Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition
Abstract
1. Introduction
2. Results
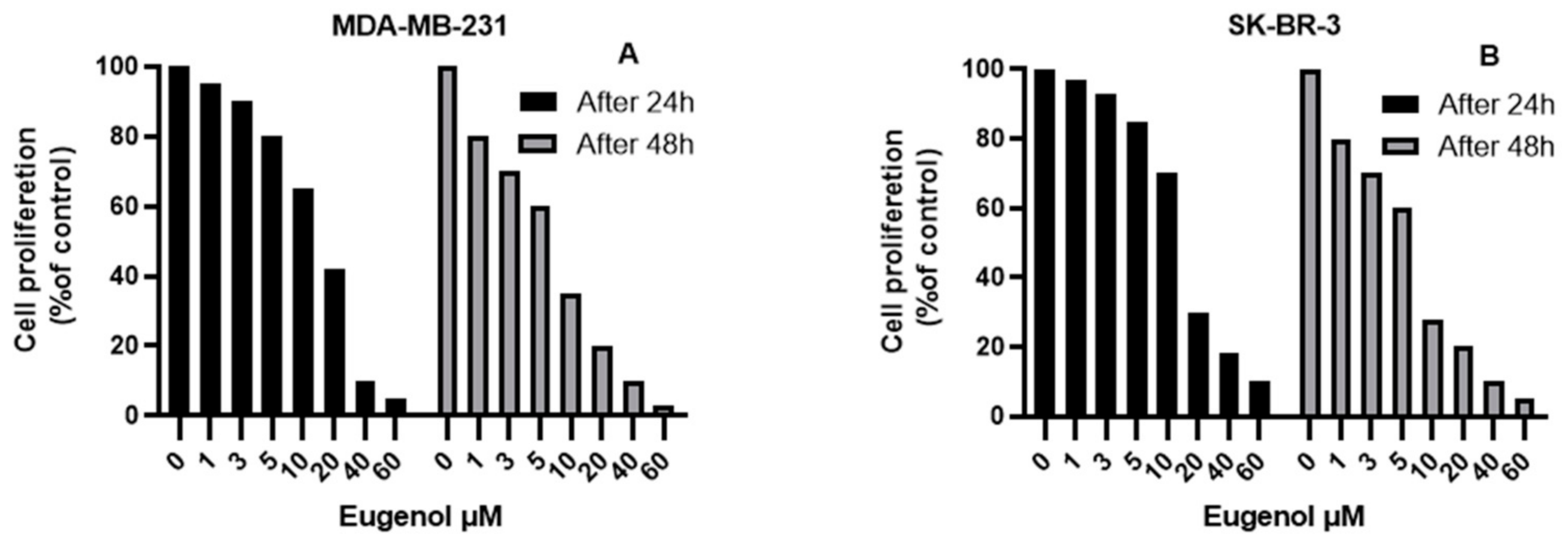
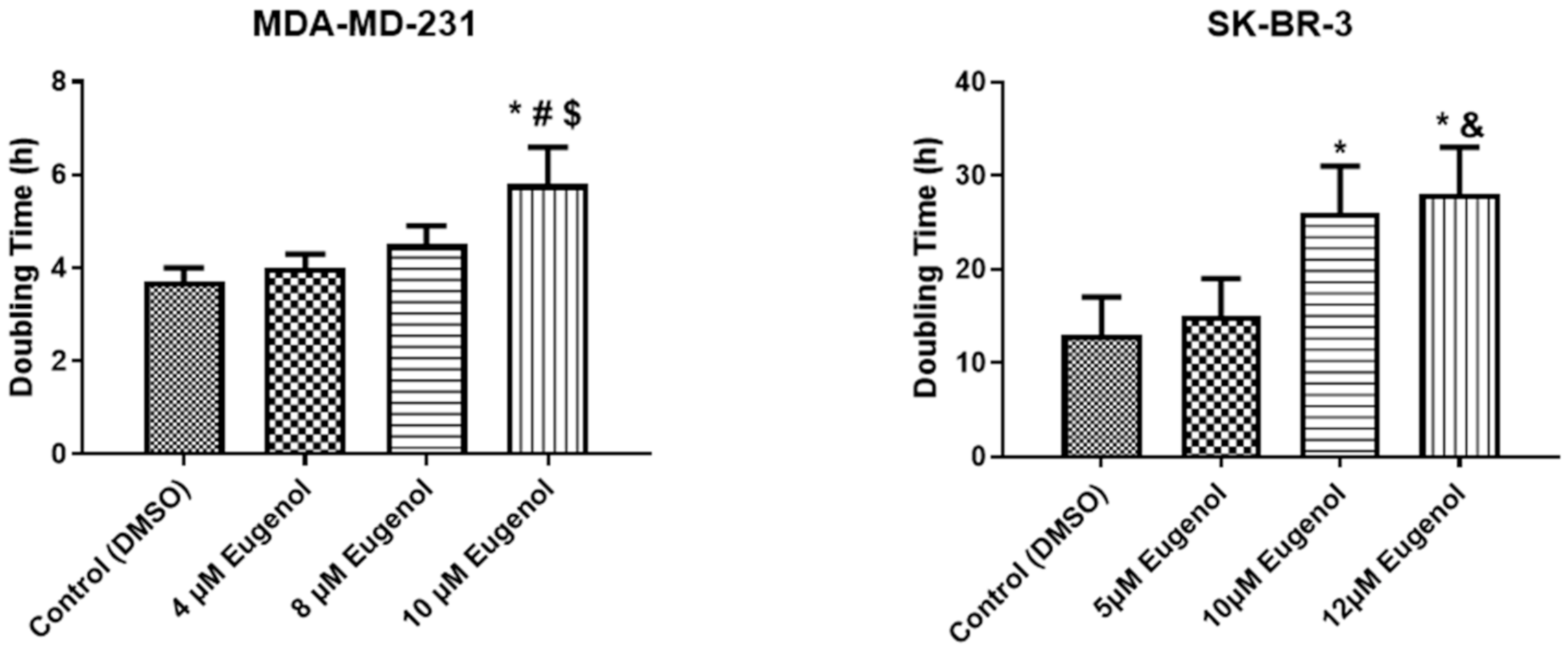
2.1. The Effect of Eugenol on Cell Viability and Analysis of Population Doubling Time (PDA)
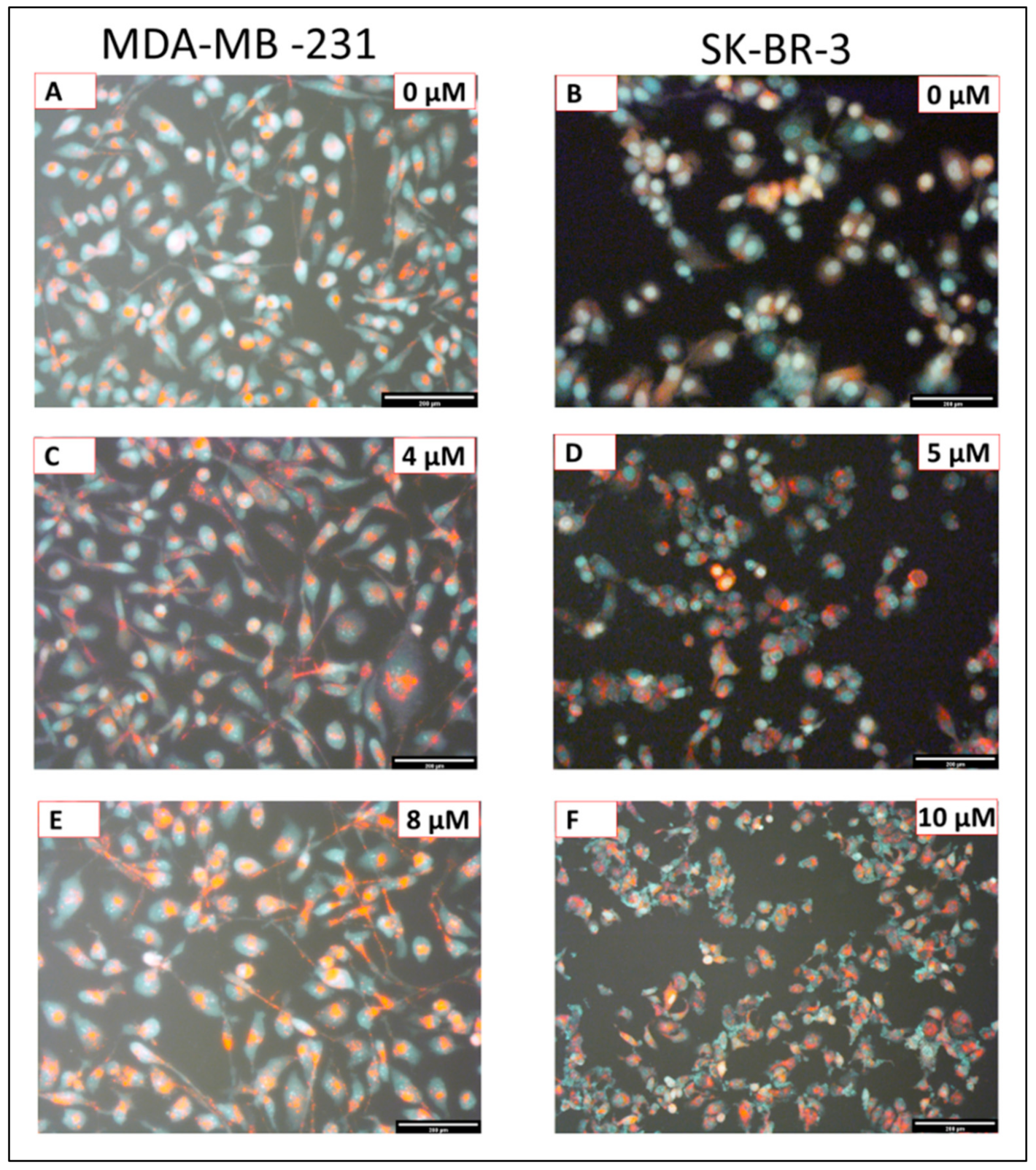
2.2. The Effect of Eugenol on Autophagy Induction
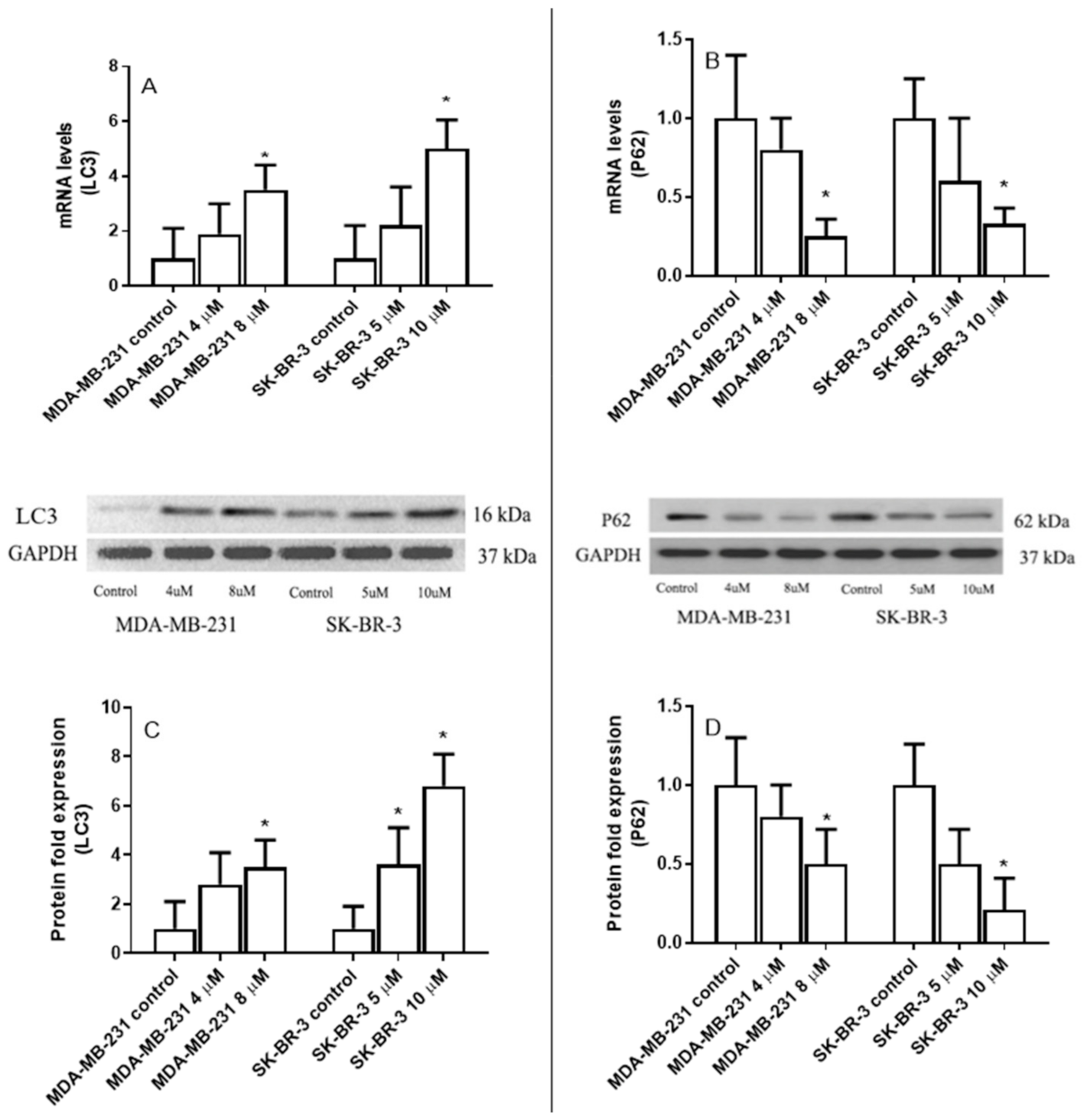
2.3. The Effect of Eugenol on Microtubule-Associated Protein 1 Light Chain 3 (LC3) and Nucleoporin 62(p62) Gene Expression Levels
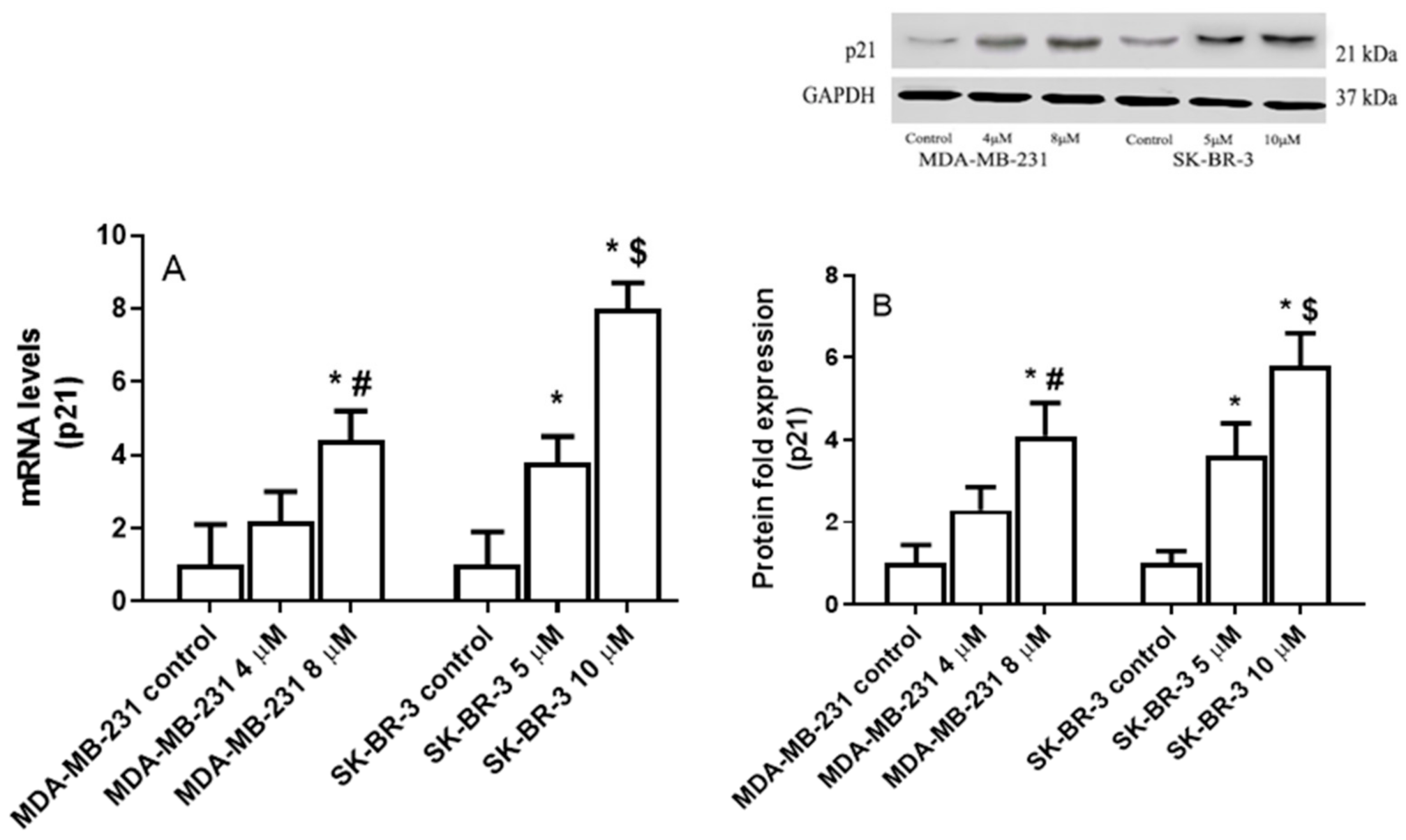
2.4. The Effect of Eugenol on Cyclin Dependent Kinase Inhibitor 1A (P21) Gene Expression Levels
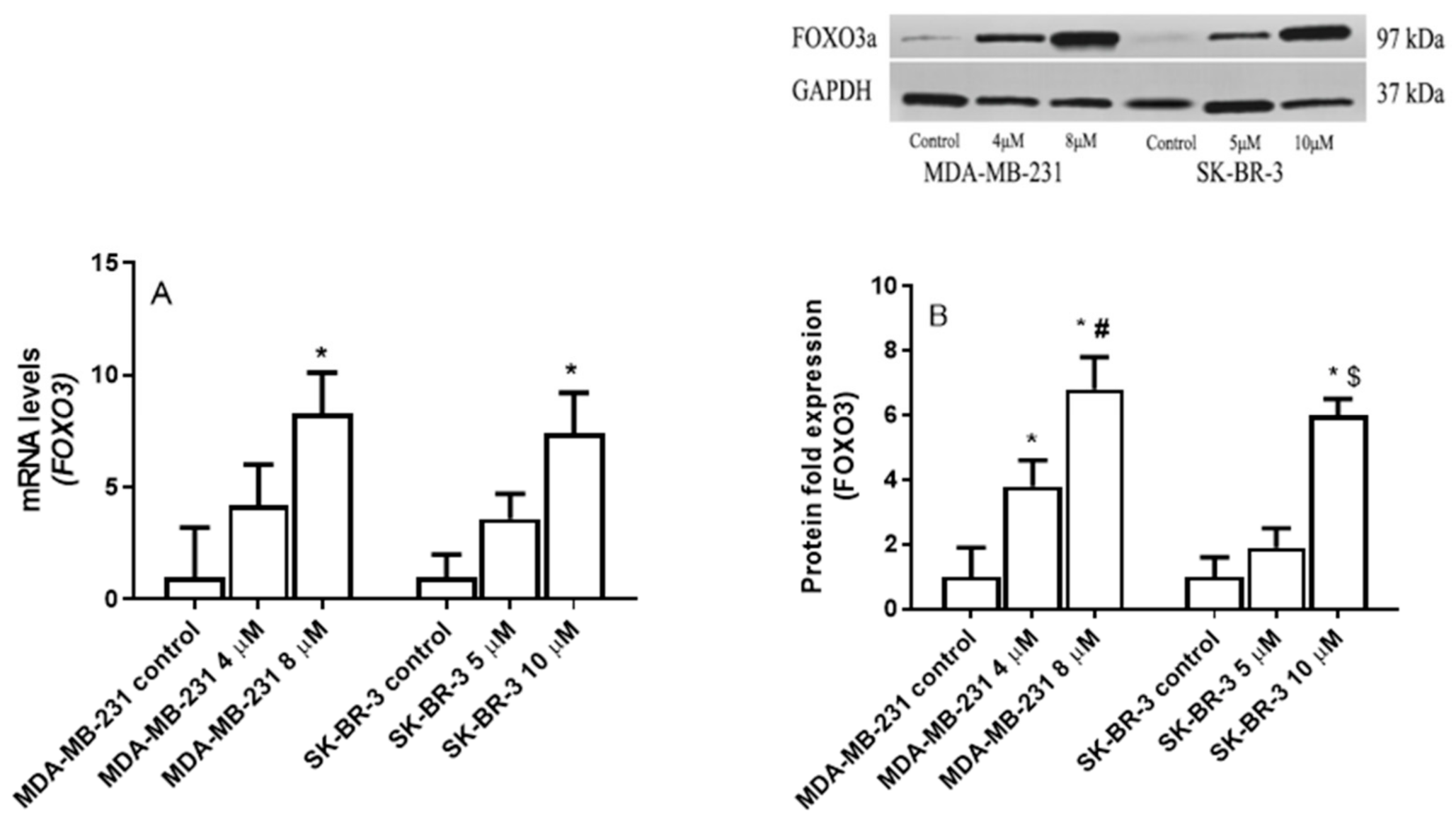
2.5. The Effect of Eugenol on Forkhead box O3 (FOXO3a) Transcription Factor Gene Expression Levels
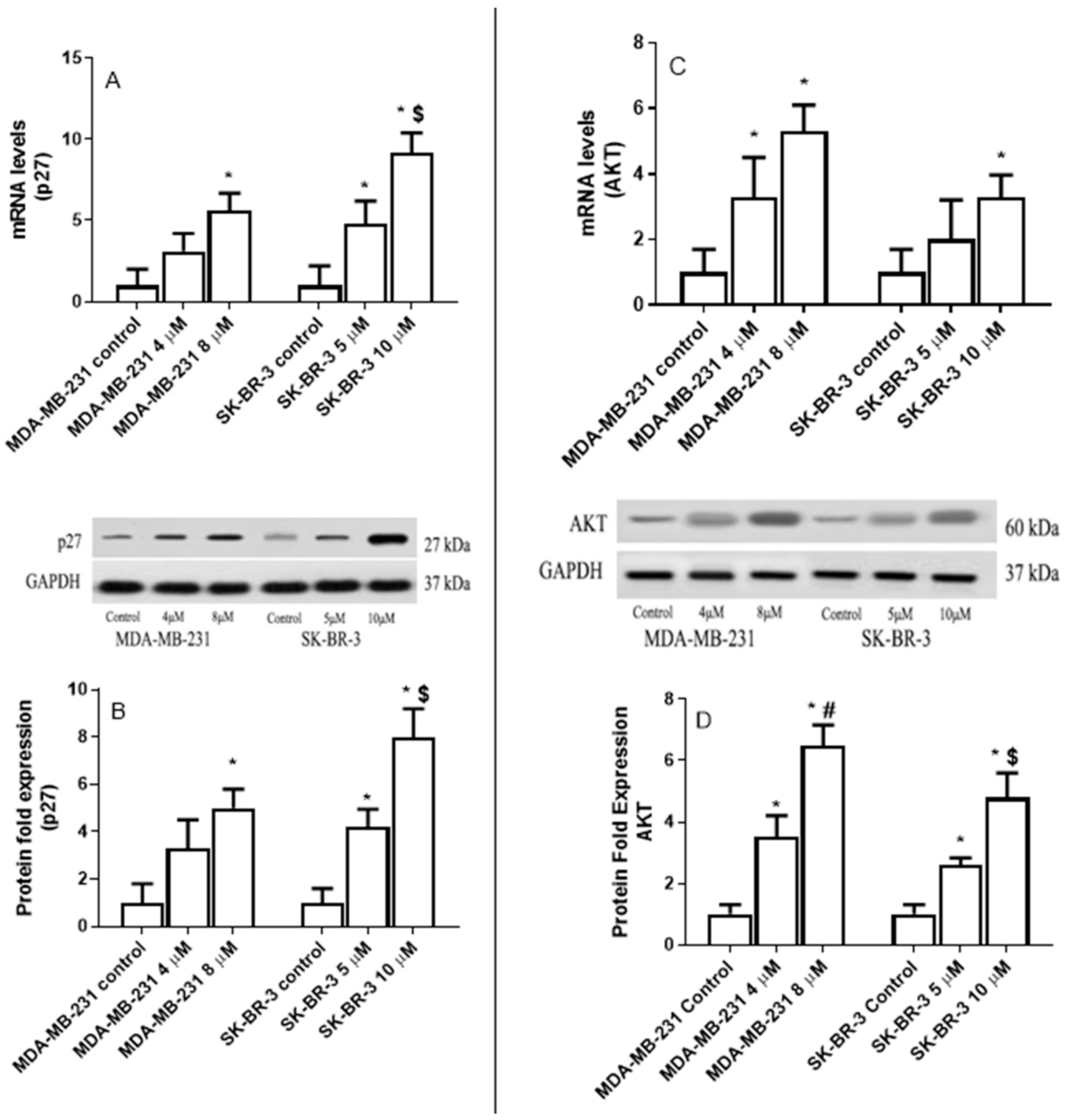
2.6. The Effect of Eugenol on Cyclin-Dependent Kinase Inhibitor (p27), and AKT Serine/Threonine Kinase 1 (AKT) Expression Levels in MDA-MB-231 and SK-BR-3 Cell Lines
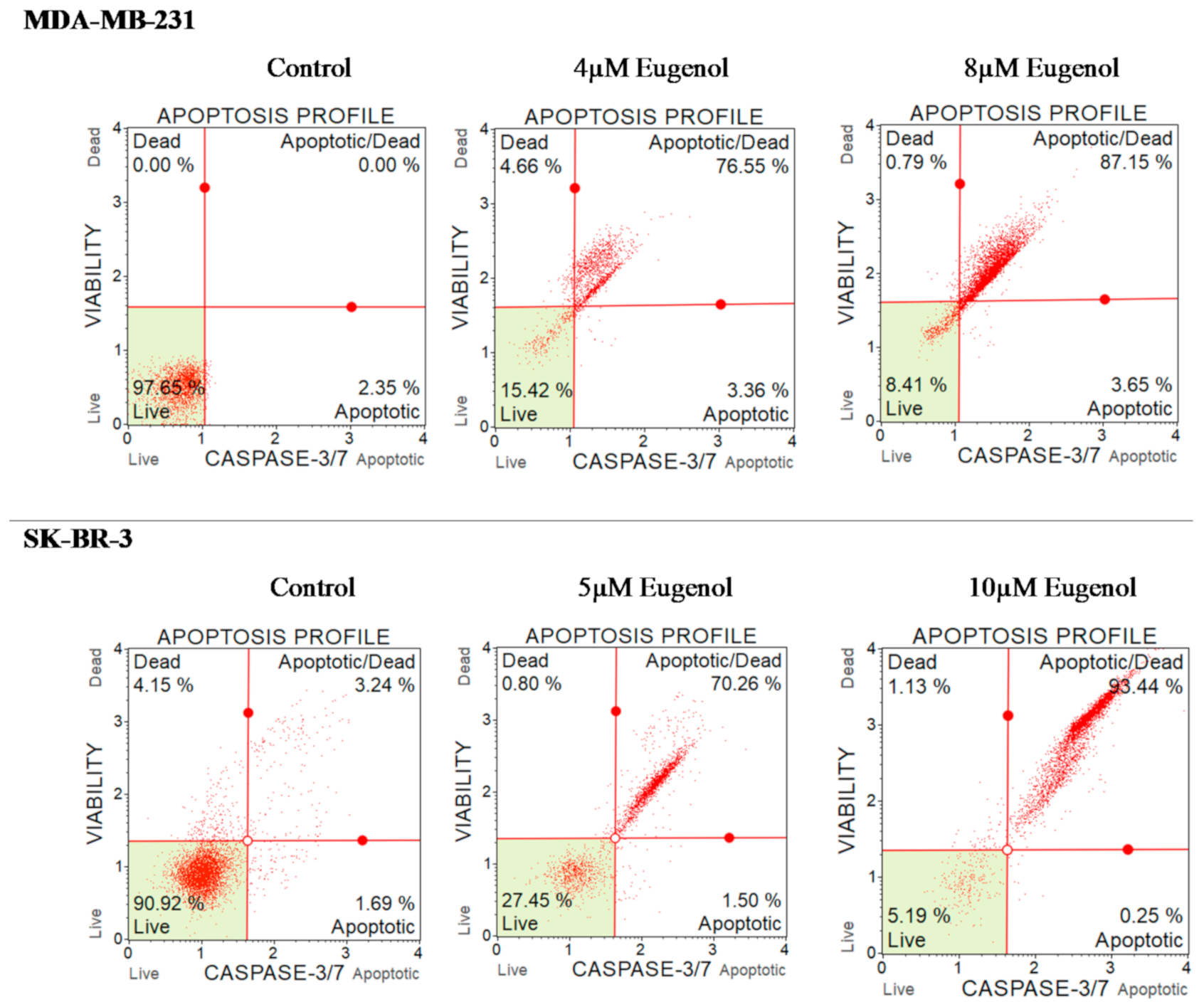
2.7. The Effect of Eugenol on Caspase-3/7 and Cell Death in MDA-MB-231 and SK-BR-3 Cell Lines
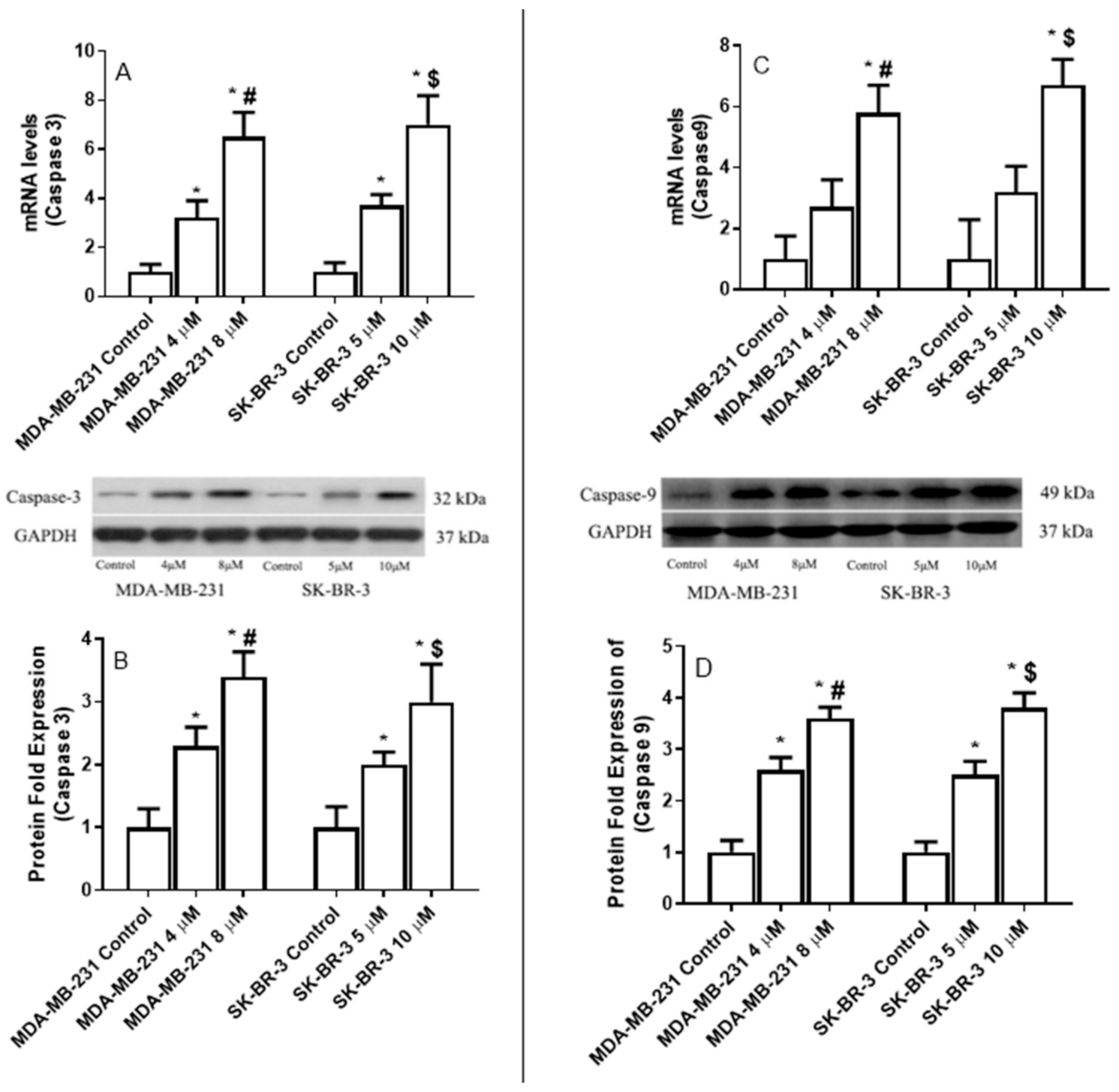
2.8. The Effect of Eugenol on the Apoptotic Gene Expression Levels
3. Discussion
4. Materials and Methods
4.1. Cell Viability Assay Using MTT
4.2. Cell Doubling Time Analysis Using a Real-Time Cell Electronic Sensing System
4.3. Autophagy Rate Determination by Acridine Orange (AO)
4.4. RNA Extraction and Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
4.5. Western Blot Analysis
4.6. Flow Cytometric Analysis of Apoptosis and Caspase-3/7 Activity
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Becker, S. A historic and scientific review of breast cancer: The next global healthcare challenge. Int. J. Gynecol. Obstet. 2015, 131, S36–S39. [Google Scholar] [CrossRef]
- Maayah, Z.H.; Ghebeh, H.; Alhaider, A.A.; El-Kadi, A.O.; Soshilov, A.A.; Denison, M.S.; Ansari, M.A.; Korashy, H.M. Metformin inhibits 7,12-dimethylbenz[a]anthracene-induced breast carcinogenesis and adduct formation in human breast cells by inhibiting the cytochrome P4501A1/aryl hydrocarbon receptor signaling pathway. Toxicol. Appl. Pharmacol. 2015, 284, 217–226. [Google Scholar] [CrossRef]
- Rychlý, B.; Sidlová, H.; Daniś, D. The 2007 World Health Organisation classification of tumours of the central nervous system, comparison with 2000 classification. Ceskoslovenska Patol. 2008, 44, 35–36. [Google Scholar]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Korlimarla, A.; Prabhu, J.S.; Remacle, J.; Rajarajan, S.; Raja, U.; Anupama, C.E.; Srinath, B.S.; Manjunath, S.; Gopinath, K.S.; Correa, M.; et al. Identification of BRCA1 Deficiency Using Multi-Analyte Estimation of BRCA1 and Its Repressors in FFPE Tumor Samples from Patients with Triple Negative Breast Cancer. PLoS ONE 2016, 11, e0153113. [Google Scholar] [CrossRef][Green Version]
- Simon, N.; Antignani, A.; Sarnovsky, R.; Hewitt, S.; Fitzgerald, D. Targeting a Cancer-Specific Epitope of the Epidermal Growth Factor Receptor in Triple-Negative Breast Cancer. J. Natl. Cancer Inst. 2016, 108, djw028. [Google Scholar] [CrossRef]
- Rastelli, F.; Biancanelli, S.; Falzetta, A.; Martignetti, A.; Casi, C.; Bascioni, R.; Giustini, L.; Crispino, S. Triple-Negative Breast Cancer: Current State of the Art. Tumori J. 2010, 96, 875–888. [Google Scholar] [CrossRef]
- Cleator, S.; Heller, W.; Coombes, R.C. Triple-negative breast cancer therapeutic options. Lancet Oncol. 2007, 8, 235–244. [Google Scholar] [CrossRef]
- Zustiak, M.P.; Pollack, J.K.; Marten, M.R.; Betenbaugh, M.J. Feast or famine: Autophagy control and engineering in eukaryotic cell culture. Curr. Opin. Biotechnol. 2008, 19, 518–526. [Google Scholar] [CrossRef]
- Gabos, Z.; Sinha, R.; Hanson, J.; Chauhan, N.; Hugh, J.; Mackey, J.R.; Abdulkarim, B. Prognostic Significance of Human Epidermal Growth Factor Receptor Positivity for the Development of Brain Metastasis After Newly Diagnosed Breast Cancer. J. Clin. Oncol. 2006, 24, 5658–5663. [Google Scholar] [CrossRef]
- Tsutsui, S.; Ohno, S.; Murakami, S.; Kataoka, A.; Kinoshita, J.; Hachitanda, Y. Prognostic significance of the coexpression of p53 protein and c-erbB2 in breast cancer. Am. J. Surg. 2003, 185, 165–167. [Google Scholar] [CrossRef]
- Ross, J.S.; Fletcher, J.A.; Linette, G.P.; Stec, J.; Clark, E.; Ayers, M.; Symmans, W.F.; Pusztai, L.; Bloom, K.J. The HER-2/ neu Gene and Protein in Breast Cancer 2003: Biomarker and Target of Therapy. Oncologist 2003, 8, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ju, D. The Role of Autophagy in Nanoparticles-Induced Toxicity and Its Related Cellular and Molecular Mechanisms. Adv. Exp. Med. Biol. 2018, 1048, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Farrow, J.M.; Yang, J.C.; Evans, C.P. Autophagy as a modulator and target in prostate cancer. Nat. Rev. Urol. 2014, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.M.; Stefanis, L.; Fredenburg, R.; Lansbury, P.T.; Sulzer, D. Impaired Degradation of Mutant -Synuclein by Chaperone-Mediated Autophagy. Science 2004, 305, 1292–1295. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; McNutt, M.A.; Zhu, W.-G. Autophagy process is associated with anti-neoplastic function. Acta Biochim. Biophys. Sin. 2011, 43, 425–432. [Google Scholar] [CrossRef]
- Mortimore, G.E.; Pösö, A.R.; Lardeux, B.R. Mechanism and regulation of protein degradation in liver. Diabetes Metab. Rev. 1989, 5, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.U.F.; Deng, Y. Role of autophagy and its significance in cellular homeostasis. Appl. Microbiol. Biotechnol. 2014, 98, 5319–5328. [Google Scholar] [CrossRef] [PubMed]
- Lamy, L.; Ngo, V.; Emre, T.; Shaffer, A.L.; Yang, Y.; Tian, E.; Nair, V.; Kruhlak, M.J.; Zingone, A.; Landgren, O.; et al. Control of Autophagic Cell Death by Caspase-10 in Multiple Myeloma. Cancer Cell 2013, 23, 435–449. [Google Scholar] [CrossRef]
- Arduino, D.M.; Esteves, A.R.; Cardoso, S.M. Mitochondria drive autophagy pathology via microtubule disassembly: A new hypothesis for Parkinson disease. Autophagy 2013, 9, 112–114. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.-I.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Colacurcio, D.J.; Pensalfini, A.; Jiang, Y.; Nixon, R.A. Dysfunction of autophagy and endosomal-lysosomal pathways: Roles in pathogenesis of Down syn-drome and Alzheimer’s Disease. Free Radic. Biol. Med. 2018, 114, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, T.; Taneike, I.; Akaishi, R.; Yoshizawa, F.; Furuya, N.; Fujimura, S.; Kadowaki, M. Amino Acids and Insulin Control Autophagic Proteolysis through Different Signaling Pathways in Relation to mTOR in Isolated Rat Hepatocytes. J. Biol. Chem. 2004, 279, 8452–8459. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhuo, N.; Li, Y.; Zhao, W.; Jiang, D. Autophagy promotes osteogenic differentiation of human bone marrow mesenchymal stem cell derived from osteoporotic vertebrae. Biochem. Biophys. Res. Commun. 2017, 488, 46–52. [Google Scholar] [CrossRef]
- Fiers, W.; Beyaert, R.; Declercq, W.; Vandenabeele, P. More than one way to die: Apoptosis, necrosis and reactive oxygen damage. Oncogene 1999, 18, 7719–7730. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial Membrane Permeabilization in Cell Death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef]
- Mathieu, V.; Berge, E.V.D.; Ceusters, J.; Konopka, T.; Cops, A.; Bruyère, C.; Pirker, C.; Berger, W.; Trieu-Van, T.; Serteyn, D.; et al. New 5-Aryl-1H-imidazoles Display in Vitro Antitumor Activity against Apoptosis-Resistant Cancer Models, Including Melanomas, through Mitochondrial Targeting. J. Med. Chem. 2013, 56, 6626–6637. [Google Scholar] [CrossRef]
- Wu, S.; Chu, Y.; Yang, Y.; Li, Y.; He, P.; Zheng, Y.; Zhang, C.; Liu, Q.; Han, L.; Huang, R. Inhibition of macrophage autophagy induced by Salmonella enterica serovar typhi plasmid. Front. Biosci. 2014, 19, 490–503. [Google Scholar] [CrossRef]
- Eisenberglerner, A.; Bialik, S.; Simon, H.-U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966–975. [Google Scholar] [CrossRef]
- An, J.; Zhou, Q.; Wu, M.; Wang, L.; Zhong, Y.; Feng, J.; Shang, Y.; Chen, Y. Interactions between oxidative stress, autophagy and apoptosis in A549 cells treated with aged black carbon. Toxicol. Vitr. 2019, 54, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Gao, M.; Xu, X.; Zhang, H.; Xu, J.; Guan, Q.; Wang, Q.; Du, J.; Li, Z.; Zuo, D.; et al. Overcoming resistance to mitochondrial apoptosis by BZML-induced mitotic catastrophe is enhanced by inhibition of autophagy in A549/Taxol cells. Cell Prolif. 2018, 51, e12450. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Huang, T.-Y.; Chen, H.-Y.; Huang, T.-C.; Lin, L.-C.; Chang, Y.-J.; Hsia, S.-M. Protective Effect of Melatonin against Oxidative Stress-Induced Apoptosis and Enhanced Autophagy in Human Retinal Pigment Epithelium Cells. Oxidative Med. Cell. Longev. 2018, 2018, 9015765. [Google Scholar] [CrossRef] [PubMed]
- Al-Sharif, I.; Remmal, A.; Aboussekhra, A. Eugenol triggers apoptosis in breast cancer cells through E2F1/survivin down-regulation. BMC Cancer 2013, 13, 600. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.L.; Hafez, M.M.; Al-Hoshani, A.; Al-Shabanah, O. Anti-metastatic and anti-proliferative activity of eugenol against triple negative and HER2 positive breast cancer cells. BMC Complement. Altern. Med. 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic Control of Autophagy. Cell 2014, 159, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Lippai, M.; Lőw, P. The Role of the Selective Adaptor p62 and Ubiquitin-Like Proteins in Autophagy. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Sanz, L.; Diaz-Meco, M.T.; Nakano, H.; Moscat, J. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1–TRAF6 pathway. EMBO J. 2000, 19, 1576–1586. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Pitti, R.; Lawrence, D.; Pham, V.C.; Lill, J.R.; Ashkenazi, A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis sig-naling. Cell 2009, 137, 721–735. [Google Scholar] [CrossRef]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.-S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.-I.; Ezaki, J.; Murata, S.; et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on hunt-ing-tin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Tasdemir, E.; Maiuri, M.C.; Morselli, E.; Criollo, A.; D’Amelio, M.; Djavaheri-Mergny, M.; Cecconi, F.; Tavernarakis, N.; Kroemer, G. A dual role of p53 in the control of autophagy. Autophagy 2008, 4, 810–814. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Ye, Z.; Li, J.-C. Oridonin Up-regulates Expression of P21 and Induces Autophagy and Apoptosis in Human Prostate Cancer Cells. Int. J. Biol. Sci. 2012, 8, 901–912. [Google Scholar] [CrossRef]
- Panzarini, E.; Inguscio, V.; Tenuzzo, B.A.; Carata, E.; Dini, L. Nanomaterials and Autophagy: New Insights in Cancer Treatment. Cancers 2013, 5, 296–319. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Daido, S.; Yamamoto, A.; Kobayashi, R.; Yokoyama, T.; Aoki, H.; Iwado, E.; Shinojima, N.; Kondo, Y.; Kondo, S. Pivotal Role of the Cyclin-dependent Kinase Inhibitor p21WAF1/CIP1 in Apoptosis and Autophagy. J. Biol. Chem. 2008, 283, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S.; Belai, A. Natural Therapies of the Inflammatory Bowel Disease: The Case of Rutin and its Aglycone, Quercetin. Mini-Rev. Med. Chem. 2018, 18, 234–243. [Google Scholar] [CrossRef]
- Wan, X.; Zheng, X.; Pang, X.; Zhang, Z.; Zhang, Q. Incorporation of lapatinib into human serum albumin nanoparticles with enhanced anti-tumor effects in HER2-positive breast cancer. Colloids Surf. B Biointerfaces 2015, 136, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.M.; Roskams, A.J. Caspase-3 and caspase-9 mediate developmental apoptosis in the mouse olfactory system. J. Comp. Neurol. 2004, 474, 136–148. [Google Scholar] [CrossRef]
- Zhou, J.; Liao, W.; Yang, J.; Ma, K.; Li, X.; Wang, Y.; Wang, D.; Wang, L.; Zhang, Y.; Yin, Y.; et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy 2012, 8, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Edinger, A.L.; Thompson, C.B. Defective autophagy leads to cancer. Cancer Cell 2003, 4, 422–424. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.-L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G.J.C. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Mammucari, C.; Schiaffino, S.; Sandri, M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy 2008, 4, 524–526. [Google Scholar] [CrossRef]
- Mammucari, C.; Milan, G.; Romanello, V.; Masiero, E.; Rudolf, R.; Del Piccolo, P.; Burden, S.J.; Di Lisi, R.; Sandri, C.; Zhao, J.; et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007, 6, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Brault, J.J.; Schild, A.; Cao, P.; Sandri, M.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007, 6, 472–483. [Google Scholar] [CrossRef]
- Hafez, M.M.; Hamed, S.S.; El-Khadragy, M.F.; Hassan, Z.K.; Al Rejaie, S.S.; Sayed-Ahmed, M.M.; Al-Harbi, N.O.; Al-Hosaini, K.A.; Al-Harbi, M.M.; Alhoshani, A.R.; et al. Effect of ginseng extract on the TGF-β1 signaling pathway in CCl4-induced liver fibrosis in rats. BMC Complement. Altern. Med. 2017, 13, 1507–1510. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, M.L.; Al-Shabanah, O.; Hassan, Z.K.; Hafez, M.M. Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition. Int. J. Mol. Sci. 2021, 22, 9243. https://doi.org/10.3390/ijms22179243
Abdullah ML, Al-Shabanah O, Hassan ZK, Hafez MM. Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition. International Journal of Molecular Sciences. 2021; 22(17):9243. https://doi.org/10.3390/ijms22179243
Chicago/Turabian StyleAbdullah, Mashan L., Othman Al-Shabanah, Zeinab K. Hassan, and Mohamed M. Hafez. 2021. "Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition" International Journal of Molecular Sciences 22, no. 17: 9243. https://doi.org/10.3390/ijms22179243
APA StyleAbdullah, M. L., Al-Shabanah, O., Hassan, Z. K., & Hafez, M. M. (2021). Eugenol-Induced Autophagy and Apoptosis in Breast Cancer Cells via PI3K/AKT/FOXO3a Pathway Inhibition. International Journal of Molecular Sciences, 22(17), 9243. https://doi.org/10.3390/ijms22179243





