Measuring and Interpreting Nuclear Transport in Neurodegenerative Disease—The Example of C9orf72 ALS
Abstract
1. Introduction
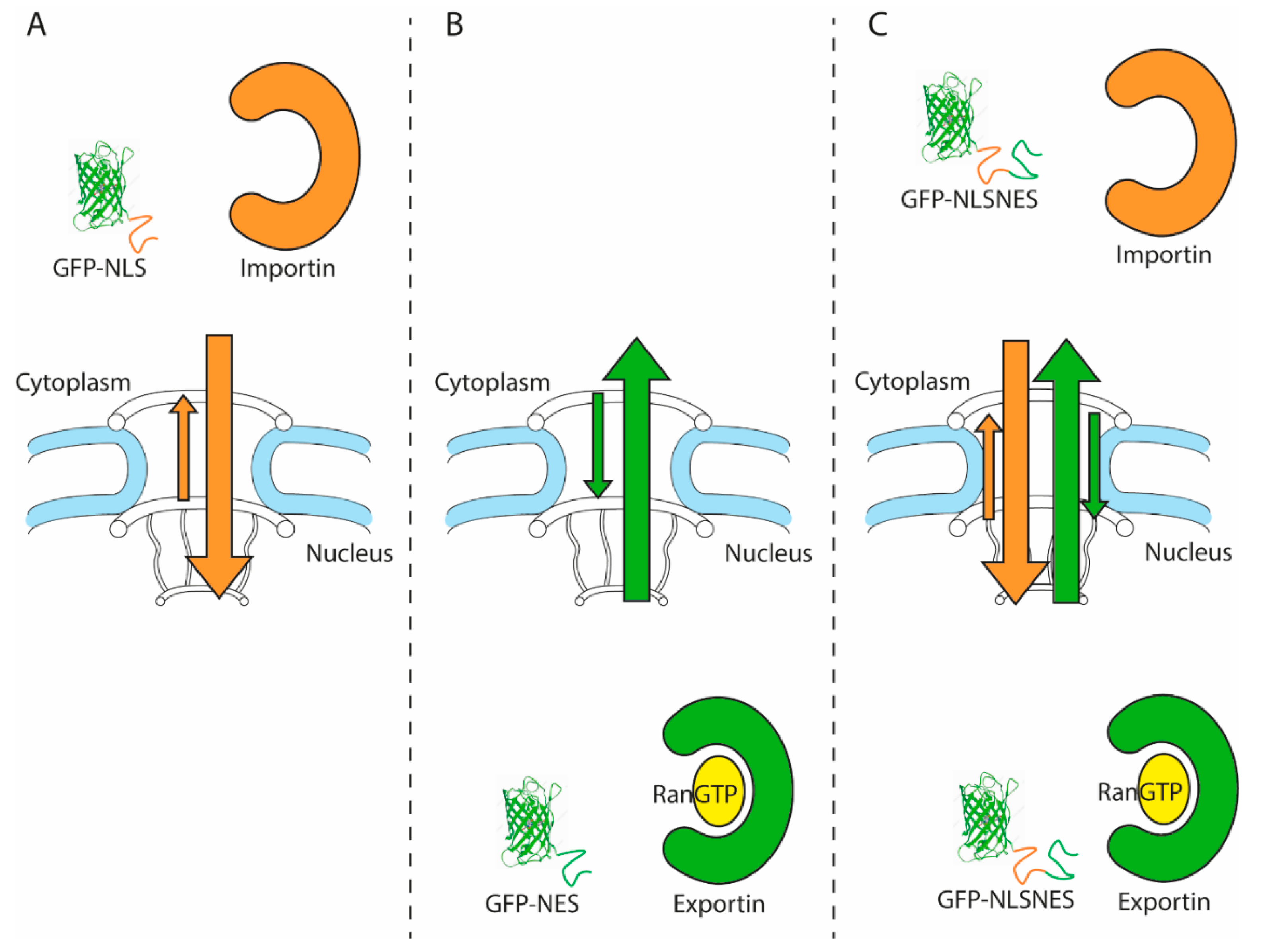
1.1. Nucleocytoplasmic Transport
1.2. Amyotrophic Lateral Sclerosis (ALS)
1.3. Nucleocytoplasmic Transport and C9orf72 ALS
1.4. Commonly Used Nuclear Transport Assays
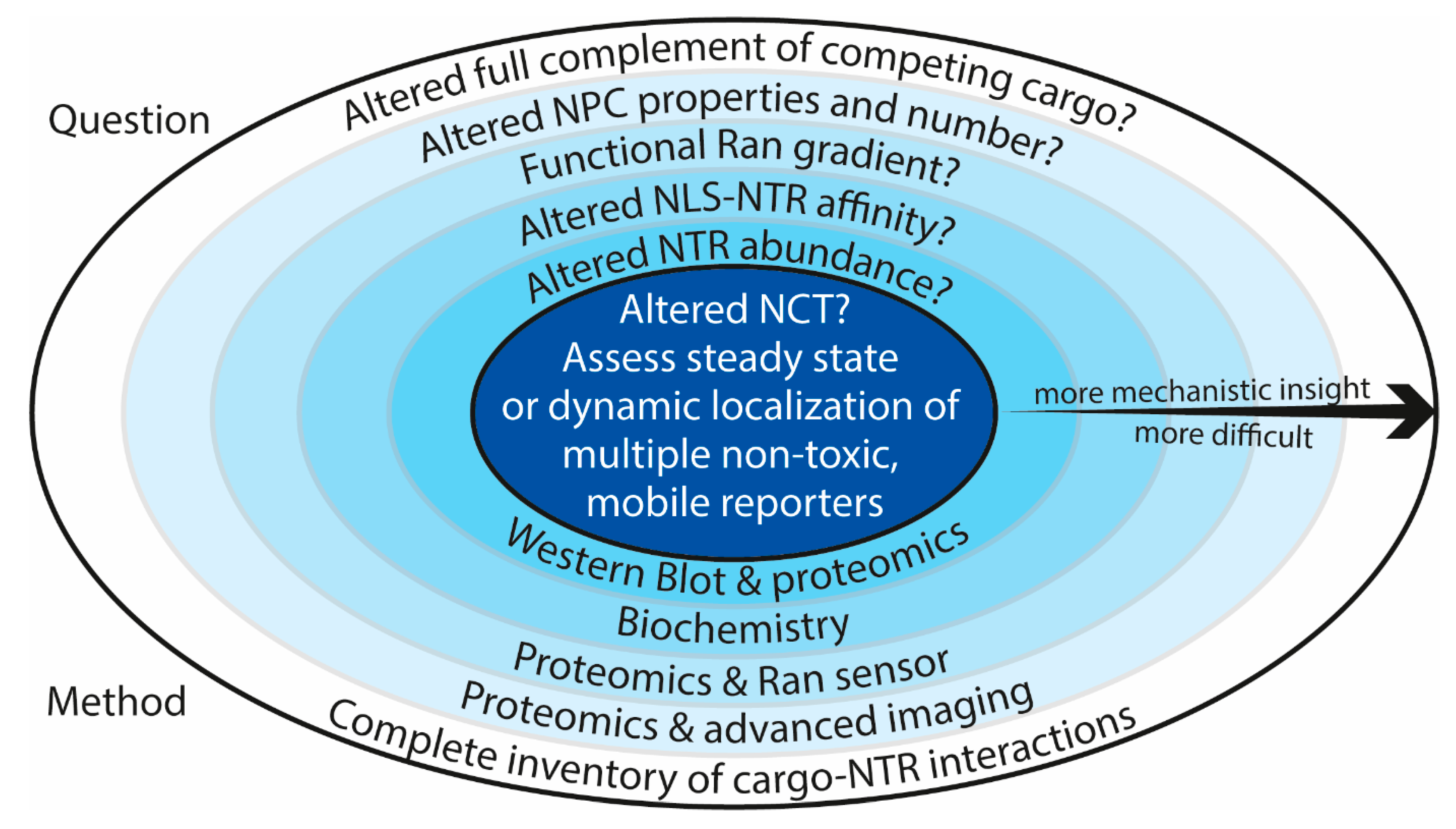
2. Nuclear Transport Up-Close: Complications when Interpreting Nuclear Transport Data
| I. Transport of Cargo Depends on a Localization Sequence that is Recognized by an NTR | |
| NTR | NLS/NES |
| Impα/β | Sv40/cNLS: PKKKRKV [90] cMyc: PAAKRVKLD [100,101] Nucleoplasmin: KRPAATKKAGQAKKKK [102] |
| TNPO1 | PY-NLS: R-X2-5-PY motif [103] e.g., FUS: RG-rich46RQDRRERPY [104] |
| Kap104 | RG-rich motif [91] |
| Kap121 | Lycine-rich sequence [92] |
| Attempts to discover more NLSs [93] | |
| CRM1 | Φ-X2-3-Φ-x2-3-Φ-x-Φ; where Φ is I, M, F, V, and mostly L [105,106] PKI: e.g., MSLNELALKLAGLDI [58] HIV-1 Rev: LQLPPLERLTL |
| II. Redundancy in NTRs for One Type of NLS | |
| NTR1 NLSi-Cargo1 NTR2 NLSi-Cargo1 | Different NTRs can recognize the same NLS, and thus transport the same cargo; e.g., Trn1, Trn2A, and Trn2B all bind the PY-NLS, and transport FUS [107]/HuR [108], as well as unique cargoes. |
| NTR1a NLSi-Cargo1 NTR1b NLSi-Cargo2 | Different isoforms of one NTR may bind the same NLS, but transport other cargoes; e.g., importin-α isoforms bind the classical NLS, but, e.g., STAT1 is bound by α5, but not by α1 [109]. Importin-α3 only recognizes the NLS with appropriate N-terminal flanking residues [110]. |
| III. Redundancy in NTRs for One Type of Cargo | |
| NTR1 NLSi -Cargo1 NTR2 NLSii-Cargo1 | A group of cargoes, with distinct NLSs, can be recognized by different NTRs; e.g., Kap121 can substitute Kap123 for import of ribosomal proteins [111,112] |
| IV. Combinations of Localization Sequences | |
| NTR1 NLSi-NLSii-Cargo1 NTR2 NLSi-NLSii-Cargo1 | One cargo may have two different NLSs; e.g., Nxf1 contains a cNLS for importin 4/11/α/β and a PY-NLS for Trn1/Trn2 [113,114,115,116,117]. |
| NTR1 NLS-NES-Cargo1 NTR2 NLS-NES-Cargo1 | One cargo may contain an NLS and an NES; cyclin B [118,119] and HIV Rev [105,120] contain an impβ1 NLS and a CRM1 NES. |
| NTR1 NES-adaptor Cargo1 | Ribosomal pre60S particles bind to an adaptor protein (NMD3), which contains the NES [121,122]. |
3. Is Nuclear Transport Altered in C9-ALS?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ALS | amyotrophic lateral sclerosis |
| C9-ALS | C9orf72-related ALS |
| DPRs | dipeptide repeat proteins |
| FG | phenylalanine glycine repeat |
| FLIP | fluorescence loss in photobleaching |
| FRAP | fluorescence recovery after photobleaching |
| FUS | fused in sarcoma |
| GA, GP, GR, PA, PR | glycine-alanine, glycine-proline, glycine-arginine, proline-alanine, proline-arginine repeats |
| LMB | leptomycin B |
| NES | nuclear export sequence |
| NLS | nuclear localization sequence |
| NP/NMP1 | nucleophosmin |
| NPC | nuclear pore complex |
| NTR | nuclear transport receptor |
| Nups | nucleoporins |
| PKI | cAMP-dependent protein kinase inhibitor |
| PTM | post-translational modification |
| RAN translation | repeat-associated non-AUG translation |
| Ran | Ras-related nuclear protein |
| RanGAP | Ran GTPase activating protein |
| RanGEF | Ran guanine nucleotide exchange factorRBP RNA binding protein |
| ROS | reactive oxygen species |
| Sv40 | simian virus 40 |
| Rev | human immunodeficiency virus type 1 Rev protein |
| TDP-43 | TAR-DNA binding protein 43 |
| TNPO1 | Kapβ2 |
| TRN1 | transportin 1 |
References
- Popken, P.; Ghavami, A.; Onck, P.R.; Poolman, B.; Veenhoff, L.M. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell 2015, 26, 1386–1394. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Rout, M.P.; Aitchison, J.D.; Suprapto, A.; Hjertaas, K.; Zhao, Y.; Chait, B.T. The Yeast Nuclear Pore Complex. J. Cell Biol. 2000, 148, 635–652. [Google Scholar] [CrossRef]
- Cronshaw, J.M.; Krutchinsky, A.N.; Zhang, W.; Chait, B.T.; Matunis, M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002, 158, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Terry, L.J.; Wente, S.R. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 2007, 178, 1121–1132. [Google Scholar] [CrossRef]
- Strawn, L.A.; Shen, T.; Shulga, N.; Goldfarb, D.S.; Wente, S.R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 2004, 6, 197–206. [Google Scholar] [CrossRef]
- Marg, A.; Shan, Y.; Meyer, T.; Meissner, T.; Brandenburg, M.; Vinkemeier, U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 2004, 165, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Allen, N.P.; Huang, L.; Burlingame, A.; Rexach, M. Proteomic Analysis of Nucleoporin Interacting Proteins. J. Biol. Chem. 2001, 276, 29268–29274. [Google Scholar] [CrossRef]
- Dilworth, D.J.; Suprapto, A.; Padovan, J.C.; Chait, B.T.; Wozniak, R.W.; Rout, M.; Aitchison, J.D. Nup2p Dynamically Associates with the Distal Regions of the Yeast Nuclear Pore Complex. J. Cell Biol. 2001, 153, 1465–1478. [Google Scholar] [CrossRef] [PubMed]
- Marelli, M.; Aitchison, J.D.; Wozniak, R.W. Specific Binding of the Karyopherin Kap121p to a Subunit of the Nuclear Pore Complex Containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998, 143, 1813–1830. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.R.; Tang, J.H.; Yassif, J.; Graf, M.; Huang, W.Y.C.; Groves, J.T.; Weis, K.; Liphardt, J.T. Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. eLife 2015, 4, e04052. [Google Scholar] [CrossRef]
- Galy, V.; Gadal, O.; Fromont-Racine, M.; Romano, A.; Jacquier, A.; Nehrbass, U. Nuclear Retention of Unspliced mRNAs in Yeast Is Mediated by Perinuclear Mlp1. Cell 2004, 116, 63–73. [Google Scholar] [CrossRef]
- Wozniak, R.W.; Rout, M.P.; Aitchison, J.D. Karyopherins and kissing cousins. Trends Cell Biol. 1998, 8, 184–188. [Google Scholar] [CrossRef]
- Titov, A.A.; Blobel, G. The Karyopherin Kap122p/Pdr6p Imports Both Subunits of the Transcription Factor Iia into the Nucleus. J. Cell Biol. 1999, 147, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Allen, N.P.; Patel, S.S.; Huang, L.; Chalkley, R.; Burlingame, A.; Lutzmann, M.; Hurt, E.C.; Rexach, M. Deciphering Networks of Protein Interactions at the Nuclear Pore Complex. Mol. Cell. Proteom. 2002, 1, 930–946. [Google Scholar] [CrossRef]
- Kose, S.; Furuta, M.; Imamoto, N. Hikeshi, a Nuclear Import Carrier for Hsp70s, Protects Cells from Heat Shock-Induced Nuclear Damage. Cell 2012, 149, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Kosyna, F.K.; Depping, R. Controlling the Gatekeeper: Therapeutic Targeting of Nuclear Transport. Cells 2018, 7, 221. [Google Scholar] [CrossRef]
- Baade, I.; Kehlenbach, R.H. The cargo spectrum of nuclear transport receptors. Curr. Opin. Cell Biol. 2019, 58, 1–7. [Google Scholar] [CrossRef]
- Pumroy, R.A.; Cingolani, G. Diversification of Importin-α Isoforms in Cellular Trafficking and Disease States. Biochem. J. 2015, 466, 13–28. [Google Scholar] [CrossRef]
- Yao, W.; Lutzmann, M.; Hurt, E. A versatile interaction platform on the Mex67–Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J. 2007, 27, 6–16. [Google Scholar] [CrossRef]
- Kutay, U.; Bischoff, F.; Kostka, S.; Kraft, R.; Görlich, D. Export of Importin α from the Nucleus Is Mediated by a Specific Nuclear Transport Factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef]
- Bischoff, F.; Görlich, D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997, 419, 249–254. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; De Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2014, 282, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Lolodi, O.; Yamazaki, H.; Otsuka, S.; Kumeta, M.; Yoshimura, S.H. Dissecting in vivo steady-state dynamics of karyopherin-dependent nuclear transport. Mol. Biol. Cell 2016, 27, 167–176. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H., Jr.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef]
- Kiernan, M.C.; Vucic, S.; Cheah, B.C.; Turner, M.; Eisen, A.; Hardiman, O.; Burrell, J.; Zoing, M.C. Amyotrophic lateral sclerosis. Lancet 2011, 377, 942–955. [Google Scholar] [CrossRef]
- Renton, A.E.; Chio, A.; Traynor, B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014, 17, 17–23. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Sánchez, J.S.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Green, K.M.; Linsalata, A.E.; Todd, P.K. RAN translation—What makes it run? Brain Res. 2016, 1647, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Ash, P.E.; Bieniek, K.; Gendron, T.F.; Caulfield, T.; Lin, W.-L.; DeJesus-Hernandez, M.; van Blitterswijk, M.; Jansen-West, K.; Paul, J.W.; Rademakers, R.; et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 2013, 77, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Weng, S.-M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzschmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.P.; Chu, C. Nuclear transport, oxidative stress, and neurodegeneration. Int. J. Clin. Exp. Pathol. 2011, 4, 215–229. [Google Scholar] [PubMed]
- Loureiro, J.; Oliveira, C.; Silveira, I. Unstable repeat expansions in neurodegenerative diseases: Nucleocytoplasmic transport emerges on the scene. Neurobiol. Aging 2016, 39, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Taylor, J.P. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 2017, 96, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lagier-Tourenne, C. Nuclear pores: The gate to neurodegeneration. Nat. Neurosci. 2018, 21, 156–158. [Google Scholar] [CrossRef]
- Ferreira, P.A. The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration. Cell. Mol. Life Sci. 2019, 76, 2247–2273. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, E.E. Nucleocytoplasmic transport. Neurology 2019, 92, 757–764. [Google Scholar] [CrossRef]
- Hutten, S.; Dormann, D. Nucleocytoplasmic Transport Defects in Neurodegeneration—Cause or Consequence? Semin. Cell Dev. Biol. 2019, 99, 151–162. [Google Scholar] [CrossRef]
- Bitetto, G.; Di Fonzo, A. Nucleo–cytoplasmic transport defects and protein aggregates in neurodegeneration. Transl. Neurodegener. 2020, 9, 1–16. [Google Scholar] [CrossRef]
- Fallini, C.; Khalil, B.; Smith, C.L.; Rossoll, W. Traffic jam at the nuclear pore: All roads lead to nucleocytoplasmic transport defects in ALS/FTD. Neurobiol. Dis. 2020, 140, 104835. [Google Scholar] [CrossRef] [PubMed]
- Diez, L.; Wegmann, S. Nuclear Transport Deficits in Tau-Related Neurodegenerative Diseases. Front. Neurol. 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, N.; Sochocka, M.; Brzecka, A.; Shimizu, T.; Gąsiorowski, K.; Szczechowiak, K.; Leszek, J. Nuclear Envelope and Nuclear Pore Complexes in Neurodegenerative Diseases—New Perspectives for Therapeutic Interventions. Mol. Neurobiol. 2021, 58, 983–995. [Google Scholar] [CrossRef]
- Xiao, S.; Macnair, L.; McGoldrick, P.; McKeever, P.M.; McLean, J.R.; Zhang, M.; Keith, J.; Zinman, L.; Rogaeva, E.; Robertson, J. Isoform-specific antibodies reveal distinct subcellular localizations of C 9orf72 in amyotrophic lateral sclerosis. Ann. Neurol. 2015, 78, 568–583. [Google Scholar] [CrossRef]
- Zhang, K.; Donnelly, C.J.; Haeusler, A.R.; Grima, J.C.; Machamer, J.B.; Steinwald, P.; Daley, E.; Miller, S.J.; Cunningham, K.; Vidensky, S.; et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nat. Cell Biol. 2015, 525, 56–61. [Google Scholar] [CrossRef]
- Freibaum, B.D.; Lu, Y.; Lopez-Gonzalez, R.; Kim, N.C.; Almeida, S.; Lee, K.-H.; Badders, N.; Valentine, M.; Miller, B.L.; Wong, P.C.; et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nat. Cell Biol. 2015, 525, 129–133. [Google Scholar] [CrossRef]
- Jovičić, A.; Mertens, J.; Boeynaems, S.; Bogaert, E.; Chai, N.; Yamada, S.B.; Paul, J.W.; Sun, S.; Herdy, J.R.; Bieri, G.; et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015, 18, 1226–1229. [Google Scholar] [CrossRef]
- Vanneste, J.; Van Den Bosch, L. The Role of Nucleocytoplasmic Transport Defects in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2021, in press. [Google Scholar]
- Zhang, Y.-J.; Gendron, T.F.; Grima, J.C.; Sasaguri, H.; Jansen-West, K.; Xu, Y.-F.; Katzman, R.B.; Gass, J.; E Murray, M.; Shinohara, M.; et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 2016, 19, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Hayes, L.R.; Duan, L.; Bowen, K.; Kalab, P.; Rothstein, J.D. C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. eLife 2020, 9, e51685. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Ito, H.; Hirano, A.; Fujita, K.; Wate, R.; Nakamura, M.; Kaneko, S.; Nakano, S.; Kusaka, H. Nuclear Contour Irregularity and Abnormal Transporter Protein Distribution in Anterior Horn Cells in Amyotrophic Lateral Sclerosis. J. Neuropathol. Exp. Neurol. 2009, 68, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.Y.; Mori, E.; Nizami, Z.F.; Lin, Y.; Kato, M.; Xiang, S.; Wu, L.C.; Ding, M.; Yu, Y.; Gall, J.G.; et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. USA. 2017, 114, E1111–E1117. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Bogaert, E.; Michiels, E.; Gijselinck, I.; Sieben, A.; Jovičić, A.; De Baets, G.; Scheveneels, W.; Steyaert, J.; Cuijt, I.; et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 2016, 6, 20877. [Google Scholar] [CrossRef] [PubMed]
- Saberi, S.; Stauffer, J.E.; Jiang, J.; Garcia, S.D.; Taylor, A.E.; Schulte, D.; Ohkubo, T.; Schloffman, C.L.; Maldonado, M.; Baughn, M.; et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 2018, 135, 459–474. [Google Scholar] [CrossRef]
- Chou, C.-C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.; Chen, Y.H.; Duong, D.; et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef]
- Solomon, D.A.; Stepto, A.; Au, W.H.; Adachi, Y.; Diaper, D.C.; Hall, R.; Rekhi, A.; Boudi, A.; Tziortzouda, P.; Lee, Y.; et al. A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain 2018, 141, 2908–2924. [Google Scholar] [CrossRef]
- Khosravi, B.; Hartmann, H.; May, S.; Möhl, C.; Ederle, H.; Michaelsen, M.; Schludi, M.H.; Dormann, D.; Edbauer, D. Cytoplasmic poly-GA aggregates impair nuclear import of TDP-43 in C9orf72 ALS/FTLD. Hum. Mol. Genet. 2016, 26, 790–800. [Google Scholar] [CrossRef]
- Vanneste, J.; Vercruysse, T.; Boeynaems, S.; Sicart, A.; Van Damme, P.; Daelemans, D.; Bosch, L.V.D. C9orf72-generated poly-GR and poly-PR do not directly interfere with nucleocytoplasmic transport. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Hutten, S.; Usluer, S.; Bourgeois, B.; Simonetti, F.; Odeh, H.M.; Fare, C.M.; Czuppa, M.; Hruska-Plochan, M.; Hofweber, M.; Polymenidou, M.; et al. Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions. Cell Rep. 2020, 33, 108538. [Google Scholar] [CrossRef]
- Shulga, N.; Roberts, P.; Gu, Z.; Spit, L.; Tab, M.M.; Nomura, M.; Goldfarb, D.S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: A role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996, 135, 329–339. [Google Scholar] [CrossRef]
- Woerner, A.C.; Frottin, F.; Hornburg, D.; Feng, L.R.; Meissner, F.; Patra, M.; Tatzelt, J.; Mann, M.; Winklhofer, K.F.; Hartl, F.U.; et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016, 351, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.; Rees, R.; Schünemann, J.; Ng, S.C.; Fünfgeld, K.; Huyton, T.; Görlich, D. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell 2018, 174, 202–217e9. [Google Scholar] [CrossRef]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nat. Cell Biol. 1997, 390, 308–311. [Google Scholar] [CrossRef]
- Nishi, K.; Yoshida, M.; Fujiwara, D.; Nishikawa, M.; Horinouchi, S.; Beppu, T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 1994, 269, 6320–6324. [Google Scholar] [CrossRef]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef] [PubMed]
- Hietanen, S.; Lain, S.; Krausz, E.; Blattner, C.; Lane, D. Activation of p53 in cervical carcinoma cells by small molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 8501–8506. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Takizawa, N.; Katoh, M.; Hoshida, K.; Kobayashi, N.; Nagata, K. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 2001, 77, 31–42. [Google Scholar] [CrossRef]
- Latonen, L.; Moore, H.M.; Bai, B.; Jäämaa, S.; Laiho, M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 2010, 30, 790–805. [Google Scholar] [CrossRef]
- Soderholm, J.F.; Bird, S.L.; Kalab, P.; Sampathkumar, Y.; Hasegawa, K.; Uehara-Bingen, M.; Weis, K.; Heald, R. Importazole, a Small Molecule Inhibitor of the Transport Receptor Importin-β. ACS Chem. Biol. 2011, 6, 700–708. [Google Scholar] [CrossRef]
- Wagstaff, K.; Sivakumaran, H.; Heaton, S.; Harrich, D.; Jans, D. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef]
- E Cansizoglu, A.; Lee, B.J.; Zhang, Z.C.; A Fontoura, B.M.; Chook, Y.M. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 2007, 14, 452–454. [Google Scholar] [CrossRef]
- Izaurralde, E.; Kutay, U.; von Kobbe, C.; Mattaj, I.; Görlich, D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997, 16, 6535–6547. [Google Scholar] [CrossRef] [PubMed]
- Haruki, H.; Nishikawa, J.; Laemmli, U.K. The Anchor-Away Technique: Rapid, Conditional Establishment of Yeast Mutant Phenotypes. Mol. Cell 2008, 31, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F.; Bizzarri, R.; Serresi, M.; Albertazzi, L.; Beltram, F. Probing Nuclear Localization Signal-Importin α Binding Equilibria in Living Cells. J. Biol. Chem. 2009, 284, 36638–36646. [Google Scholar] [CrossRef] [PubMed]
- Meinema, A.C.; Poolman, B.; Veenhoff, L.M. Quantitative Analysis of Membrane Protein Transport Across the Nuclear Pore Complex. Traffic 2013, 14, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Marr, R.S.; Gerace, L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990, 111, 807–816. [Google Scholar] [CrossRef]
- Schnell, U.; Dijk, F.; Sjollema, K.A.; Giepmans, B. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 2012, 9, 152–158. [Google Scholar] [CrossRef]
- Lohka, M.J.; Masui, Y. Roles of cytosol and cytoplasmic particles in nuclear envelope assembly and sperm pronuclear formation in cell-free preparations from amphibian eggs. J. Cell Biol. 1984, 98, 1222–1230. [Google Scholar] [CrossRef]
- Newmeyer, D.D.; Finlay, D.R.; Forbes, D.J. In vitro transport of a fluorescent nuclear protein and exclusion of non-nuclear proteins. J. Cell Biol. 1986, 103, 2091–2102. [Google Scholar] [CrossRef]
- Newmeyer, D.; Lucocq, J.; Bürglin, T.; De Robertis, E. Assembly in vitro of nuclei active in nuclear protein transport: ATP is required for nucleoplasmin accumulation. EMBO J. 1986, 5, 501–510. [Google Scholar] [CrossRef]
- Newmeyer, D.D.; Forbes, D.J. Nuclear import can be separated into distinct steps in vitro: Nuclear pore binding and translocation. Cell 1988, 52, 641–653. [Google Scholar] [CrossRef]
- Finlay, D.R.; Newmeyer, D.D.; Price, T.M.; Forbes, D.J. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 1987, 104, 189–200. [Google Scholar] [CrossRef]
- Cassany, A.; Gerace, L. Reconstitution of Nuclear Import in Permeabilized Cells. In The Nucleus; Springer International Publishing: Basel, Switzerland, 2008; Volume 464, pp. 181–205. [Google Scholar]
- Timney, B.L.; Novatt, J.; Agate, D.S.; Williams, R.; Zhang, W.; Chait, B.T.; Rout, M.P. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006, 175, 579–593. [Google Scholar] [CrossRef]
- Hodel, A.E.; Harreman, M.T.; Pulliam, K.F.; Harben, M.E.; Holmes, J.S.; Hodel, M.R.; Berland, K.M.; Corbett, A.H. Nuclear Localization Signal Receptor Affinity Correlates with in Vivo Localization in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 23545–23556. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.A.; Anderson, D.J.; Richard, E.; Hetzer, M.W. Nuclear Pores Form de Novo from Both Sides of the Nuclear Envelope. Science 2006, 312, 440–443. [Google Scholar] [CrossRef]
- Görlich, D. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 1994, 79, 767–778. [Google Scholar] [CrossRef]
- Radtke, T.; Schmalz, D.; Coutavas, E.; Soliman, T.M.; Peters, R. Kinetics of protein import into isolated Xenopus oocyte nuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 2407–2412. [Google Scholar] [CrossRef]
- Kopito, R.B.; Elbaum, M. Nucleocytoplasmic transport: A thermodynamic mechanism. HFSP J. 2009, 3, 130–141. [Google Scholar] [CrossRef]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Lee, D.C.Y.; Aitchison, J.D. Kap104p-mediated Nuclear Import. J. Biol. Chem. 1999, 274, 29031–29037. [Google Scholar] [CrossRef]
- Kobayashi, J.; Matsuura, Y. Structural Basis for Cell-Cycle-Dependent Nuclear Import Mediated by the Karyopherin Kap121p. J. Mol. Biol. 2013, 425, 1852–1868. [Google Scholar] [CrossRef]
- Mackmull, M.; Klaus, B.; Heinze, I.; Chokkalingam, M.; Beyer, A.; Russell, R.B.; Ori, A.; Beck, M. Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol. 2017, 13, 962. [Google Scholar] [CrossRef]
- Yang, W.; Musser, S.M. Nuclear import time and transport efficiency depend on importin β concentration. J. Cell Biol. 2006, 174, 951–961. [Google Scholar] [CrossRef]
- Kim, S.; Elbaum, M. A Simple Kinetic Model with Explicit Predictions for Nuclear Transport. Biophys. J. 2013, 105, 565–569. [Google Scholar] [CrossRef][Green Version]
- Ribbeck, K.; Görlich, D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001, 20, 1320–1330. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Saiwaki, T.; Yamashita, J.; Yasuda, Y.; Kotera, I.; Shibata, S.; Shigeta, M.; Hiraoka, Y.; Haraguchi, T.; Yoneda, Y. Cellular stresses induce the nuclear accumulation of importin α and cause a conventional nuclear import block. J. Cell Biol. 2004, 165, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Kodiha, M.; Bański, P.; Ho-Wo-Cheong, D.; Stochaj, U. Dissection of the molecular mechanisms that control the nuclear accumulation of transport factors importin-α and CAS in stressed cells. Cell. Mol. Life Sci. 2008, 65, 1756–1767. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.; Suzuki, T.; Yasuda, Y.; Murata, M.; Katahira, J.; Yoneda, Y. Identification of importin α1 as a novel constituent of RNA stress granules. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1803, 865–871. [Google Scholar] [CrossRef]
- Makkerh, J.P.; Dingwall, C.; Laskey, R.A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr. Biol. 1996, 6, 1025–1027. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.; Dilwortht, S.M.; Laskey, R.A.; Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64, 615–623. [Google Scholar] [CrossRef]
- Lee, B.J.; Cansizoglu, A.E.; Süel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for Nuclear Localization Sequence Recognition by Karyopherinβ2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Gal, J.; Zhang, J.; Kwinter, D.M.; Zhai, J.; Jia, H.; Jia, J.; Zhu, H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging 2011, 32, 2323.e27–2323.e40. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.; Lührmann, R. The HIV-1 Rev Activation Domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef]
- Wen, W.; Meinkotht, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- Dormann, D.; Rodde, R.; Edbauer, D.; Bentmann, E.; Fischer, I.; Hruscha, A.; Than, M.E.; Mackenzie, I.R.; Capell, A.; Schmid, B.; et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010, 29, 2841–2857. [Google Scholar] [CrossRef]
- Rebane, A.; Aab, A.; Steitz, J.A. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 2004, 10, 590–599. [Google Scholar] [CrossRef]
- Sekimoto, T.; Imamoto, N.; Nakajima, K.; Hirano, T.; Yoneda, Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997, 16, 7067–7077. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Imamoto, N.; Sekimoto, T.; Tachibana, T.; Seki, T.; Tada, S.; Enomoto, T.; Yoneda, Y. Differential Modes of Nuclear Localization Signal (NLS) Recognition by Three Distinct Classes of NLS Receptors. J. Biol. Chem. 1997, 272, 26375–26381. [Google Scholar] [CrossRef]
- Rout, M.; Blobel, G.; Aitchison, J.D. A Distinct Nuclear Import Pathway Used by Ribosomal Proteins. Cell 1997, 89, 715–725. [Google Scholar] [CrossRef]
- Sydorskyy, Y.; Dilworth, D.J.; Yi, E.C.; Goodlett, D.R.; Wozniak, R.W.; Aitchison, J.D. Intersection of the Kap123p-Mediated Nuclear Import and Ribosome Export Pathways. Mol. Cell. Biol. 2003, 23, 2042–2054. [Google Scholar] [CrossRef]
- Truant, R.; Kang, Y.; Cullen, B.R. The Human Tap Nuclear RNA Export Factor Contains a Novel Transportin-dependent Nuclear Localization Signal That Lacks Nuclear Export Signal Function. J. Biol. Chem. 1999, 274, 32167–32171. [Google Scholar] [CrossRef]
- Guttinger, S.; Muhlhausser, P.; Koller-Eichhorn, R.; Brennecke, J.; Kutay, U. From The Cover: Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. USA 2004, 101, 2918–2923. [Google Scholar] [CrossRef] [PubMed]
- Imasaki, T.; Shimizu, T.; Hashimoto, H.; Hidaka, Y.; Kose, S.; Imamoto, N.; Yamada, M.; Sato, M. Structural Basis for Substrate Recognition and Dissociation by Human Transportin 1. Mol. Cell 2007, 28, 57–67. [Google Scholar] [CrossRef]
- Zhang, Z.C.; Satterly, N.; Fontoura, B.M.A.; Chook, Y.M. Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol. Biol. Cell 2011, 22, 4657–4668. [Google Scholar] [CrossRef]
- Kimura, M.; Kose, S.; Okumura, N.; Imai, K.; Furuta, M.; Sakiyama, N.; Tomii, K.; Horton, P.; Takao, T.; Imamoto, N. Identification of Cargo Proteins Specific for the Nucleocytoplasmic Transport Carrier Transportin by Combination of an in Vitro Transport System and Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-based Quantitative Proteomics. Mol. Cell. Proteom. 2013, 12, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.D.; Yang, J.; Truant, R.; Kornbluth, S. Nuclear Import of Cdk/Cyclin Complexes: Identification of Distinct Mechanisms for Import of Cdk2/Cyclin E and Cdc2/Cyclin B1. J. Cell Biol. 1999, 144, 213–224. [Google Scholar] [CrossRef]
- Yang, J.; Bardes, E.S.; Moore, J.D.; Brennan, J.; Powers, M.A.; Kornbluth, S. Control of Cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998, 12, 2131–2143. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B.R. The Arginine-Rich Domains Present in Human Immunodeficiency Virus Type 1 Tat and Rev Function as Direct Importin β-Dependent Nuclear Localization Signals. Mol. Cell. Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.H.-N.; Kallstrom, G.; Johnson, A.W. Nmd3p Is a Crm1p-Dependent Adapter Protein for Nuclear Export of the Large Ribosomal Subunit. J. Cell Biol. 2000, 151, 1057–1066. [Google Scholar] [CrossRef]
- Gadal, O.; Strauß, D.; Kessl, J.; Trumpower, B.; Tollervey, D.; Hurt, E. Nuclear Export of 60S Ribosomal Subunits Depends on Xpo1p and Requires a Nuclear Export Sequence-Containing Factor, Nmd3p, That Associates with the Large Subunit Protein Rpl10p. Mol. Cell. Biol. 2001, 21, 3405–3415. [Google Scholar] [CrossRef]
- Xiao, C.-Y.; Jans, P.; Jans, D.A. Negative charge at the protein kinase CK2 site enhances recognition of the SV40 large T-antigen NLS by importin: Effect of conformation. FEBS Lett. 1998, 440, 297–301. [Google Scholar] [CrossRef]
- Hu, W.; Jans, D.A. Efficiency of Importin α/β-Mediated Nuclear Localization Sequence Recognition and Nuclear Import. J. Biol. Chem. 1999, 274, 15820–15827. [Google Scholar] [CrossRef]
- Engelsma, D.; Bernad, R.; Calafat, J.; Fornerod, M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004, 23, 3643–3652. [Google Scholar] [CrossRef]
- Feldherr, C.; Akin, D. Stimulation of nuclear import by simian virus 40-transformed cell extracts is dependent on protein kinase activity. Mol. Cell. Biol. 1995, 15, 7043–7049. [Google Scholar] [CrossRef] [PubMed]
- Kehlenbach, R.; Gerace, L. Phosphorylation of the Nuclear Transport Machinery Down-regulates Nuclear Protein Import in Vitro. J. Biol. Chem. 2000, 275, 17848–17856. [Google Scholar] [CrossRef] [PubMed]
- Hübner, S.; Xiao, C.-Y.; Jans, D.A. The Protein Kinase CK2 Site (Ser111/112) Enhances Recognition of the Simian Virus 40 Large T-antigen Nuclear Localization Sequence by Importin. J. Biol. Chem. 1997, 272, 17191–17195. [Google Scholar] [CrossRef]
- Nardozzi, J.D.; Lott, K.; Cingolani, G. Phosphorylation meets nuclear import: A review. Cell Commun. Signal. 2010, 8, 1–17. [Google Scholar] [CrossRef]
- A Jans, D. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 1995, 311, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.; Jans, D. Regulation of Nuclear Transport: Central Role in Development and Transformation? Traffic 2005, 6, 173–186. [Google Scholar] [CrossRef]
- And, D.A.J.; Hubner, S. Regulation of Protein Transport to the Nucleus: Central Role of Phosphorylation. Physiol. Rev. 1996, 76, 651–685. [Google Scholar]
- Smith, W.A.; Schurter, B.T.; Wong-Staal, F.; David, M. Arginine Methylation of RNA Helicase A Determines Its Subcellular Localization. J. Biol. Chem. 2004, 279, 22795–22798. [Google Scholar] [CrossRef]
- Dormann, D.; Madl, T.; Valori, C.F.; Bentmann, E.; Tahirovic, S.; Abou-Ajram, C.; Kremmer, E.; Ansorge, O.; A Mackenzie, I.R.; Neumann, M.C.; et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012, 31, 4258–4275. [Google Scholar] [CrossRef]
- Lohrum, M.A.E.; Woods, D.B.; Ludwig, R.L.; Bálint, E.; Vousden, K.H. C-Terminal Ubiquitination of p53 Contributes to Nuclear Export. Mol. Cell. Biol. 2001, 21, 8521–8532. [Google Scholar] [CrossRef]
- Plafker, S.M.; Plafker, K.S.; Weissman, A.M.; Macara, I.G. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 2004, 167, 649–659. [Google Scholar] [CrossRef]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination Regulates PTEN Nuclear Import and Tumor Suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Madison, D.L.; Yaciuk, P.; Kwok, R.P.S.; Lundblad, J.R. Acetylation of the Adenovirus-transforming Protein E1A Determines Nuclear Localization by Disrupting Association with Importin-α. J. Biol. Chem. 2002, 277, 38755–38763. [Google Scholar] [CrossRef] [PubMed]
- Santiago, A.; Li, D.; Zhao, L.Y.; Godsey, A.; Liao, D. p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1. Mol. Biol. Cell 2013, 24, 2739–2752. [Google Scholar] [CrossRef]
- Putker, M.; Madl, T.; Vos, H.R.; de Ruiter, H.; Visscher, M.; van den Berg, M.C.W.; Kaplan, M.; Korswagen, H.C.; Boelens, R.; Vermeulen, M.; et al. Redox-Dependent Control of FOXO/DAF-16 by Transportin-1. Mol. Cell 2013, 49, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Twyffels, L.; Gueydan, C.; Kruys, V. Transportin-1 and Transportin-2: Protein nuclear import and beyond. FEBS Lett. 2014, 588, 1857–1868. [Google Scholar] [CrossRef]
- Blank, V.; Kourilsky, P.; Israël, A. Cytoplasmic retention, DNA binding and processing of the NF-kappa B p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991, 10, 4159–4167. [Google Scholar] [CrossRef] [PubMed]
- Traenckner, E.; Wilk, S.; Baeuerle, P. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J. 1994, 13, 5433–5441. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.; Zhang, Z.; Davies, K.; Kalpana, G.V. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: Implications for tumorigenesis. EMBO J. 2002, 21, 31–42. [Google Scholar] [CrossRef]
- Zhu, J.; McKeon, F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nat. Cell Biol. 1999, 398, 256–260. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-KappaB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Jeyasekharan, A.; Liu, Y.; Hattori, H.; Pisupati, V.; Jonsdottir, A.B.; Rajendra, E.; Lee, M.; Sundaramoorthy, E.; Schlachter, S.; Kaminski, C.; et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013, 20, 1191–1198. [Google Scholar] [CrossRef]
- Fineberg, K.; Fineberg, T.; Graessmann, A.; Luedtke, N.W.; Tor, Y.; Lixin, R.; Jans, D.A.; Loyter, A. Inhibition of Nuclear Import Mediated by the Rev-Arginine Rich Motif by RNA Molecules†. Biochemistry 2003, 42, 2625–2633. [Google Scholar] [CrossRef]
- Stommel, J.M.; Marchenko, N.D.; Jimenez, G.S.; Moll, U.M.; Hope, T.J.; Wahl, G.M. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999, 18, 1660–1672. [Google Scholar] [CrossRef]
- Kuge, S.; Arita, M.; Murayama, A.; Maeta, K.; Izawa, S.; Inoue, Y.; Nomoto, A. Regulation of the Yeast Yap1p Nuclear Export Signal Is Mediated by Redox Signal-Induced Reversible Disulfide Bond Formation. Mol. Cell. Biol. 2001, 21, 6139–6150. [Google Scholar] [CrossRef]
- Nadler, S.G.; Tritschler, D.; Haffar, O.K.; Blake, J.; Bruce, A.G.; Cleaveland, J.S. Differential Expression and Sequence-specific Interaction of Karyopherin α with Nuclear Localization Sequences. J. Biol. Chem. 1997, 272, 4310–4315. [Google Scholar] [CrossRef] [PubMed]
- Köhler, M.; Speck, C.; Christiansen, M.; Bischoff, F.R.; Prehn, S.; Haller, H.; Görlich, D.; Hartmann, E. Evidence for Distinct Substrate Specificities of Importin α Family Members in Nuclear Protein Import. Mol. Cell. Biol. 1999, 19, 7782–7791. [Google Scholar] [CrossRef]
- Coyne, A.N.; Baskerville, V.; Zaepfel, B.L.; Dickson, D.W.; Rigo, F.; Bennett, F.; Lusk, C.P.; Rothstein, J.D. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 2021, 13, eabe1923. [Google Scholar] [CrossRef]
- Rempel, I.L.; Crane, M.M.; Thaller, D.J.; Mishra, A.; Jansen, D.P.; Janssens, G.; Popken, P.; Akşit, A.; Kaeberlein, M.; Van Der Giessen, E.; et al. Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife 2019, 8, e48186. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; van der Velde, H.; Fornerod, M.; Pickersgill, H. Nup358/RanBP2 Attaches to the Nuclear Pore Complex via Association with Nup88 and Nup214/CAN and Plays a Supporting Role in CRM1-Mediated Nuclear Protein Export. Mol. Cell. Biol. 2004, 24, 2373–2384. [Google Scholar] [CrossRef]
- Bernad, R.; Engelsma, D.; Sanderson, H.; Pickersgill, H.; Fornerod, M. Nup214-Nup88 Nucleoporin Subcomplex Is Required for CRM1-mediated 60 S Preribosomal Nuclear Export. J. Biol. Chem. 2006, 281, 19378–19386. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.; Kehlenbach, R. Nup214 Is Required for CRM1-Dependent Nuclear Protein Export In Vivo. Mol. Cell. Biol. 2006, 26, 6772–6785. [Google Scholar] [CrossRef] [PubMed]
- Roloff, S.; Spillner, C.; Kehlenbach, R.H. Several Phenylalanine-Glycine Motives in the Nucleoporin Nup214 Are Essential for Binding of the Nuclear Export Receptor CRM1. J. Biol. Chem. 2013, 288, 3952–3963. [Google Scholar] [CrossRef]
- Waldmann, I.; Spillner, C.; Kehlenbach, R.H. The nucleoporin-like protein NLP1 (hCG1) promotes CRM1-dependent nuclear protein export. J. Cell Sci. 2012, 125, 144–154. [Google Scholar] [CrossRef]
- Takeda, A.; Sarma, N.J.; Abdul-Nabi, A.M.; Yaseen, N.R. Inhibition of CRM1-mediated Nuclear Export of Transcription Factors by Leukemogenic NUP98 Fusion Proteins. J. Biol. Chem. 2010, 285, 16248–16257. [Google Scholar] [CrossRef]
- Oka, M.; Asally, M.; Yasuda, Y.; Ogawa, Y.; Tachibana, T.; Yoneda, Y. The Mobile FG Nucleoporin Nup98 Is a Cofactor for Crm1-dependent Protein Export. Mol. Biol. Cell 2010, 21, 1885–1896. [Google Scholar] [CrossRef]
- Kim, S.; Elbaum, M. Enzymatically Driven Transport: A Kinetic Theory for Nuclear Export. Biophys. J. 2013, 105, 1997–2005. [Google Scholar] [CrossRef][Green Version]
- Zeitler, B.; Weis, K. The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo. J. Cell Biol. 2004, 167, 583–590. [Google Scholar] [CrossRef]
- Pyhtila, B.; Rexach, M. A Gradient of Affinity for the Karyopherin Kap95p along the Yeast Nuclear Pore Complex. J. Biol. Chem. 2003, 278, 42699–42709. [Google Scholar] [CrossRef] [PubMed]
- Shulga, N.; Mosammaparast, N.; Wozniak, R.; Goldfarb, D.S. Yeast Nucleoporins Involved in Passive Nuclear Envelope Permeability. J. Cell Biol. 2000, 149, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Makhnevych, T.; Lusk, C.P.; Anderson, A.M.; Aitchison, J.D.; Wozniak, R.W. Cell Cycle Regulated Transport Controlled by Alterations in the Nuclear Pore Complex. Cell 2003, 115, 813–823. [Google Scholar] [CrossRef]
- Fan, F.; Liu, C.-P.; Korobova, O.; Heyting, C.; Offenberg, H.H.; Trump, G.; Arnheim, N. cDNA Cloning and Characterization ofNpap60:A Novel Rat Nuclear Pore-Associated Protein with an Unusual Subcellular Localization during Male Germ Cell Differentiation. Genomics 1997, 40, 444–453. [Google Scholar] [CrossRef]
- Olsson, M.; Schéele, S.; Ekblom, P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 2004, 292, 359–370. [Google Scholar] [CrossRef]
- Ori, A.; Banterle, N.; Iskar, M.; Andres-Pons, A.; Escher, C.; Bui, H.K.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648. [Google Scholar] [CrossRef]
- Gomez-Cavazos, J.S.; Hetzer, M.W. The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. J. Cell Biol. 2015, 208, 671–681. [Google Scholar] [CrossRef]
- Kane, M.; Rebensburg, S.V.; A Takata, M.; Zang, T.M.; Yamashita, M.; Kvaratskhelia, M.; Bieniasz, P.D. Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. eLife 2018, 7, e35738. [Google Scholar] [CrossRef]
- Lupu, F.; Alves, A.; Anderson, K.; Doye, V.; Lacy, E. Nuclear Pore Composition Regulates Neural Stem/Progenitor Cell Differentiation in the Mouse Embryo. Dev. Cell 2008, 14, 831–842. [Google Scholar] [CrossRef]
- D’Angelo, M.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-Dependent Deterioration of Nuclear Pore Complexes Causes a Loss of Nuclear Integrity in Postmitotic Cells. Cell 2009, 136, 284–295. [Google Scholar] [CrossRef]
- Prieve, M.G.; Guttridge, K.L.; Munguia, J.E.; Waterman, M.L. The Nuclear Localization Signal of Lymphoid Enhancer Factor-1 Is Recognized by Two Differentially Expressed Srp1-Nuclear Localization Sequence Receptor Proteins. J. Biol. Chem. 1996, 271, 7654–7658. [Google Scholar] [CrossRef]
- Tsuji, L.; Takumi, T.; Imamoto, N.; Yoneda, Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997, 416, 30–34. [Google Scholar] [CrossRef]
- Köhler, M.; Ansieau, S.; Prehn, S.; Leutz, A.; Haller, H.; Hartmann, E. Cloning of two novel human importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett. 1997, 417, 104–108. [Google Scholar] [CrossRef]
- Nachury, M.; Ryder, U.W.; Lamond, A.; Weis, K. Cloning and characterization of hSRP1, a tissue-specific nuclear transport factor. Proc. Natl. Acad. Sci. USA 1998, 95, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Dahl, E.; Kristiansen, G.; Gottlob, K.; Klaman, I.; Ebner, E.; Hinzmann, B.; Hermann, K.; Pilarsky, C.; Dürst, M.; Klinkhammer-Schalke, M.; et al. Molecular Profiling of Laser-Microdissected Matched Tumor and Normal Breast Tissue Identifies Karyopherin α2 as a Potential Novel Prognostic Marker in Breast Cancer. Clin. Cancer Res. 2006, 12, 3950–3960. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, U.; Gallo, M.; Polymeropoulos, M.H.; Pastan, I. The human CAS (cellular apoptosis susceptibility) gene mapping on chromosome 20q13 is amplified in BT474 breast cancer cells and part of aberrant chromosomes in breast and colon cancer cell lines. Genome Res. 1996, 6, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Behrens, P.; Brinkmann, U.; Fogt, F.; Wernert, N.; Wellmann, A. Implication of the Proliferation and Apoptosis Associated CSE1L/CAS Gene for Breast Cancer Development. Anticancer. Res. 2001, 21, 2413–2417. [Google Scholar]
- Wellmann, A.; Flemming, P.; Behrens, P.; Wuppermann, K.; Lang, H.; Oldhafer, K.; Pastan, I.; Brinkmann, U. High expression of the proliferation and apoptosis associated CSE1L/CAS gene in hepatitis and liver neoplasms: Correlation with tumor progression. Int. J. Mol. Med. 2001, 7, 489–494. [Google Scholar] [CrossRef]
- Van der Watt, P.J.; Maske, C.P.; Hendricks, D.T.; Parker, M.I.; Denny, L.; Govender, D.; Birrer, M.J.; Leaner, V.D. The Karyopherin proteins, Crm1 and Karyopherin β1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int. J. Cancer 2009, 124, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, J.; Kojima, K.; Hail, N.; Tabe, Y.; Andreeff, M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol. Ther. 2015, 153, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Saulino, D.M.; Younes, P.S.; Bailey, J.M.; Younes, M. CRM1/XPO1 expression in pancreatic adenocarcinoma correlates with survivin expression and the proliferative activity. Oncotarget 2018, 9, 21289–21295. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Lu, C.; Chen, L.; Keohavong, P. Overexpression of CRM1: A Characteristic Feature in a Transformed Phenotype of Lung Carcinogenesis and a Molecular Target for Lung Cancer Adjuvant Therapy. J. Thorac. Oncol. 2015, 10, 815–825. [Google Scholar] [CrossRef]
- Noske, A.; Weichert, W.; Niesporek, S.; Röske, A.; Buckendahl, A.-C.; Koch, I.; Sehouli, J.; Dietel, M.; Denkert, C. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer 2008, 112, 1733–1743. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Yue, L.; Qiu, W.-S.; Wang, L.-W.; Zhou, X.-H.; Sun, Y.-J. Prognostic value of CRM1in pancreas cancer. Clin. Investig. Med. 2009, 32, E315–E321. [Google Scholar] [CrossRef]
- Shen, P.A.; Wang, M.Y.; Zhao, M.Y.; Zou, M.L.; Sun, M.L.; Cheng, M.C. Expression of crm1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery 2009, 65, 153–160. [Google Scholar] [CrossRef]
- Ho, Y.; Yao, Y.; Dong, Y.; Lin, F.; Zhao, H.; Shen, Z.; Chen, P.; Sun, Y.-J.; Tang, L.-N.; Zheng, S.-E. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol. Rep. 1994, 21, 229–235. [Google Scholar] [CrossRef]
- Fang, X.-D.; Chen, T.; Tran, K.; Parker, C.S. Developmental regulation of the heat shock response by nuclear transport factor karyopherin-α3. Development 2001, 128, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Giarrè, M.; Török, I.; Schmitt, R.; Gorjánácz, M.; Kiss, I.; Mechler, B.M. Patterns of Importin-α Expression during Drosophila Spermatogenesis. J. Struct. Biol. 2002, 140, 279–290. [Google Scholar] [CrossRef]
- Koehler, M.; Fiebeler, A.; Hartwig, M.; Thiel, S.; Prehn, S.; Kettritz, R.; Luft, F.; Hartmann, E. Differential Expression of Classical Nuclear Transport Factors During Cellular Proliferation and Differentiation. Cell. Physiol. Biochem. 2002, 12, 335–344. [Google Scholar] [CrossRef]
- Yasuhara, N.; Shibazaki, N.; Tanaka, S.; Nagai, M.; Kamikawa, Y.; Oe, S.; Asally, M.; Kamachi, Y.; Kondoh, H.; Yoneda, Y. Triggering neural differentiation of ES cells by subtype switching of importin-α. Nat. Cell Biol. 2006, 9, 72–79. [Google Scholar] [CrossRef]
- Wen, X.; Tan, W.; Westergard, T.; Krishnamurthy, K.; Markandaiah, S.S.; Shi, Y.; Lin, S.; Shneider, N.; Monaghan, J.; Pandey, U.B.; et al. Antisense Proline-Arginine RAN Dipeptides Linked to C9ORF72-ALS/FTD Form Toxic Nuclear Aggregates that Initiate In Vitro and In Vivo Neuronal Death. Neuron 2014, 84, 1213–1225. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, H.; Xia, Q.; Li, K.; Li, K.; Jiang, X.; Xu, G.; Wang, G.; Ying, Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015, 24, 2426–2441. [Google Scholar] [CrossRef]
- Swaminathan, A.; Bouffard, M.; Liao, M.; Ryan, S.; Callister, J.B.; Pickering-Brown, S.; Armstrong, G.; Drapeau, P. Expression of C9orf72-related dipeptides impairs motor function in a vertebrate model. Hum. Mol. Genet. 2018, 27, 1754–1762. [Google Scholar] [CrossRef]
- Schludi, M.H.; German Consortium for Frontotemporal Lobar Degeneration; May, S.; Grässer, F.A.; Rentzsch, K.; Kremmer, E.; Küpper, C.; Klopstock, T.; Arzberger, T.; Edbauer, D.; et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015, 130, 537–555. [Google Scholar] [CrossRef]
- Zu, T.; Liu, Y.; Bañez-Coronel, M.; Reid, T.; Pletnikova, O.; Lewis, J.; Miller, T.M.; Harms, M.B.; Falchook, A.E.; Subramony, S.H.; et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA 2013, 110, E4968–E4977. [Google Scholar] [CrossRef] [PubMed]
- Rudich, P.; Snoznik, C.; Watkins, S.; Monaghan, J.; Pandey, U.B.; Lamitina, S.T. Nuclear localized C9orf72-associated arginine-containing dipeptides exhibit age-dependent toxicity in C. elegans. Hum. Mol. Genet. 2017, 26, 4916–4928. [Google Scholar] [CrossRef]
- Lee, K.-H.; Zhang, P.; Kim, H.J.; Mitrea, D.M.; Sarkar, M.; Freibaum, B.D.; Cika, J.; Coughlin, M.; Messing, J.; Molliex, A.; et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 2016, 167, 774–788e17. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Xiang, S.; Kato, M.; Wu, L.; Theodoropoulos, P.; Wang, T.; Kim, J.; Yun, J.; Xie, Y.; McKnight, S.L. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 2014, 345, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Callister, J.B.; Ryan, S.; Sim, J.; Rollinson, S.; Pickering-Brown, S.M. Modelling C9orf72 dipeptide repeat proteins of a physiologically relevant size. Hum. Mol. Genet. 2016, 25, 5069–5082. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, D.; Abdallah, A.; Li, Z.; Lu, Y.; Almeida, S.; Gao, F.-B. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 2015, 130, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Kanekura, K.; Yagi, T.; Cammack, A.J.; Mahadevan, J.; Kuroda, M.; Harms, M.B.; Miller, T.M.; Urano, F. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum. Mol. Genet. 2016, 25, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Jansen-West, K.; Xu, Y.-F.; Gendron, T.F.; Bieniek, K.; Lin, W.-L.; Sasaguri, H.; Caulfield, T.; Hubbard, J.; Daughrity, L.; et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014, 128, 505–524. [Google Scholar] [CrossRef] [PubMed]
- May, S.; Hornburg, D.; Schludi, M.H.; Arzberger, T.; Rentzsch, K.; Schwenk, B.M.; Grässer, F.A.; Mori, K.; Kremmer, E.; Banzhaf-Strathmann, J.; et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014, 128, 485–503. [Google Scholar] [CrossRef]
- Gill, A.L.; Wang, M.Z.; Levine, B.; Premasiri, A.; Vieira, F.G. Primary Neurons and Differentiated NSC-34 Cells Are More Susceptible to Arginine-Rich ALS Dipeptide Repeat Protein-Associated Toxicity than Non-Differentiated NSC-34 and CHO Cells. Int. J. Mol. Sci. 2019, 20, 6238. [Google Scholar] [CrossRef]
- Boeynaems, S.; Bogaert, E.; Kovacs, D.; Konijnenberg, A.; Timmerman, E.; Volkov, A.; Guharoy, M.; De Decker, M.; Jaspers, T.; Ryan, V.; et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 2017, 65, 1044–1055e5. [Google Scholar] [CrossRef]
- Seibel, N.M.; Eljouni, J.; Nalaskowski, M.M.; Hampe, W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 2007, 368, 95–99. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Gendron, T.F.; Ebbert, M.T.W.; O’Raw, A.; Yue, M.; Jansen-West, K.; Zhang, X.; Prudencio, M.; Chew, J.; Cook, C.N.; et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 2018, 24, 1136–1142. [Google Scholar] [CrossRef]
- MacKenzie, I.R.A.; Frick, P.; Grässer, F.A.; Gendron, T.F.; Petrucelli, L.; Cashman, N.R.; Edbauer, D.; Kremmer, E.; Prudlo, J.; Troost, D.; et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015, 130, 845–861. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, R.; Lu, Y.; Gendron, T.F.; Karydas, A.; Tran, H.; Yang, D.; Petrucelli, L.; Miller, B.L.; Almeida, S.; Gao, F.-B. Poly(GR) in C9ORF72 -Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron 2016, 92, 383–391. [Google Scholar] [CrossRef]
- Yamakawa, M.; Ito, D.; Honda, T.; Kubo, K.-I.; Noda, M.; Nakajima, K.; Suzuki, N. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum. Mol. Genet. 2015, 24, 1630–1645. [Google Scholar] [CrossRef]
- Gupta, R.; Lan, M.; Mojsilovic-Petrovic, J.; Choi, W.H.; Safren, N.; Barmada, S.; Lee, M.J.; Kalb, R. The Proline/Arginine Dipeptide from Hexanucleotide Repeat Expanded C9ORF72 Inhibits the Proteasome. Eneuro 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.J.; Haney, M.S.; Morgens, D.W.; Jovičić, A.; Couthouis, J.; Li, A.; Ousey, J.; Ma, R.; Bieri, G.; Tsui, C.K.; et al. CRISPR–Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet. 2018, 50, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Mizielinska, S.; Grönke, S.; Niccoli, T.; Ridler, C.E.; Clayton, E.L.; Devoy, A.; Moens, T.; Norona, F.E.; Woollacott, I.O.C.; Pietrzyk, J.; et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 2014, 345, 1192–1194. [Google Scholar] [CrossRef] [PubMed]
| Summary of Experimental Setup and Results of Nuclear Transport Studies in C9-ALS Models | |||||
|---|---|---|---|---|---|
| Reporter (Localization in Wildtype) | NTR | Model | Effect of Indicated C9-ALS DNA Repeats or DPRs on Transport Reporter | Reference | |
| Changed | Unchanged | ||||
| (A) NLSSv40-tdTomato-NESRev (nuclear) | Impα/β complex CRM1 | (G4C2)n expression in C9-ALS iPSNs | Recovery of nuclear fluorescence after FRAP is decreased | Zhang 2015 [45] | |
| (B) NLSSv40-NESPKI-GFP (nuclear) | (G4C2)30 expression in Drosophila salivary gland cells | Nuclear intensity is decreased | |||
| (C) NLSRanGAP-ΔNESPKI-GFP (nuclear) | |||||
| (D) NLSSv40-NESPKI-GFP (nuclear excluded) | (G4C2)30 expression in motor neurons | Nuclear exclusion is increased | |||
| (E) NLSSv40-ΔNESPKI-GFP (cellular diffuse) | |||||
| (F) GFP-NLSSv40-NESPKI (nuclear excluded) | Impα/β complex CRM1 | PR20 peptide injection in U2OS cells | Nuclear intensity after export inhibition by Leptomycin B is decreased (PR20 dose-dependent) | Shi 2017 [52] | |
| (G) BSA-NLS(Sv40)n (nuclear) | Impα/β complex | DPR peptide injection in permeabilized HeLa cells | Nuclear intensity is decreased (PR20 dose-dependent) | Nuclear intensity is unchanged (PG20/PA20) | |
| (H) RFP-NLSTDP (nuclear) | Impα/β complex | DPR expression in HeLa cells | Cytoplasmic intensity (C/T) is increased (more so with GA149-GFP, than with GA149-GFP-NLS) | Cytoplasmic intensity (C/T) is unchanged (GFP-GR149/PR175-GFP) † | Khosravi 2017 [57] |
| (I) RFP-NLSPY (hnRNPA1) (nuclear) | TNPO1 | Cytoplasmic intensity (C/T) is unchanged (GA149/GR149/PR175) | |||
| NESRev-tdTomato-NLSSv40 | Impα/β complex CRM1 | C9-ALS fibroblasts | Nuclear accumulation (N/C) is decreased | Chou 2018 [55] | |
| (J) NLSSv40-mNeon Green2x-NESPKI (cytoplasmic) | Impα/β complex CRM1 | DPR peptide injection in HeLa Kyoto cells | Nuclear accumulation (N/total) after export inhibition by Leptomycin B is unchanged (PR20/GR20) | Vanneste 2019 [58] | |
| (K) NLSPY (FUS)-mNeon Green2x-NESPKI * (nuclear) | TNPO1 CRM1 | Nuclear accumulation (N/total) after import inhibition by MGM9 is unchanged (PR20/GR20) | |||
| (L) NLSSv40-mNeon Green2x-NESPKI (nuclear) | Impα/β complex CRM1 | mCherry-DPR lentiviral expression in Hela Kyoto cells | †† Nuclear accumulation (N/total) after export inhibition by Leptomycin B is increased (PR100) | Nuclear accumulation (N/total) after export inhibition by Leptomycin B is unchanged (PA100 /GR100 /GA100) | |
| (M) NLSc-myc-GFP2x-NESikβ2 (nuclear) | N/total unchanged (PA100/GA100/GR100/ PR100) | ||||
| (N) NLSSv40-mNeon Green2x-NESPKI (cytoplasmic) | mCherry-DPR lentiviral expression in SH-Sy5y neuronal cells and iPSC motor neurons | Nuclear accumulation (N/total) after export inhibition by Leptomycin B is decreased (GA100) | Nuclear accumulation (N/total) after export inhibition by Leptomycin B is unchanged (PA100 /GR100 /PR100) | ||
| (O) IBB-domainKPNA2-FRET sensor (nuclear) | Impβ | DPR Peptide injection in permeabilized primary mouse cortical neurons | Nuclear accumulation (N/C) is decreased (dose-dependent and more so with PR20/GR20 than with PR10/GR10), and minimal decrease with GA10/PA10 | Nuclear accumulation (N/C) is unchanged (GP10) | Hayes 2020 [50] |
| IBB-domainKPNA2-FRET sensor (nuclear) | Impβ | DPR Peptide injection in permeabilized HeLa cells | Nuclear accumulation (N/C) is decreased (more so with PR20/GR20 than with PR10/GR10) ††† | Nuclear accumulation (N/C) is unchanged (PA10/GA10 /GP10) | |
| GST-GFP-NLSSv40 (nuclear) | Impα/β complex | Nuclear accumulation (N/C) is decreased (more so with PR20/GR20 than with PR10/GR10) | Nuclear accumulation (N/C) is unchanged (PA10/GA10 /GP10) | ||
| (P) YFP-M9hnRNPA1-CFP (nuclear) | TNPO1 | Nuclear accumulation (N/C) is decreased (more so with PR20/GR20 than with PR10/GR10) | Nuclear accumulation (N/C) is unchanged (PA10/GA10 /GP10) | ||
| GCR2-GFP2-MBP-NLSSV40 (nuclear ‡) | Impα/β complex | DPR peptide addition to medium HeLa cells | Nuclear accumulation (N/C) is decreased (GR25) | Hutten 2020 [59] | |
| GCR2-GFP2-NLSSV40 (nuclear ‡) | Nuclear accumulation (N/C) is decreased (GR25) ‡‡ | ||||
 | Cell permeabilization + Allows addition of specific concentration of cargo/NTR [76,83] - Permeabilization can relocate or extract soluble proteins [77] including transport factors/ATP [76]; might deplete NTRs from NPCs; and reduces concentrations of competitive cargo, which increases import rates [84] |
 | In vivo N/C ratios; use of inhibitors + N/C ratio is a robust read-out largely independent of reporter levels [85] - Expression/injection of reporter might be toxic; use of inhibitors to measure rates of nuclear entry and exit might be toxic |
 | In vivo FRAP/FLIP + Direct measurement of kinetics of nuclear entry and exit; area under the curve indicates mobile fraction of the reporter - Expression/injection of reporter might be toxic; photobleaching induces ROS |
 | Xenopus egg extracts + Proteins can be added to/immunodepleted from the extract - No native cytosolic fraction ·Allows studying nuclear pore complex assembly, function [86], transport [87,88], and cargo competition [89]. |
| Cell Type | PR | PA | GP | GR | GA |
|---|---|---|---|---|---|
| Patients Frontal cortex | (Rare) Cytoplasmic aggregation [197,198] | (Rare) Cytoplasmic aggregation [198] | Cytoplasmic, paranucleolar aggregation [197] | Cytoplasmic aggregation [197] | Cytoplasmic, (paranuclear) inclusions [205] |
| Paranucleolar aggregation [197] | Paranucleolar aggregation [197,205] | ||||
| (Rare) [198] Sparse: Cytoplasmic inclusions, or intra-nuclear inclusions [211] | Sparse: Cytoplasmic inclusions, cytoplasmic granular, or intranuclear inclusions [211] | Numerous: Cytoplasmic inclusions, cytoplasmic granular, or intranuclear inclusions [211] | (Rare) [198] Moderate: Cytoplasmic inclusions or intranuclear inclusions [211] | Numerous: Cytoplasmic inclusions, cytoplasmic granular, or intranuclear inclusions [211] | |
| Patients spinal chord | Nuclear and extra-nuclear inclusions [194] | ||||
| iPSC neurons | Nucleolar [194] [194] | Cytoplasmic diffuse [194,203,212] | Perinuclear [212] or cytoplasmic inclusions [194,203] | ||
| Neuro2a cells | Cytoplasmic and nucleolar [213] | Cellular diffuse [213] | Cellular diffuse [213] | Cytoplasmically diffuse [213] | Cytosolic and nuclear inclusion bodies [213] |
| [200] | [200] | [200] | [200] | [200] | |
| Rat neurons | [194,214] | [194] | [194] | Length dependent toxicity [194] | [194] |
| Nucleolar aggre-gation, cytoplas-mic diffuse [197,214] | Cytoplasmic aggregation [206] | ||||
| Mouse neurons | Cytoplasmic diffuse plus inclusions [195,213] | Cellular diffuse [195,213] | Cellular diffuse [195,213] | Cellular diffuse [195,213] | Cytoplasmic aggregation [205,213] |
| [195] | |||||
| NCS34 cells (mouse motor neurons) | Nuclear aggregation [194,204,207] | Cytoplasmic aggregation [194,207] | Cellular diffuse [194,207] | Nuclear aggregation [194,204] | Cytoplasmic aggregation [194,204] |
| [207] | |||||
| Differentiated SH-SY5Y cells (neuroblast) | Nuclear [202] | Diffuse cytosolic [202] | Diffuse cytoplasmic, plus nucleolar [202] | Stellate-like cytoplasmic inclusions [202] | |
| HEK293 cells (cancer cells) | Cellular diffuse, nucleolar/cytoplasmic inclusions [205,206] | Cellular diffuse [205,206,213] | Cellular diffuse [205,206,213] | Cytoplasmic aggregation [206,213] | Cytoplasmic, occasionally nuclear aggregation [49,205,206] |
| Cytoplasmic aggregates, plus nucleolar [213] | Nuclear aggregation [205] | ||||
| Nucle(ol)ar [195] | Cellular diffuse [195] | Cellular diffuse [195] | Nucle(ol)ar [195] | Cellular [195,213] | |
| HeLa cells (cancer cells) | Nucleolar [200] | Cellular diffuse [200] | Cellular diffuse, preferentially nuclear [200] | Nucleolar [200] | Paranuclear aggregation [200] Cytoplasmic inclusions [203] |
| Cytoplasmic diffuse [203] | |||||
| Cytoplasmic inclusions [213] | Diffuse cellular [213] | ||||
| K562 cells (cancer cells) | [215] | [215] | |||
| U2OS cells (cancer cells) | Nucleoli [201] | Nucleoli [201] | |||
| CHO cells (Chinese ham-ster ovary) | [207] | [207] | [207] | [207] | |
| COS cells (fibroblast-like monkey kidney cells) | Cellular diffuse [213] | Cytoplasmic inclusions [213] | Cellular diffuse [213] | ||
| C. elegans (muscle) | Nucleolar [199] | Cellular diffuse, preferentially nuclear [199] | Cytoplasmic diffuse and nucleolar [199] | Cytoplasmic aggregation [199] | |
| D. melanogaster | [53,193,199,202,211] | [53,194,200,216] | [46,200] | Cytoplasmic diffuse [203] | [46,53,194,200,203] |
| S. cerevisiae | [47] | [47] | [47] | [47] | |
| Zebrafish embryo’s | [196] | [196] | [196] | [196] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semmelink, M.F.W.; Steen, A.; Veenhoff, L.M. Measuring and Interpreting Nuclear Transport in Neurodegenerative Disease—The Example of C9orf72 ALS. Int. J. Mol. Sci. 2021, 22, 9217. https://doi.org/10.3390/ijms22179217
Semmelink MFW, Steen A, Veenhoff LM. Measuring and Interpreting Nuclear Transport in Neurodegenerative Disease—The Example of C9orf72 ALS. International Journal of Molecular Sciences. 2021; 22(17):9217. https://doi.org/10.3390/ijms22179217
Chicago/Turabian StyleSemmelink, Marije F. W., Anton Steen, and Liesbeth M. Veenhoff. 2021. "Measuring and Interpreting Nuclear Transport in Neurodegenerative Disease—The Example of C9orf72 ALS" International Journal of Molecular Sciences 22, no. 17: 9217. https://doi.org/10.3390/ijms22179217
APA StyleSemmelink, M. F. W., Steen, A., & Veenhoff, L. M. (2021). Measuring and Interpreting Nuclear Transport in Neurodegenerative Disease—The Example of C9orf72 ALS. International Journal of Molecular Sciences, 22(17), 9217. https://doi.org/10.3390/ijms22179217






