Zinc Favors Triple-Negative Breast Cancer’s Microenvironment Modulation and Cell Plasticity
Abstract
:1. Introduction
2. Results
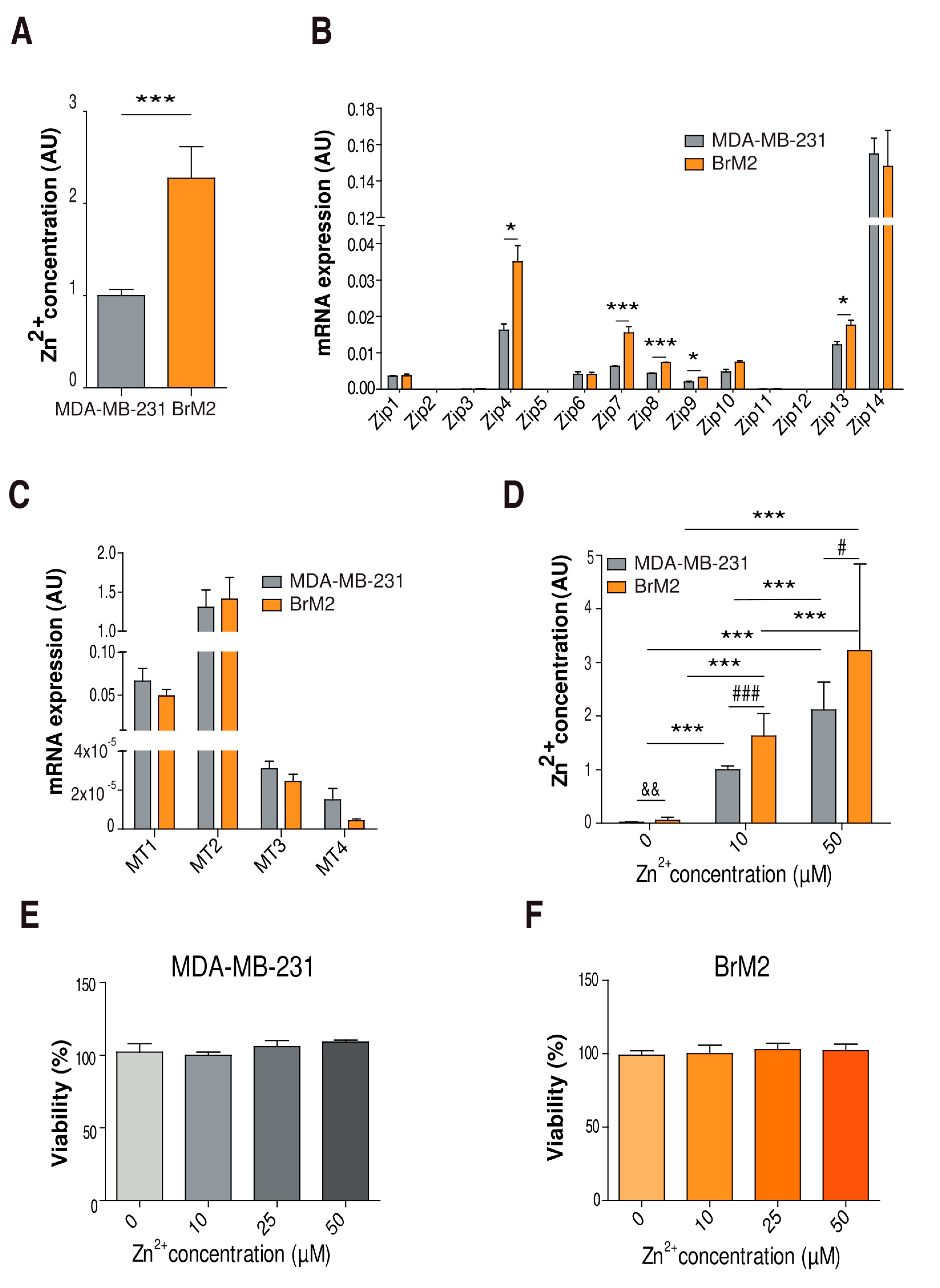
2.1. Characterization of Cellular Zinc Content in TNBC Cell Lines
2.2. TNBC Zinc Modulation in Cell Migration, Motility, and Invasion
2.3. Implication of Zinc Homeostasis in TNBC Brain Microenvironment Modulation
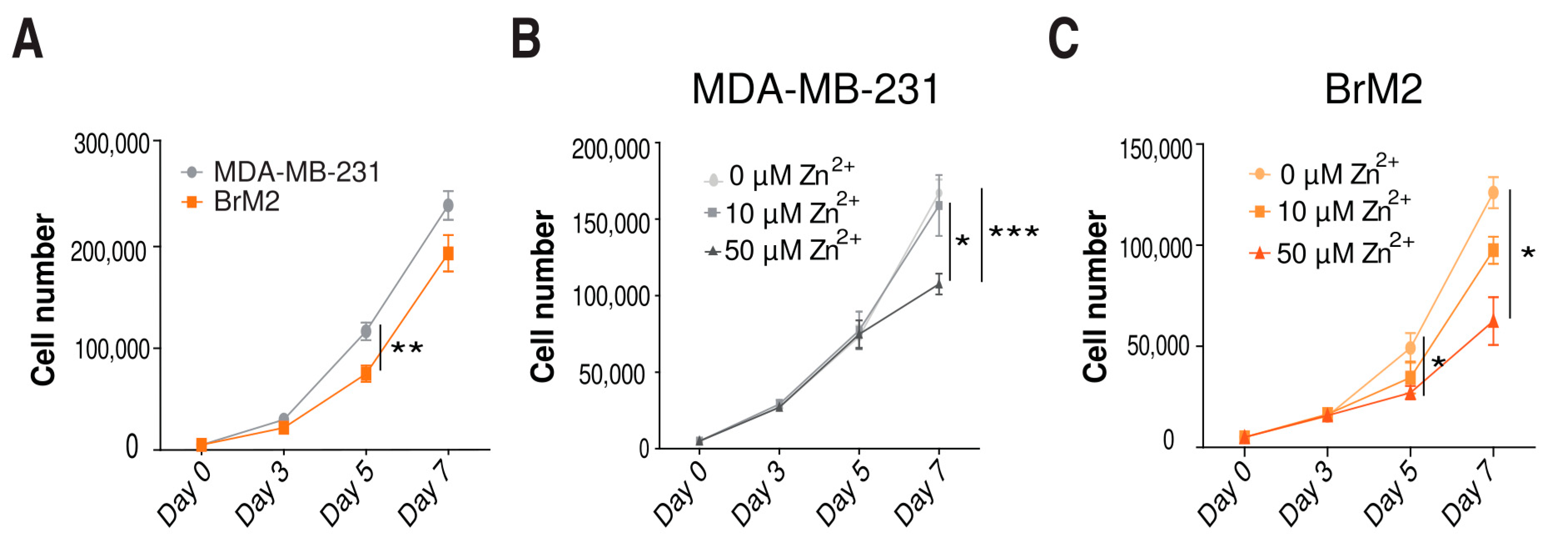
2.4. Role of Zinc in Cell Proliferation
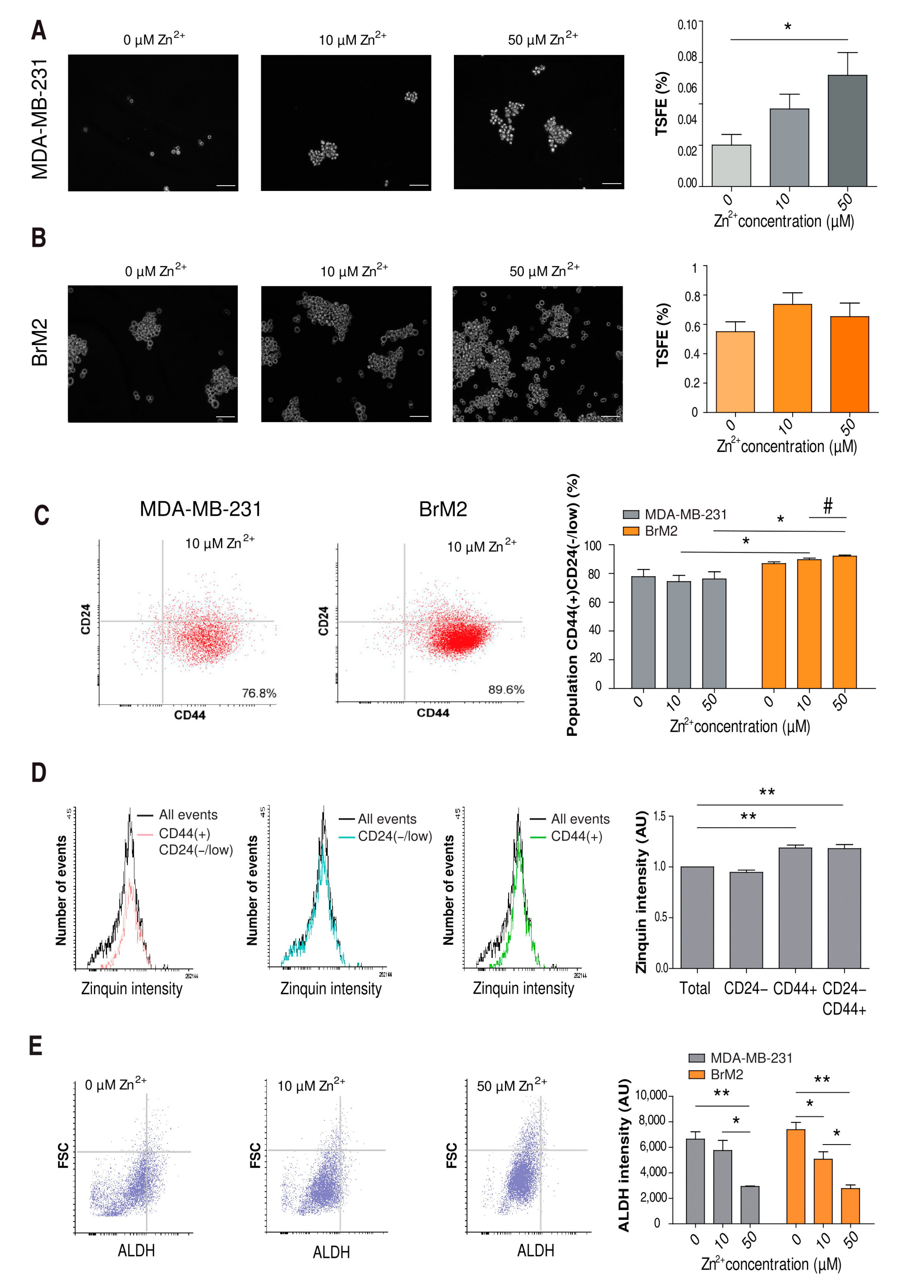
2.5. Influence of Zinc on CSC Tumorigenicity
2.6. Influence of Zinc on CSC Tumorigenicity
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Zinc Measurements
4.3. Cell Viability and Proliferation Assays
4.4. Real-Time RT PCR
4.5. Western Blotting
4.6. Tumorsphere Formation and Analysis
4.7. Migration Assay
4.8. Invasion Assay
4.9. Immunostaining
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-Negative Breast Cancer Molecular Subtyping and Treatment Progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Niu, Y. Triple Negative Breast Cancer: Special Histological Types and Emerging Therapeutic Methods. Cancer Biol. Med. 2020, 17, 293–306. [Google Scholar] [CrossRef]
- Lah, T.T.; Novak, M.; Breznik, B. Brain Malignancies: Glioblastoma and Brain Metastases. Semin. Cancer Biol. 2020, 60, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Witzel, I.; Oliveira-Ferrer, L.; Pantel, K.; Müller, V.; Wikman, H. Breast Cancer Brain Metastases: Biology and New Clinical Perspectives. Breast Cancer Res. 2016, 18, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, D.R.; Hurst, D.R. Defining the Hallmarks of Metastasis. Cancer Res. 2019, 79, 3011–3027. [Google Scholar] [CrossRef] [PubMed]
- Spano, D.; Zollo, M. Tumor Microenvironment: A Main Actor in the Metastasis Process. Clin. Exp. Metastasis 2012, 29, 381–395. [Google Scholar] [CrossRef]
- Valiente, M.; Obenauf, A.C.; Jin, X.; Chen, Q.; Zhang, X.H.-F.; Lee, D.J.; Chaft, J.E.; Kris, M.G.; Huse, J.T.; Brogi, E.; et al. Serpins Promote Cancer Cell Survival and Vascular Co-Option in Brain Metastasis. Cell 2014, 156, 1002–1016. [Google Scholar] [CrossRef] [Green Version]
- da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Muñoz, P. Cancer Cell Plasticity: Impact on Tumor Progression and Therapy Response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef]
- Clara, J.A.; Monge, C.; Yang, Y.; Takebe, N. Targeting Signalling Pathways and the Immune Microenvironment of Cancer Stem Cells—A Clinical Update. Nat. Rev. Clin. Oncol. 2020, 17, 204–232. [Google Scholar] [CrossRef]
- Kambe, T.; Tsuji, T.; Hashimoto, A.; Itsumura, N. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol. Rev. 2015, 95, 749–784. [Google Scholar] [CrossRef]
- Aydemir, T.B.; Sitren, H.S.; Cousins, R.J. The Zinc Transporter Zip14 Influences c-Met Phosphorylation and Hepatocyte Proliferation during Liver Regeneration in Mice. Gastroenterology 2012, 142, 1536–1546.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi, W.; Kimura, S.; Iwanaga, T.; Furusawa, Y.; Irié, T.; Izumi, H.; Watanabe, T.; Hijikata, A.; Hara, T.; Ohara, O.; et al. Zinc Transporter SLC39A7/ZIP7 Promotes Intestinal Epithelial Self-Renewal by Resolving ER Stress. PLoS Genet. 2016, 12, e1006349. [Google Scholar] [CrossRef]
- Ollig, J.; Kloubert, V.; Taylor, K.M.; Rink, L. B Cell Activation and Proliferation Increase Intracellular Zinc Levels. J. Nutr. Biochem. 2019, 64, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Miyagi, C.; Fukada, T.; Kagara, N.; Che, Y.-S.; Hirano, T. Zinc Transporter LIVI Controls Epithelial-Mesenchymal Transition in Zebrafish Gastrula Organizer. Nature 2004, 429, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Hiscox, S.; Nicholson, R.I.; Hogstrand, C.; Kille, P. Protein Kinase CK2 Triggers Cytosolic Zinc Signaling Pathways by Phosphorylation of Zinc Channel ZIP7. Sci. Signal. 2012, 5, ra11. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.M.; Muraina, I.A.; Brethour, D.; Schmitt-Ulms, G.; Nimmanon, T.; Ziliotto, S.; Kille, P.; Hogstrand, C. Zinc Transporter ZIP10 Forms a Heteromer with ZIP6 Which Regulates Embryonic Development and Cell Migration. Biochem. J. 2016, 473, 2531–2544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.; Beyersmann, D. Transient Peaks in Zinc and Metallothionein Levels during Differentiation of 3T3L1 Cells. Arch. Biochem. Biophys. 1999, 364, 91–98. [Google Scholar] [CrossRef]
- Apostolova, M.D.; Ivanova, I.A.; Cherian, M.G. Signal Transduction Pathways, and Nuclear Translocation of Zinc and Metallothionein during Differentiation of Myoblasts. Biochem. Cell Biol. 2000, 78, 27–37. [Google Scholar] [CrossRef]
- Wu, C.; Pot, C.; Apetoh, L.; Thalhamer, T.; Zhu, B.; Murugaiyan, G.; Xiao, S.; Lee, Y.; Rangachari, M.; Yosef, N.; et al. Metallothioneins Negatively Regulate IL-27-Induced Type 1 Regulatory T-Cell Differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 7802–7807. [Google Scholar] [CrossRef] [Green Version]
- Miyai, T.; Hojyo, S.; Ikawa, T.; Kawamura, M.; Irié, T.; Ogura, H.; Hijikata, A.; Bin, B.-H.; Yasuda, T.; Kitamura, H.; et al. Zinc Transporter SLC39A10/ZIP10 Facilitates Antiapoptotic Signaling during Early B-Cell Development. Proc. Natl. Acad. Sci. USA 2014, 111, 11780–11785. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhao, H.; Xu, Z.; Cheng, X. Zinc Dysregulation in Cancers and Its Potential as a Therapeutic Target. Cancer Biol Med. 2020, 17, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Jouybari, L.; Kiani, F.; Akbari, A.; Sanagoo, A.; Sayehmiri, F.; Aaseth, J.; Chartrand, M.S.; Sayehmiri, K.; Chirumbolo, S.; Bjørklund, G. A Meta-Analysis of Zinc Levels in Breast Cancer. J. Trace Elem. Med. Biol. 2019, 56, 90–99. [Google Scholar] [CrossRef]
- Riesop, D.; Hirner, A.V.; Rusch, P.; Bankfalvi, A. Zinc Distribution within Breast Cancer Tissue: A Possible Marker for Histological Grading? J. Cancer Res. Clin. Oncol. 2015, 141, 1321–1331. [Google Scholar] [CrossRef]
- Taylor, K.M.; Morgan, H.E.; Smart, K.; Zahari, N.M.; Pumford, S.; Ellis, I.O.; Robertson, J.F.R.; Nicholson, R.I. The Emerging Role of the LIV-1 Subfamily of Zinc Transporters in Breast Cancer. Mol. Med. 2007, 13, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Brethour, D.; Mehrabian, M.; Williams, D.; Wang, X.; Ghodrati, F.; Ehsani, S.; Rubie, E.A.; Woodgett, J.R.; Sevalle, J.; Xi, Z.; et al. A ZIP6-ZIP10 Heteromer Controls NCAM1 Phosphorylation and Integration into Focal Adhesion Complexes during Epithelial-to-Mesenchymal Transition. Sci. Rep. 2017, 7, 40313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziliotto, S.; Gee, J.M.W.; Ellis, I.O.; Green, A.R.; Finlay, P.; Gobbato, A.; Taylor, K.M. Activated Zinc Transporter ZIP7 as an Indicator of Anti-Hormone Resistance in Breast Cancer. Metallomics 2019, 11, 1579–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, P.D.; Zhang, X.H.-F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes That Mediate Breast Cancer Metastasis to the Brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef]
- Pardo-Pastor, C.; Rubio-Moscardo, F.; Vogel-González, M.; Serra, S.A.; Afthinos, A.; Mrkonjic, S.; Destaing, O.; Abenza, J.F.; Fernández-Fernández, J.M.; Trepat, X.; et al. Piezo2 Channel Regulates RhoA and Actin Cytoskeleton to Promote Cell Mechanobiological Responses. Proc. Natl. Acad. Sci. USA 2018, 115, 1925–1930. [Google Scholar] [CrossRef] [Green Version]
- Nimmanon, T.; Ziliotto, S.; Ogle, O.; Burt, A.; Gee, J.M.W.; Andrews, G.K.; Kille, P.; Hogstrand, C.; Maret, W.; Taylor, K.M. The ZIP6/ZIP10 Heteromer Is Essential for the Zinc-Mediated Trigger of Mitosis. Cell. Mol. Life Sci. 2021, 78, 1781–1798. [Google Scholar] [CrossRef]
- De Angelis, M.L.; Francescangeli, F.; Zeuner, A. Breast Cancer Stem Cells as Drivers of Tumor Chemoresistance, Dormancy and Relapse: New Challenges and Therapeutic Opportunities. Cancers 2019, 11, 1569. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Li, H.; Wang, H.; Shi, X.; Fan, Y.; Ding, X.; Lin, C.; Zhan, Q.; Qian, H.; Xu, B. Enriched CD44(+)/CD24(-) Population Drives the Aggressive Phenotypes Presented in Triple-Negative Breast Cancer (TNBC). Cancer Lett. 2014, 353, 153–159. [Google Scholar] [CrossRef]
- Lee, N.-H.; Park, S.-R.; Lee, J.W.; Lim, S.; Lee, S.-H.; Nam, S.; Kim, D.Y.; Hah, S.Y.; Hong, I.-S.; Lee, H.-Y. SERPINB2 Is a Novel Indicator of Cancer Stem Cell Tumorigenicity in Multiple Cancer Types. Cancers 2019, 11, 499. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of Their Normal Counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Pérez, G.; López-Moncada, F.; Indo, S.; Torres, M.J.; Castellón, E.A.; Contreras, H.R. Knockdown of ZEB1 Reverses Cancer Stem Cell Properties in Prostate Cancer Cells. Oncol. Rep. 2021, 45, 58. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Bharadwaj, U.; Zhai, Q.J.; Ahern, C.H.; Fisher, W.E.; Brunicardi, F.C.; Logsdon, C.D.; Chen, C.; Yao, Q. Down-Regulation of ZIP4 by RNA Interference Inhibits Pancreatic Cancer Growth and Increases the Survival of Nude Mice with Pancreatic Cancer Xenografts. Clin. Cancer Res. 2009, 15, 5993–6001. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Guo, H.-J.; Xie, H.-Y.; Li, J.; Zhuang, R.-Z.; Ling, Q.; Zhou, L.; Wei, X.-Y.; Liu, Z.-K.; Ding, S.-M.; et al. ZIP4, a Novel Determinant of Tumor Invasion in Hepatocellular Carcinoma, Contributes to Tumor Recurrence after Liver Transplantation. Int. J. Biol. Sci. 2014, 10, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Yang, J.; Zhang, Y.; Zhou, Z.; Cui, X.; Zhang, L.; Fung, K.-M.; Zheng, W.; Allard, F.D.; Yee, E.U.; et al. ZIP4 Promotes Pancreatic Cancer Progression by Repressing ZO-1 and Claudin-1 through a ZEB1-Dependent Transcriptional Mechanism. Clin. Cancer Res. 2018, 24, 3186–3196. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef] [Green Version]
- Kagara, N.; Tanaka, N.; Noguchi, S.; Hirano, T. Zinc and Its Transporter ZIP10 Are Involved in Invasive Behavior of Breast Cancer Cells. Cancer Sci. 2007, 98, 692–697. [Google Scholar] [CrossRef]
- Taylor, K.M.; Vichova, P.; Jordan, N.; Hiscox, S.; Hendley, R.; Nicholson, R.I. ZIP7-Mediated Intracellular Zinc Transport Contributes to Aberrant Growth Factor Signaling in Antihormone-Resistant Breast Cancer Cells. Endocrinology 2008, 149, 4912–4920. [Google Scholar] [CrossRef] [Green Version]
- Hogstrand, C.; Kille, P.; Ackland, M.L.; Hiscox, S.; Taylor, K.M. A Mechanism for Epithelial-Mesenchymal Transition and Anoikis Resistance in Breast Cancer Triggered by Zinc Channel ZIP6 and STAT3 (signal Transducer and Activator of Transcription 3). Biochem. J. 2013, 455, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Suk Kim, H.; Ki Choi, S.; Hye Hwang, E.; Woo, J.; Suk Ryu, H.; Kim, K.; Moon, A.; Kyung Moon, W. microRNA-200c/141 Upregulates SerpinB2 to Promote Breast Cancer Cell Metastasis and Reduce Patient Survival. Oncotarget 2017, 8, 32769–32782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akkoc, Y.; Peker, N.; Akcay, A.; Gozuacik, D. Autophagy and Cancer Dormancy. Front. Oncol. 2021, 11, 627023. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Y.; Liu, Z.; Bharadwaj, U.; Wang, H.; Wang, X.; Zhang, S.; Liuzzi, J.P.; Chang, S.-M.; Cousins, R.J.; et al. Aberrant Expression of Zinc Transporter ZIP4 (SLC39A4) Significantly Contributes to Human Pancreatic Cancer Pathogenesis and Progression. Proc. Natl. Acad. Sci. USA 2007, 104, 18636–18641. [Google Scholar] [CrossRef] [Green Version]
- Fan, Q.; Zhang, W.; Emerson, R.E.; Xu, Y. ZIP4 Is a Novel Cancer Stem Cell Marker in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 3692. [Google Scholar] [CrossRef]



| Primers | Forward Sequence (5′→3′) | Reverse Sequence (3′→5′) |
|---|---|---|
| Zip1 | GATTGGGGAAGACACTTGACTGCT | GAAAGAGGAAGGGGATTTGTTTGG |
| Zip2 | CCCTTGTCCTCTTGCTGTCACTCT | AGCTCCCGTGGAAGAATTTCTAGG |
| Zip3 | GTGGAGATATGAGGACCCCCTGTT | GATGAACTCAGCGCTAACCGATCT |
| Zip4 | AGACTGAGCCCAGAGTTGAGGCTA | TGTCGCAGAGTGCTACGTAGAGGA |
| Zip5 | GAGCAGGAGCAGAACCATTACCTG | CAATGAGTGGTCCAGCAACAGAAG |
| Zip6 | CATAGCCATGAAGAACCAGCAATG | GAGAATCAAAGTGGGAGGGCTCTT |
| Zip7 | ACTGAAGGAGGAGCAGTGGACAGT | AGGCCCTAATGCCAAAGTAACCAT |
| Zip8 | CCTCGGATTGATTTTGACTCCACT | AGCAGGATTTGCATAGCATGTCAC |
| Zip9 | GCCTAAAGAACTGGAAAGCCCACT | GTGTTTCACTTGCTTGGTGGTGTT |
| Zip10 | TAGCCGTCTTCTGTCATGACTGC | TCATAGAGGGCAATCACCAGCATA |
| Zip11 | TCTCCTAAGCATTTTGGTGGCCTA | TCTCTTCTTTCCACAGGGCTCACT |
| Zip12 | CAACCACTCAAGAAGCCTCATCAA | AAGTACTGCCTGGTGAAAGCCAAG |
| Zip13 | AAGAAGATCGGGCTCCTGACAAC | GAGAACAGCACCATTACCACGATG |
| Zip14 | CATTTGGTTTCAACCCTCTGGAAG | TTTCAGCCAGTAGCAAGCACTCTG |
| SerpinB2 | GTTCATGCAGCAGATCCAGA | CGCAGACTTCTCACCAAACA |
| Metallothionein-1 | ATCTGCAAAGGGACGTCAGA | ACGGGTCAGGGTTGTACAAA |
| Metallothionein-2 | TCCTGCAAGAAAAGCTGCTG | TCTTTACATCTGGGAGCGGG |
| Metallothionein-3 | ACACACAGTCCTTGGCACAC | AAGTGCGAGGGATGCAAAT′. |
| Metallothionein-4 | GTGTCTGCATGTCTGGAGGA | TCTGAGCCTCCTTTGCAGAT |
| ZEB1 | GATGATGAATGCGAGTCAGATGC | ACAGCAGTGTCTTGTTGTTGT |
| ZEB1-AS1 | CCGTGGGCACTGCTGAAT | CTGCTGGCAAGCGGAACT |
| Oct4 | ACATCAAAGCTCTGCAGAAAGAACT | CTGAATACCTTCCCAAATAGAACCC |
| Sox2 | AAATGGGAGGGGTGCAAAAGAGGAG | CAGCTGTCATTTGCTGTGGGTGATG |
| Nanog | ACATGCAACCTGAAGACGTGTG | CATGGAAACCAGAACACGTGG |
| GAPDH | GGAGTCCACTGGCGTCTTC | TGGCTCCCCCCTGCAAATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vogel-González, M.; Musa-Afaneh, D.; Rivera Gil, P.; Vicente, R. Zinc Favors Triple-Negative Breast Cancer’s Microenvironment Modulation and Cell Plasticity. Int. J. Mol. Sci. 2021, 22, 9188. https://doi.org/10.3390/ijms22179188
Vogel-González M, Musa-Afaneh D, Rivera Gil P, Vicente R. Zinc Favors Triple-Negative Breast Cancer’s Microenvironment Modulation and Cell Plasticity. International Journal of Molecular Sciences. 2021; 22(17):9188. https://doi.org/10.3390/ijms22179188
Chicago/Turabian StyleVogel-González, Marina, Dunia Musa-Afaneh, Pilar Rivera Gil, and Rubén Vicente. 2021. "Zinc Favors Triple-Negative Breast Cancer’s Microenvironment Modulation and Cell Plasticity" International Journal of Molecular Sciences 22, no. 17: 9188. https://doi.org/10.3390/ijms22179188
APA StyleVogel-González, M., Musa-Afaneh, D., Rivera Gil, P., & Vicente, R. (2021). Zinc Favors Triple-Negative Breast Cancer’s Microenvironment Modulation and Cell Plasticity. International Journal of Molecular Sciences, 22(17), 9188. https://doi.org/10.3390/ijms22179188







