary conserved function in meiotic recombination across distant plant species. Plant J. 2020, 103, 2052–2068. [Google Scholar] [CrossRef]
- Benyahya, F.; Nadaud, I.; Da Ines, O.; Rimbert, H.; White, C.; Sourdille, P. SPO11. 2 is essential for programmed double strand break formation during meiosis in bread wheat (Triticum aestivum L.). Plant J. 2020, 104, 30–43. [Google Scholar] [CrossRef]
- Cole, C.; Barber, J.D.; Barton, G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008, 36, W197–W201. [Google Scholar] [CrossRef]
- Town, C.D.; Cheung, F.; Maiti, R.; Crabtree, J.; Haas, B.J.; Wortman, J.R.; Hine, E.E.; Althoff, R.; Arbogast, T.S.; Tallon, L.J. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell Online 2006, 18, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Pedersen, B.; Freeling, M. Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 2010, 6, e1000949. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Bell, C.D.; Kim, S.; Soltis, P.S. Origin and early evolution of angiosperms. Ann. N. Y. Acad. Sci. 2008, 1133, 3–25. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.-C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Bleuyard, J.-Y.; White, C.I. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004, 23, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Bleuyard, J.-Y.; Gallego, M.E.; White, C.I. Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair 2006, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bray, C.M.; West, C.E. DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005, 168, 511–528. [Google Scholar] [CrossRef]
- Kozak, J.; West, C.E.; White, C.; Da Costa-Nunes, J.A.; Angelis, K.J. Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair 2009, 8, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Shingu, Y.; Mikawa, T.; Onuma, M.; Hirayama, T.; Shibata, T. A DNA-binding surface of SPO11-1, an Arabidopsis SPO11 orthologue required for normal meiosis. FEBS J. 2010, 277, 2360–2374. [Google Scholar] [CrossRef]
- Li, W.; Yang, X.; Lin, Z.; Timofejeva, L.; Xiao, R.; Makaroff, C.A.; Ma, H. The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 2005, 138, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Puizina, J.; Siroky, J.; Mokros, P.; Schweizer, D.; Riha, K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell Online 2004, 16, 1968–1978. [Google Scholar] [CrossRef]
- Romanienko, P.J.; Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 2000, 6, 975–987. [Google Scholar] [CrossRef]
- Prieler, S.; Penkner, A.; Borde, V.; Klein, F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes Dev. 2005, 19, 255–269. [Google Scholar] [CrossRef]
- Ku, J.-C.; Ronceret, A.; Golubovskaya, I.; Lee, D.H.; Wang, C.; Timofejeva, L.; Kao, Y.-H.; Gomez Angoa, A.K.; Kremling, K.; Williams-Carrier, R. Dynamic localization of SPO11-1 and conformational changes of meiotic axial elements during recombination initiation of maize meiosis. PLoS Genet. 2020, 16, e1007881. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatilepPZP family ofAgrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
- Bonnet, S.; Knoll, A.; Hartung, F.; Puchta, H. Different functions for the domains of the Arabidopsis thaliana RMI1 protein in DNA cross-link repair, somatic and meiotic recombination. Nucleic Acids Res. 2013, 41, 9349–9360. [Google Scholar] [CrossRef]
- Pawlowski, W.P.; Grelon, M.; Armstrong, S. Plant Meiosis: Methods and Protocols; Springer Nature Customer Service Center LLC: New York, NY, USA, 2013. [Google Scholar]
- Armstrong, S.J. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002, 115, 3645–3655. [Google Scholar] [CrossRef]
- Sanchez-Moran, E.; Osman, K.; Higgins, J.D.; Pradillo, M.; Cunado, N.; Jones, G.H.; Franklin, F.C.H. ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenet. Genome Res. 2008, 120, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005, 19, 2488–2500. [Google Scholar] [CrossRef] [PubMed]

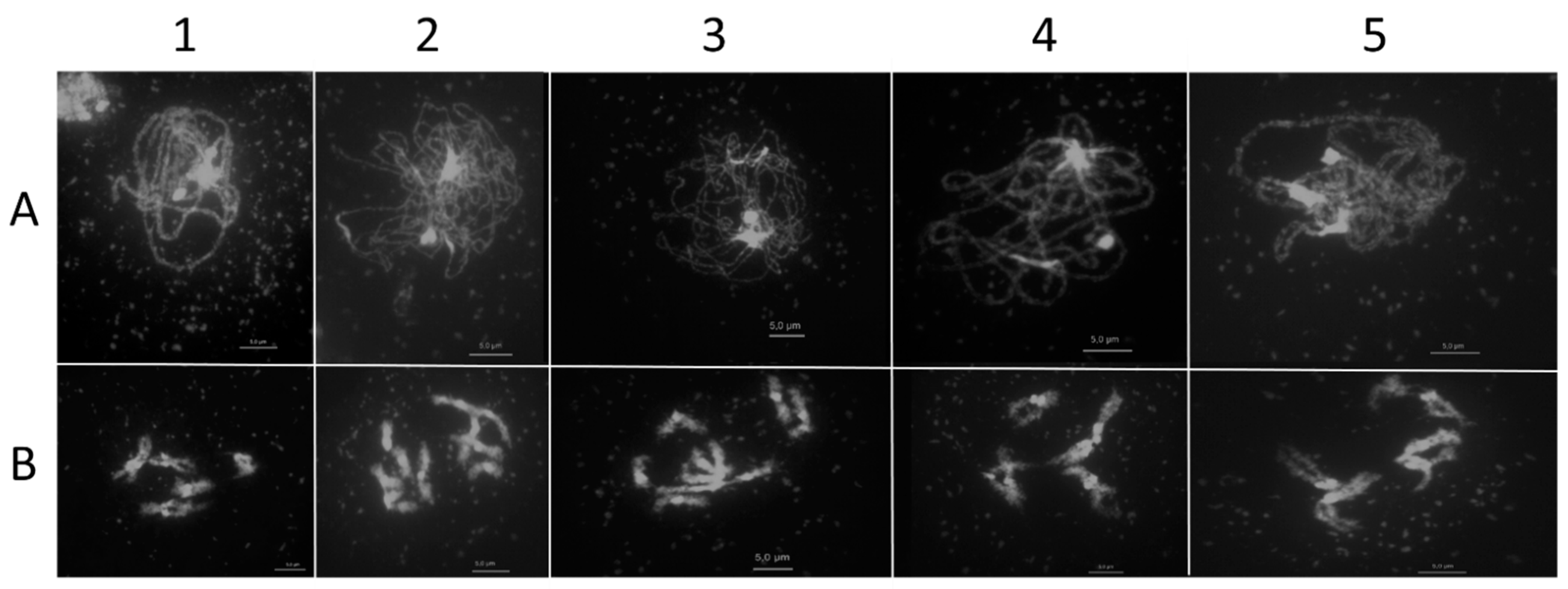
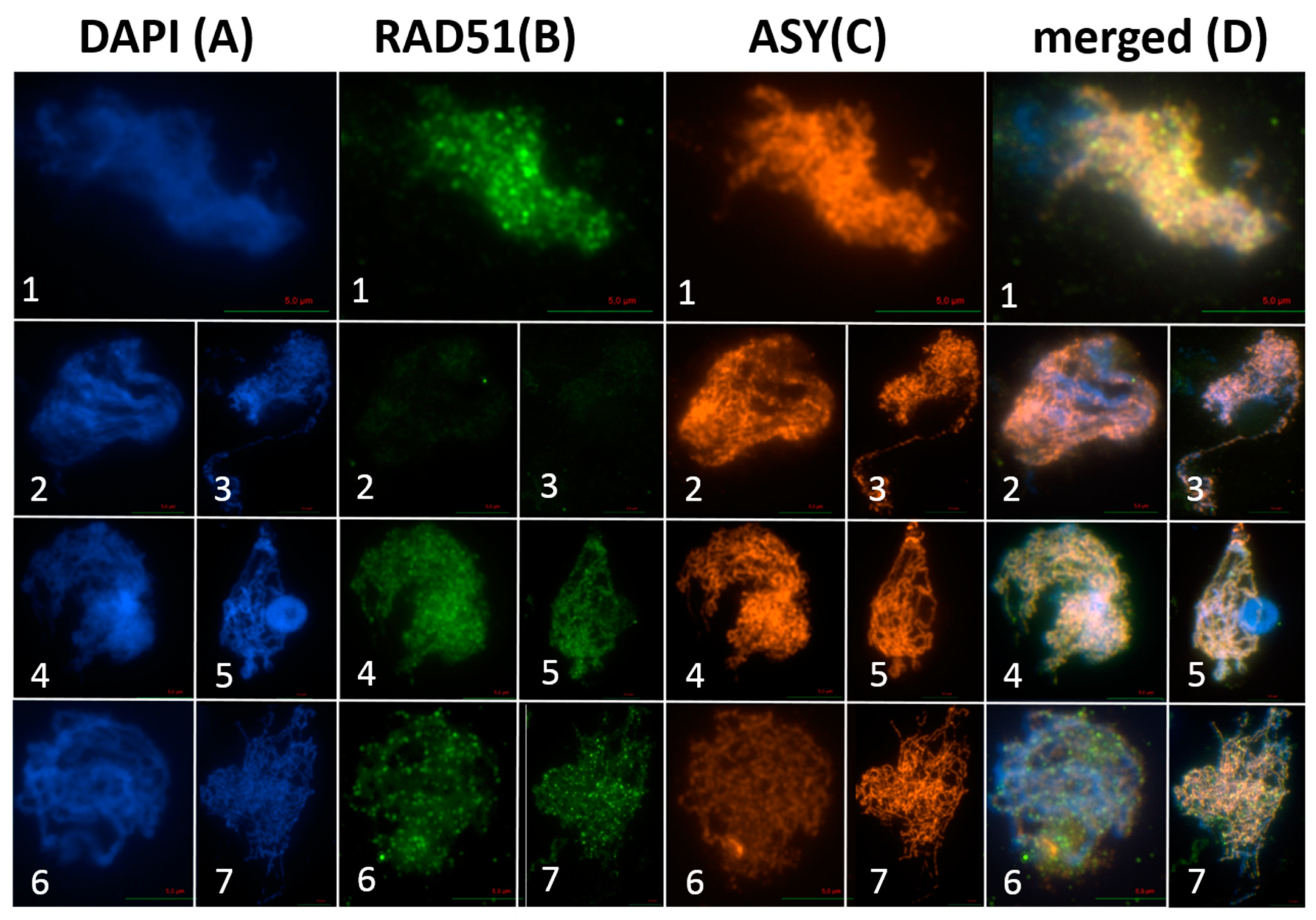

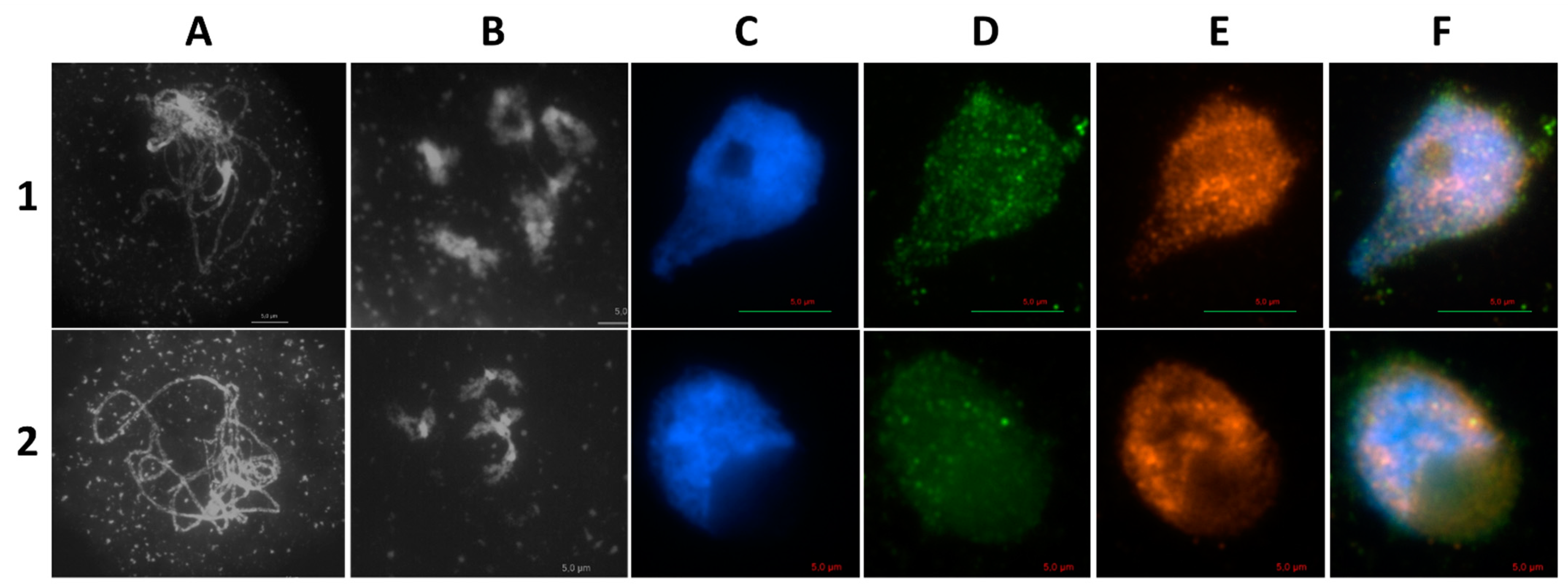
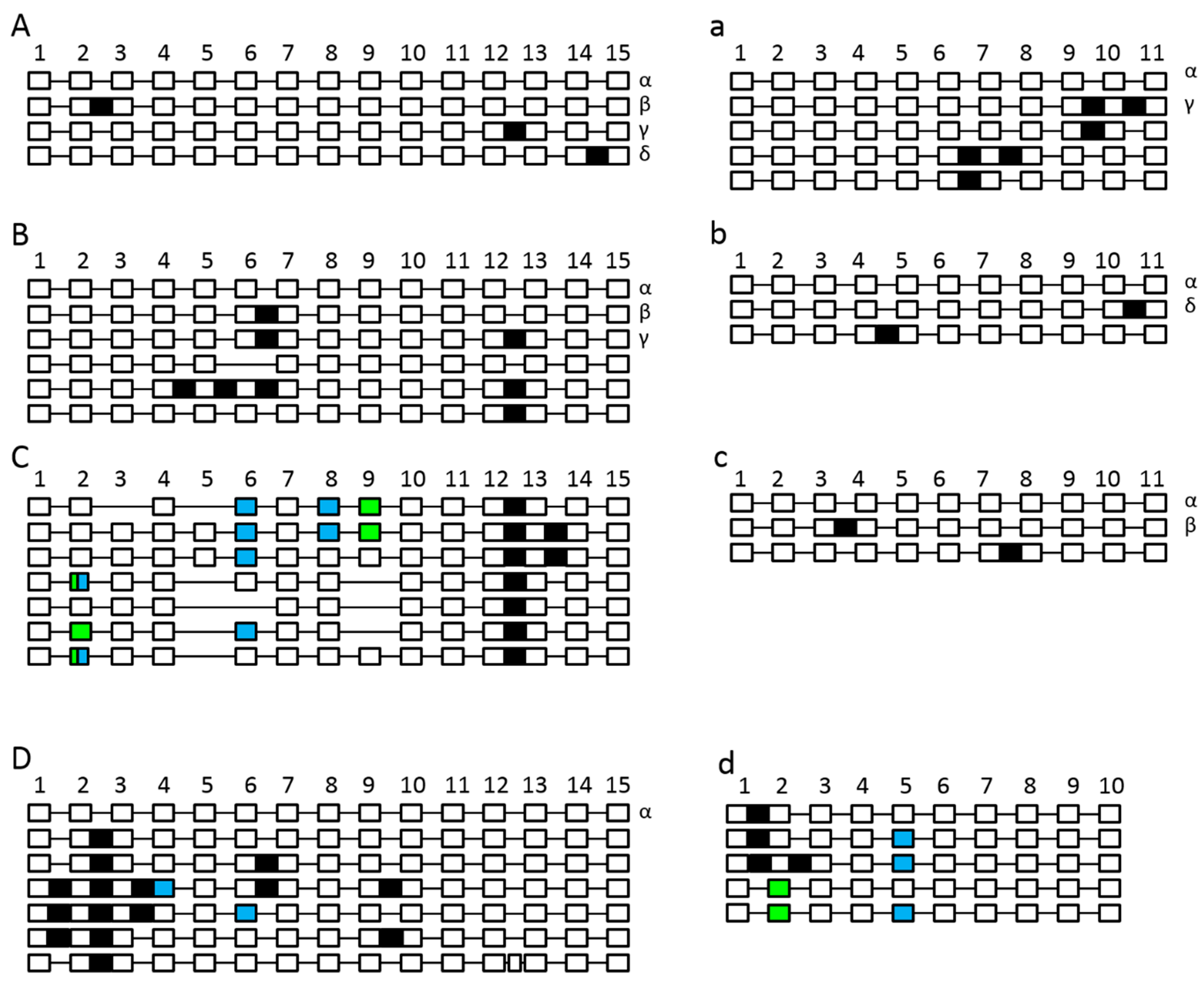

| Construct | Complementing spo11-1-3 Lines | Complementing spo11-2-3 Lines |
|---|---|---|
| AthSPO11-1g | 6/8 | - |
| Ath SPO11-2g | - | 7/8 |
| Ath SPO1swap1 | 0/19 | 0/11 |
| Ath SPO1swap2 | 0/14 | 0/6 |
| Ath SPO1swap3 | 0/17 | 0/8 |
| Ath SPO2swap1 | 0/11 | 0/8 |
| Ath SPO2swap2 | 0/12 | 0/23 |
| Ath SPO2swap3 | 0/4 | 0/9 |
| Ath SPO1swap4 | 4/21 | 0/8 |
| Ath SPO2swap4 | 0/12 | 0/24 |
| Ath SPO1Δlex | 0/30 | - |
| Ath SPO2Δlex | - | 0/21 |
| Organism | Gene | gDNA (bp) | CDS (bp) | Protein (aa) | Ath SPO11-1 (%) | Ath SPO11-2 (%) |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | SPO11-1 | 2633 | 1089 | 362 | 100 | 30.5 |
| SPO11-2 | 2029 | 1152 | 383 | 30.5 | 100 | |
| Brassica rapa | SPO11-1 | 2492 | 1089 | 362 | 90.3 | 30.1 |
| SPO11-2 | 1870 | 1143 | 380 | 31.8 | 92.3 | |
| Carica papaya | SPO11-1 | 4491 | 1086 | 361 | 72.8 | 32.2 |
| SPO11-2 | 2163 | 1149 | 382 | 30.5 | 73.8 | |
| Oryza sativa | SPO11-1 | 3710 | 1146 | 381 | 58.7 | 31.4 |
| SPO11-2 | 2710 | 1158 | 385 | 29.1 | 62.8 |
| Line | Construct | Number of Complementing Lines | Average Seed (%) Set Hm-Lines |
|---|---|---|---|
| spo11-1-3 | BraSPO11-1g | 4/4 | - |
| spo11-2-3 | BraSPO11-2g | 11/11 | 101.4 ± 13.4 (n = 18) |
| spo11-1-3 | CpaSPO11-1g | 0/15 | 2.49 ± 0.54 (n = 15) |
| spo11-2-3 | CpaSPO11-2g | 0/22 | 2.6 ± 0.63 (n = 19) |
| spo11-1-3 | OsaSPO11-1g | 0/11 | 6.47 ±1.34 (n = 15) |
| spo11-2-3 | OsaSPO11-2g | 0/11 | 3.6 ± 1.36 (n = 7) |
| spo11-1-3 | AthSPO11-1c | 14/17 | 72.53 ± 12.53 (n = 8) |
| spo11-2-3- | AthSPO11-2c | 16/16 | 92.6 ± 11 (n = 7) |
| spo11-1-3 | BraSPO11-1c | 14/14 | 84.2 ±19.74 (n = 6) |
| spo11-2-3 | BraSPO11-2c | 13/15 | 100.6 ± 22 (n = 7) |
| spo11-1-3 | CpaSPO11-1c | 3/8 | 19.04 ± 4.5 (n = 11) |
| spo11-2-3 | CpaSPO11-2c | 0/8 | 0.72 ± 0.12 (n = 13) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sprink, T.; Hartung, F. Heterologous Complementation of SPO11-1 and -2 Depends on the Splicing Pattern. Int. J. Mol. Sci. 2021, 22, 9346. https://doi.org/10.3390/ijms22179346
Sprink T, Hartung F. Heterologous Complementation of SPO11-1 and -2 Depends on the Splicing Pattern. International Journal of Molecular Sciences. 2021; 22(17):9346. https://doi.org/10.3390/ijms22179346
Chicago/Turabian StyleSprink, Thorben, and Frank Hartung. 2021. "Heterologous Complementation of SPO11-1 and -2 Depends on the Splicing Pattern" International Journal of Molecular Sciences 22, no. 17: 9346. https://doi.org/10.3390/ijms22179346
APA StyleSprink, T., & Hartung, F. (2021). Heterologous Complementation of SPO11-1 and -2 Depends on the Splicing Pattern. International Journal of Molecular Sciences, 22(17), 9346. https://doi.org/10.3390/ijms22179346







