The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action
Abstract
1. Introduction
2. Basics of Molecular Action of Nuclear Hormone Receptors
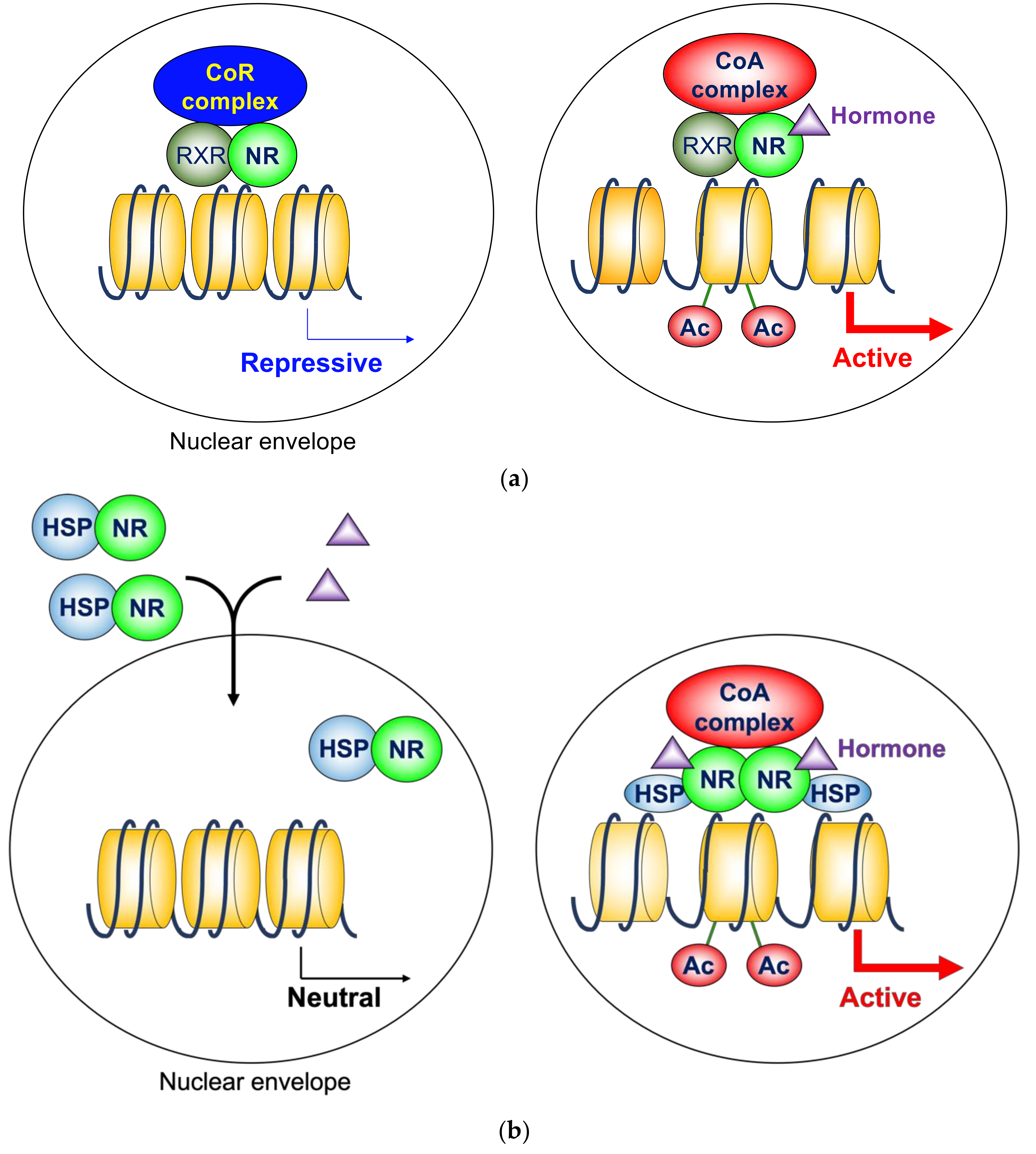
2.1. Heterodimeric Nuclear Hormone Receptors
2.2. Homodimeric Nuclear Hormone Receptors
2.3. Orphan Nuclear Hormone Receptors
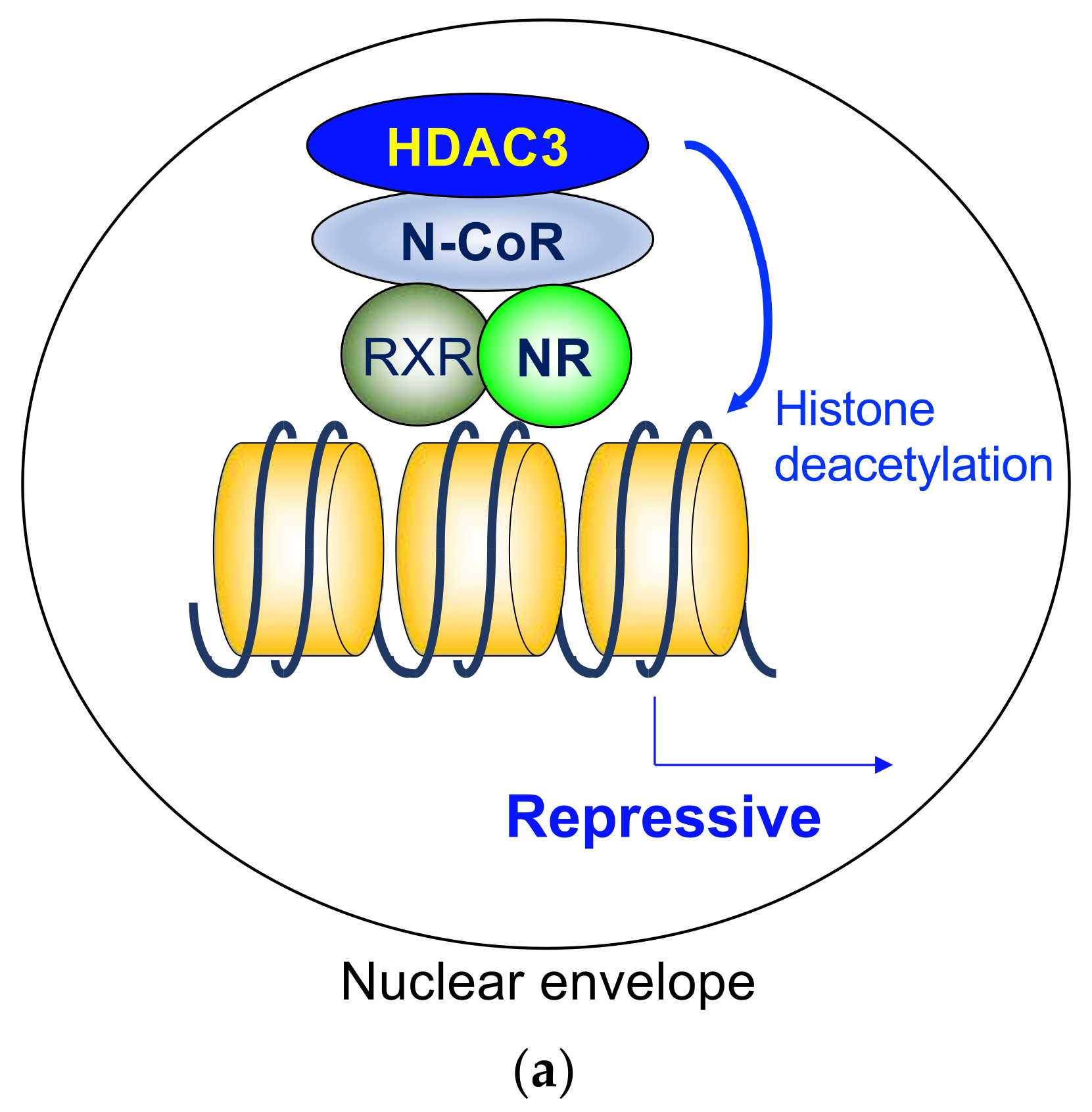
3. Histone Deacetylase 3 in Nuclear Hormone Receptor Corepressor Complex
3.1. Basics of Histone Deacetylase 3
3.2. Histone Deacetylase 3 and Nuclear Hormone Receptor Corepressors
3.3. Physiological Roles of Histone Deacetylase 3
4. Histone Deacetylase 3 in Nuclear Hormone Receptor Action
4.1. Ligand-Independent Repression
4.2. Gene Repression by Orphan Nuclear Hormone Receptors
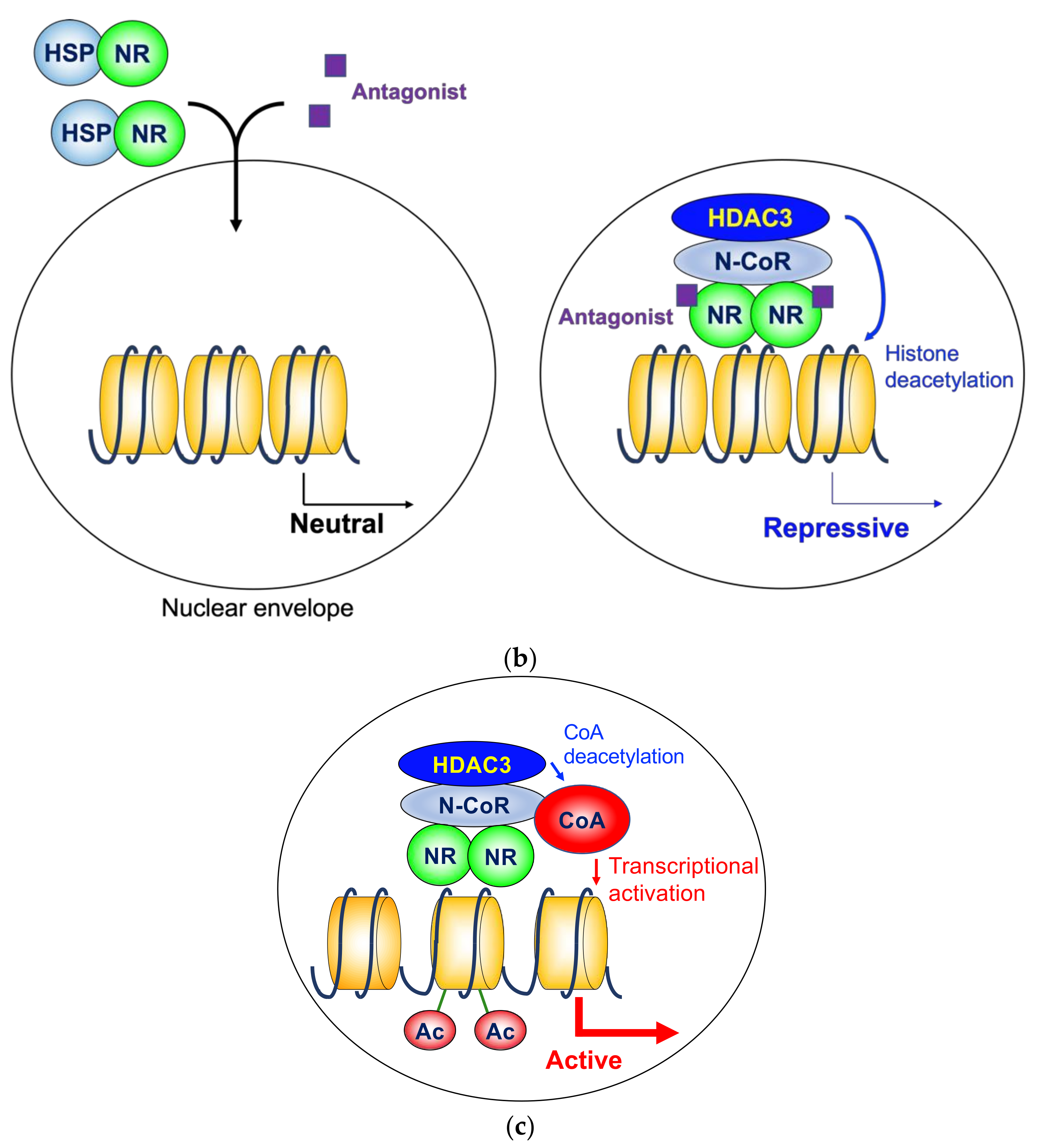
4.3. Nuclear Hormone Receptor Antagonist Action
4.4. Ligand-Induced Repression
4.5. Activation of Transcriptional Coactivator by Histone Deacetylase 3
5. Non-Canonical Mechanisms of Histone Deacetylase 3 Action for Other Transcription Factors
6. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| APL | Acute promyelocytic leukemia |
| AR | Androgen receptor |
| DAX1 | Dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 |
| DAD | Deacetylase activating domain |
| ER | Estrogen receptor |
| ERRα | Estrogen-related receptor α |
| GPS2 | G protein pathway suppressor 2 |
| GR | Glucocorticoid receptor |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HRE | Hormone response element |
| HSP | Heat shock protein |
| KAT | Lysine acetyltransferase |
| KDAC | Lysine deacetylase |
| LBD | Ligand binding domain |
| MR | Mineralocorticoid receptor |
| NAD+ | Nicotinamide adenine dinucleotide |
| N-CoR | Nuclear receptor corepressor |
| NF-κB | Nuclear factor kappa-B |
| NR | Nuclear hormone receptor |
| PGC1α | Peroxisome proliferator-activated receptor-γ coactivator 1α |
| PML | Promyelocytic leukemia |
| PR | Progesterone receptor |
| RAR | Retinoid A receptor |
| RXR | Retinoid X receptor |
| SMRT | Silencing mediator for retinoid and thyroid-hormone receptors |
| SRY | Sex-determining region Y |
| TBL1X | Transducin beta-like 1 X-linked |
| TBL1XR1 | TBL1X receptor 1 |
| TR | Thyroid hormone receptor |
| VDR | Vitamin D receptor |
References
- Dumitrescu, A.M.; Liao, X.H.; Best, T.B.; Brockmann, K.; Refetoff, S. A novel syndrome combining thyroid and neurological abnormalities is associated with mutations in a monocarboxylate transporter gene. Am. J. Hum. Genet. 2004, 74, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Carter-SU, C. Principles of Hormone Action. In Williams Textbook of Endocrinology, 14th ed.; Melmed, S., Auchus, R.J., Goldfine, A.B., Koenig, R.J., Rosen, C.J., Eds.; Elsevir: Philadelphia, PA, USA, 2020; pp. 13–41. [Google Scholar]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Committee, N.R.N. A unified nomenclature system for the nuclear receptor superfamily. Cell 1999, 97, 161–163. [Google Scholar] [CrossRef]
- Krust, A.; Green, S.; Argos, P.; Kumar, V.; Walter, P.; Bornert, J.M.; Chambon, P. The chicken oestrogen receptor sequence: Homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986, 5, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Chandra, V.; Rastinejad, F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu. Rev. Physiol. 2010, 72, 247–272. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M. DNA binding specificity of steroid receptors. Cell 1989, 57, 1065–1068. [Google Scholar] [CrossRef]
- Gronemeyer, H.; Moras, D. Nuclear receptors. How to finger DNA. Nature 1995, 375, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Tora, L.; White, J.; Brou, C.; Tasset, D.; Webster, N.; Scheer, E.; Chambon, P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell 1989, 59, 477–487. [Google Scholar] [CrossRef]
- Bourguet, W.; Ruff, M.; Chambon, P.; Gronemeyer, H.; Moras, D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature 1995, 375, 377–382. [Google Scholar] [CrossRef]
- Moras, D.; Gronemeyer, H. The nuclear receptor ligand-binding domain: Structure and function. Curr. Opin. Cell Biol. 1998, 10, 384–391. [Google Scholar] [CrossRef]
- Lonard, D.M.; O’Malley, B.W. Nuclear receptor coregulators: Modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 2012, 8, 598–604. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell. Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Bird, A. Histone deacetylases: Silencers for hire. Trends Biochem. Sci. 2000, 25, 121–126. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, K.; Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005, 121, 375–385. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Trinchera, M. Epigenetic Bases of Aberrant Glycosylation in Cancer. Int. J. Mol. Sci. 2017, 18, 998. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr. Rev. 1994, 15, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Yen, P.M. Classical nuclear hormone receptor activity as a mediator of complex biological responses: A look at health and disease. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 517–528. [Google Scholar] [CrossRef]
- Baniahmad, A.; Steiner, C.; Köhne, A.C.; Renkawitz, R. Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell 1990, 61, 505–514. [Google Scholar] [CrossRef]
- Burcin, M.; Köhne, A.C.; Runge, D.; Steiner, C.; Renkawitz, R. Factors influencing nuclear receptors in transcriptional repression. Semin. Cancer Biol. 1994, 5, 337–346. [Google Scholar] [PubMed]
- Hu, X.; Lazar, M.A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 2000, 11, 6–10. [Google Scholar] [CrossRef]
- Hörlein, A.J.; Näär, A.M.; Heinzel, T.; Torchia, J.; Gloss, B.; Kurokawa, R.; Ryan, A.; Kamei, Y.; Söderström, M.; Glass, C.K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 1995, 377, 397–404. [Google Scholar] [CrossRef]
- Chen, J.D.; Evans, R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 1995, 377, 454–457. [Google Scholar] [CrossRef]
- Jepsen, K.; Hermanson, O.; Onami, T.M.; Gleiberman, A.S.; Lunyak, V.; McEvilly, R.J.; Kurokawa, R.; Kumar, V.; Liu, F.; Seto, E.; et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 2000, 102, 753–763. [Google Scholar] [CrossRef]
- Jepsen, K.; Solum, D.; Zhou, T.; McEvilly, R.J.; Kim, H.J.; Glass, C.K.; Hermanson, O.; Rosenfeld, M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 2007, 450, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K.; Rosenfeld, M.G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000, 14, 121–141. [Google Scholar] [PubMed]
- Perissi, V.; Jepsen, K.; Glass, C.K.; Rosenfeld, M.G. Deconstructing repression: Evolving models of co-repressor action. Nat. Rev. Genet. 2010, 11, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Cavaillès, V.; Dauvois, S.; Danielian, P.S.; Parker, M.G. Interaction of proteins with transcriptionally active estrogen receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 10009–10013. [Google Scholar] [CrossRef] [PubMed]
- Oñate, S.A.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 1995, 270, 1354–1357. [Google Scholar] [CrossRef]
- Hong, H.; Kohli, K.; Garabedian, M.J.; Stallcup, M.R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 1997, 17, 2735–2744. [Google Scholar] [CrossRef]
- Anzick, S.L.; Kononen, J.; Walker, R.L.; Azorsa, D.O.; Tanner, M.M.; Guan, X.Y.; Sauter, G.; Kallioniemi, O.P.; Trent, J.M.; Meltzer, P.S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 1997, 277, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Kamei, Y.; Xu, L.; Heinzel, T.; Torchia, J.; Kurokawa, R.; Gloss, B.; Lin, S.C.; Heyman, R.A.; Rose, D.W.; Glass, C.K.; et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 1996, 85, 403–414. [Google Scholar] [CrossRef]
- Torchia, J.; Rose, D.W.; Inostroza, J.; Kamei, Y.; Westin, S.; Glass, C.K.; Rosenfeld, M.G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 1997, 387, 677–684. [Google Scholar] [CrossRef]
- Renaud, J.P.; Rochel, N.; Ruff, M.; Vivat, V.; Chambon, P.; Gronemeyer, H.; Moras, D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 1995, 378, 681–689. [Google Scholar] [CrossRef]
- Shabtai, Y.; Nagaraj, N.K.; Batmanov, K.; Cho, Y.W.; Guan, Y.; Jiang, C.; Remsberg, J.; Forrest, D.; Lazar, M.A. A coregulator shift, rather than the canonical switch, underlies thyroid hormone action in the liver. Genes Dev. 2021, 35, 367–378. [Google Scholar] [CrossRef]
- Pratt, W.B.; Toft, D.O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997, 18, 306–360. [Google Scholar] [CrossRef]
- Leclercq, G.; Lacroix, M.; Laïos, I.; Laurent, G. Estrogen receptor alpha: Impact of ligands on intracellular shuttling and turnover rate in breast cancer cells. Curr. Cancer Drug Targets 2006, 6, 39–64. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elbi, C.; Walker, D.A.; Romero, G.; Sullivan, W.P.; Toft, D.O.; Hager, G.L.; DeFranco, D.B. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA 2004, 101, 2876–2881. [Google Scholar] [CrossRef]
- Xin, Q.L.; Qiu, J.T.; Cui, S.; Xia, G.L.; Wang, H.B. Transcriptional activation of nuclear estrogen receptor and progesterone receptor and its regulation. Sheng Li Xue Bao 2016, 68, 435–454. [Google Scholar]
- Mullican, S.E.; Dispirito, J.R.; Lazar, M.A. The orphan nuclear receptors at their 25-year reunion. J. Mol. Endocrinol. 2013, 51, T115–T140. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell. Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Yao, Y.L.; Sun, J.M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef]
- Emiliani, S.; Fischle, W.; Van Lint, C.; Al-Abed, Y.; Verdin, E. Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. USA 1998, 95, 2795–2800. [Google Scholar] [CrossRef]
- Dangond, F.; Hafler, D.A.; Tong, J.K.; Randall, J.; Kojima, R.; Utku, N.; Gullans, S.R. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem. Biophys. Res. Commun. 1998, 242, 648–652. [Google Scholar] [CrossRef]
- Bhaskara, S.; Chyla, B.J.; Amann, J.M.; Knutson, S.K.; Cortez, D.; Sun, Z.W.; Hiebert, S.W. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell 2008, 30, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kao, G.D.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Qin, J.; Phelan, C.; Lazar, M.A. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006, 20, 2566–2579. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Kurasawa, Y.; Wong, J.; Yu-Lee, L.Y. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc. Natl. Acad. Sci. USA 2008, 105, 4179–4184. [Google Scholar] [CrossRef] [PubMed]
- Eot-Houllier, G.; Fulcrand, G.; Watanabe, Y.; Magnaghi-Jaulin, L.; Jaulin, C. Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes Dev. 2008, 22, 2639–2644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bhaskara, S.; Knutson, S.K.; Jiang, G.; Chandrasekharan, M.B.; Wilson, A.J.; Zheng, S.; Yenamandra, A.; Locke, K.; Yuan, J.L.; Bonine-Summers, A.R.; et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010, 18, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.R.; Fischer, M.A.; Stengel, K.R.; Zhao, Y.; Kaiser, J.F.; Wells, C.E.; Hunt, A.; Bhaskara, S.; Luzwick, J.W.; Sampathi, S.; et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J. Clin. Investig. 2013, 123, 3112–3123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Hsieh, J. HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc. Natl. Acad. Sci. USA 2014, 111, 13541–13546. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.D.; Perissi, V.; Staszewski, L.M.; Yang, W.M.; Krones, A.; Glass, C.K.; Rosenfeld, M.G.; Seto, E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 2000, 97, 7202–7207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kalkum, M.; Chait, B.T.; Roeder, R.G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 2002, 9, 611–623. [Google Scholar] [CrossRef]
- Guenther, M.G.; Lane, W.S.; Fischle, W.; Verdin, E.; Lazar, M.A.; Shiekhattar, R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000, 14, 1048–1057. [Google Scholar] [PubMed]
- Li, J.; Lin, Q.; Wang, W.; Wade, P.; Wong, J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002, 16, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.G.; Chan, D.W.; Huang, Z.Q.; Li, J.; Fondell, J.D.; Qin, J.; Wong, J. Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003, 22, 1336–1346. [Google Scholar] [CrossRef]
- Guenther, M.G.; Barak, O.; Lazar, M.A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001, 21, 6091–6101. [Google Scholar] [CrossRef]
- Watson, P.J.; Fairall, L.; Santos, G.M.; Schwabe, J.W. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 2012, 481, 335–340. [Google Scholar] [CrossRef]
- You, S.H.; Lim, H.W.; Sun, Z.; Broache, M.; Won, K.J.; Lazar, M.A. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 2013, 20, 182–187. [Google Scholar] [CrossRef]
- Ishizuka, T.; Lazar, M.A. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol. Endocrinol. 2005, 19, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ozawa, Y.; Lee, H.; Wen, Y.D.; Tan, T.H.; Wadzinski, B.E.; Seto, E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005, 19, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Karagianni, P.; Wong, J. HDAC3: Taking the SMRT-N-CoRrect road to repression. Oncogene 2007, 26, 5439–5449. [Google Scholar] [CrossRef]
- Yoon, H.G.; Choi, Y.; Cole, P.A.; Wong, J. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 2005, 25, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Perissi, V.; Aggarwal, A.; Glass, C.K.; Rose, D.W.; Rosenfeld, M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 2004, 116, 511–526. [Google Scholar] [CrossRef]
- Heinen, C.A.; Losekoot, M.; Sun, Y.; Watson, P.J.; Fairall, L.; Joustra, S.D.; Zwaveling-Soonawala, N.; Oostdijk, W.; van den Akker, E.L.; Alders, M.; et al. Mutations in TBL1X Are Associated With Central Hypothyroidism. J. Clin. Endocrinol. Metab. 2016, 101, 4564–4573. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.; Schneider, S.; Wagner, E.F.; Zhang, X.; Seto, E.; Bohmann, D. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 2003, 22, 3686–3695. [Google Scholar] [CrossRef] [PubMed]
- Emmett, M.J.; Lazar, M.A. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell. Biol. 2019, 20, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.L.; Potthoff, M.J.; Haberland, M.; Qi, X.; Matsuzaki, S.; Humphries, K.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Investig. 2008, 118, 3588–3597. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, S.L.; Janardhan, H.P.; Smee, K.M.; Bachman, M.; Sun, Z.; Lazar, M.A.; Trivedi, C.M. Histone deacetylase 3 modulates Tbx5 activity to regulate early cardiogenesis. Hum. Mol. Genet. 2014, 23, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Poleshko, A.; Shah, P.P.; Gupta, M.; Babu, A.; Morley, M.P.; Manderfield, L.J.; Ifkovits, J.L.; Calderon, D.; Aghajanian, H.; Sierra-Pagán, J.E.; et al. Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 2017, 171, 573–587.e514. [Google Scholar] [CrossRef]
- Knutson, S.K.; Chyla, B.J.; Amann, J.M.; Bhaskara, S.; Huppert, S.S.; Hiebert, S.W. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008, 27, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Liu, T.; Sun, Z.; Bugge, A.; Mullican, S.E.; Alenghat, T.; Liu, X.S.; Lazar, M.A. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011, 331, 1315–1319. [Google Scholar] [CrossRef]
- Sun, Z.; Miller, R.A.; Patel, R.T.; Chen, J.; Dhir, R.; Wang, H.; Zhang, D.; Graham, M.J.; Unterman, T.G.; Shulman, G.I.; et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 2012, 18, 934–942. [Google Scholar] [CrossRef]
- Mullican, S.E.; Gaddis, C.A.; Alenghat, T.; Nair, M.G.; Giacomin, P.R.; Everett, L.J.; Feng, D.; Steger, D.J.; Schug, J.; Artis, D.; et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011, 25, 2480–2488. [Google Scholar] [CrossRef]
- Navabi, N.; Whitt, J.; Wu, S.E.; Woo, V.; Moncivaiz, J.; Jordan, M.B.; Vallance, B.A.; Way, S.S.; Alenghat, T. Epithelial Histone Deacetylase 3 Instructs Intestinal Immunity by Coordinating Local Lymphocyte Activation. Cell Rep. 2017, 19, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gupta, M.; Trivedi, C.M.; Singh, M.K.; Li, L.; Epstein, J.A. Murine craniofacial development requires Hdac3-mediated repression of Msx gene expression. Dev. Biol. 2013, 377, 333–344. [Google Scholar] [CrossRef]
- Feigenson, M.; Shull, L.C.; Taylor, E.L.; Camilleri, E.T.; Riester, S.M.; van Wijnen, A.J.; Bradley, E.W.; Westendorf, J.J. Histone Deacetylase 3 Deletion in Mesenchymal Progenitor Cells Hinders Long Bone Development. J. Bone Miner. Res. 2017, 32, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Norwood, J.; Franklin, J.M.; Sharma, D.; D’Mello, S.R. Histone deacetylase 3 is necessary for proper brain development. J. Biol. Chem. 2014, 289, 34569–34582. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Frank, D.B.; Morley, M.P.; Zhou, S.; Wang, X.; Lu, M.M.; Lazar, M.A.; Morrisey, E.E. HDAC3-Dependent Epigenetic Pathway Controls Lung Alveolar Epithelial Cell Remodeling and Spreading via miR-17-92 and TGF-β Signaling Regulation. Dev. Cell 2016, 36, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Emmett, M.J.; Lim, H.W.; Jager, J.; Richter, H.J.; Adlanmerini, M.; Peed, L.C.; Briggs, E.R.; Steger, D.J.; Ma, T.; Sims, C.A.; et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 2017, 546, 544–548. [Google Scholar] [CrossRef]
- Hong, S.; Zhou, W.; Fang, B.; Lu, W.; Loro, E.; Damle, M.; Ding, G.; Jager, J.; Zhang, S.; Zhang, Y.; et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med. 2017, 23, 223–234. [Google Scholar] [CrossRef]
- Remsberg, J.R.; Ediger, B.N.; Ho, W.Y.; Damle, M.; Li, Z.; Teng, C.; Lanzillotta, C.; Stoffers, D.A.; Lazar, M.A. Deletion of histone deacetylase 3 in adult beta cells improves glucose tolerance via increased insulin secretion. Mol. Metab. 2017, 6, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, R.; Banerjee, S.; Amin, S.A.; Adhikari, N.; Jha, T. Histone deacetylase 3 (HDAC3) inhibitors as anticancer agents: A review. Eur. J. Med. Chem. 2020, 192, 112171. [Google Scholar] [CrossRef]
- Bates, S.E. Epigenetic Therapies for Cancer. N. Engl. J. Med. 2020, 383, 650–663. [Google Scholar] [CrossRef]
- Wondisford, F.E. Thyroid hormone action: Insight from transgenic mouse models. J. Investig. Med. 2003, 51, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Koibuchi, N. Animal models to study thyroid hormone action in cerebellum. Cerebellum 2009, 8, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Lazar, M.A. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 2003, 23, 5122–5131. [Google Scholar] [CrossRef] [PubMed]
- Alenghat, T.; Yu, J.; Lazar, M.A. The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 2006, 25, 3966–3974. [Google Scholar] [CrossRef] [PubMed]
- Urvalek, A.M.; Gudas, L.J. Retinoic acid and histone deacetylases regulate epigenetic changes in embryonic stem cells. J. Biol. Chem. 2014, 289, 19519–19530. [Google Scholar] [CrossRef]
- Malinen, M.; Saramäki, A.; Ropponen, A.; Degenhardt, T.; Väisänen, S.; Carlberg, C. Distinct HDACs regulate the transcriptional response of human cyclin-dependent kinase inhibitor genes to Trichostatin A and 1alpha,25-dihydroxyvitamin D3. Nucleic Acids Res. 2008, 36, 121–132. [Google Scholar] [CrossRef]
- Jimenez, J.J.; Chale, R.S.; Abad, A.C.; Schally, A.V. Acute promyelocytic leukemia (APL): A review of the literature. Oncotarget 2020, 11, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- de Thé, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef]
- Atsumi, A.; Tomita, A.; Kiyoi, H.; Naoe, T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARalpha as a component of the N-CoR co-repressor complex to repress transcription in vivo. Biochem. Biophys. Res. Commun. 2006, 345, 1471–1480. [Google Scholar] [CrossRef]
- Villa, R.; Morey, L.; Raker, V.A.; Buschbeck, M.; Gutierrez, A.; De Santis, F.; Corsaro, M.; Varas, F.; Bossi, D.; Minucci, S.; et al. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc. Natl. Acad. Sci. USA 2006, 103, 1400–1405. [Google Scholar] [CrossRef]
- Matthews, G.M.; Mehdipour, P.; Cluse, L.A.; Falkenberg, K.J.; Wang, E.; Roth, M.; Santoro, F.; Vidacs, E.; Stanley, K.; House, C.M.; et al. Functional-genetic dissection of HDAC dependencies in mouse lymphoid and myeloid malignancies. Blood 2015, 126, 2392–2403. [Google Scholar] [CrossRef] [PubMed]
- Lazar, M.A.; Hodin, R.A.; Darling, D.S.; Chin, W.W. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol. Cell. Biol. 1989, 9, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Sap, J.; Muñoz, A.; Damm, K.; Goldberg, Y.; Ghysdael, J.; Leutz, A.; Beug, H.; Vennström, B. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature 1986, 324, 635–640. [Google Scholar] [CrossRef]
- Weinberger, C.; Thompson, C.C.; Ong, E.S.; Lebo, R.; Gruol, D.J.; Evans, R.M. The c-erb-A gene encodes a thyroid hormone receptor. Nature 1986, 324, 641–646. [Google Scholar] [CrossRef]
- Kim, Y.H.; Marhon, S.A.; Zhang, Y.; Steger, D.J.; Won, K.J.; Lazar, M.A. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 2018, 359, 1274–1277. [Google Scholar] [CrossRef]
- Olivieri, D.; Castelli, E.; Kawamura, Y.K.; Papasaikas, P.; Lukonin, I.; Rittirsch, M.; Hess, D.; Smallwood, S.A.; Stadler, M.B.; Peters, A.H.F.M.; et al. Cooperation between HDAC3 and DAX1 mediates lineage restriction of embryonic stem cells. EMBO J. 2021, 40, e106818. [Google Scholar] [CrossRef]
- Cui, S.; Kolodziej, K.E.; Obara, N.; Amaral-Psarris, A.; Demmers, J.; Shi, L.; Engel, J.D.; Grosveld, F.; Strouboulis, J.; Tanabe, O. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic β-type globin promoters in differentiated adult erythroid cells. Mol. Cell. Biol. 2011, 31, 3298–3311. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Yu, R.T.; Evans, R.M.; Shi, Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl. Acad. Sci. USA 2007, 104, 15282–15287. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Nawaz, Z.; Tsai, S.Y.; O’Malley, B.W.; Tsai, M.J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT). Mol. Endocrinol. 1997, 11, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.M.; Guo, C.; Zhang, J. Nuclear receptor corepressor complexes in cancer: Mechanism, function and regulation. Am. J. Clin. Exp. Urol. 2014, 2, 169–187. [Google Scholar] [PubMed]
- Patel, H.K.; Bihani, T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol. Ther. 2018, 186, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Bagchi, M.K. Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J. Biol. Chem. 2004, 279, 15050–15058. [Google Scholar] [CrossRef]
- Berrevoets, C.A.; Umar, A.; Brinkmann, A.O. Antiandrogens: Selective androgen receptor modulators. Mol. Cell Endocrinol. 2002, 198, 97–103. [Google Scholar] [CrossRef]
- Ishii, S.; Yamada, M.; Satoh, T.; Monden, T.; Hashimoto, K.; Shibusawa, N.; Onigata, K.; Morikawa, A.; Mori, M. Aberrant dynamics of histone deacetylation at the thyrotropin-releasing hormone gene in resistance to thyroid hormone. Mol. Endocrinol. 2004, 18, 1708–1720. [Google Scholar] [CrossRef][Green Version]
- Dong, X.; Lutz, W.; Schroeder, T.M.; Bachman, L.A.; Westendorf, J.J.; Kumar, R.; Griffin, M.D. Regulation of relB in dendritic cells by means of modulated association of vitamin D receptor and histone deacetylase 3 with the promoter. Proc. Natl. Acad. Sci. USA 2005, 102, 16007–16012. [Google Scholar] [CrossRef]
- Giguère, V.; Yang, N.; Segui, P.; Evans, R.M. Identification of a new class of steroid hormone receptors. Nature 1988, 331, 91–94. [Google Scholar] [CrossRef]
- Villena, J.A.; Hock, M.B.; Chang, W.Y.; Barcas, J.E.; Giguère, V.; Kralli, A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 1418–1423. [Google Scholar] [CrossRef]
- Schreiber, S.N.; Knutti, D.; Brogli, K.; Uhlmann, T.; Kralli, A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha). J. Biol. Chem. 2003, 278, 9013–9018. [Google Scholar] [CrossRef]
- Lerin, C.; Rodgers, J.T.; Kalume, D.E.; Kim, S.H.; Pandey, A.; Puigserver, P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006, 3, 429–438. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Somech, R.; Shaklai, S.; Geller, O.; Amariglio, N.; Simon, A.J.; Rechavi, G.; Gal-Yam, E.N. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005, 118, 4017–4025. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, S.L.; Janardhan, H.P.; Trivedi, C.M. Histone Deacetylase 3 Coordinates Deacetylase-independent Epigenetic Silencing of Transforming Growth Factor-β1 (TGF-β1) to Orchestrate Second Heart Field Development. J. Biol. Chem. 2015, 290, 27067–27089. [Google Scholar] [CrossRef]
- Nguyen, H.C.B.; Adlanmerini, M.; Hauck, A.K.; Lazar, M.A. Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature 2020, 584, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.F.; Fischle, W.; Verdin, E.; Greene, W.C. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 2001, 293, 1653–1657. [Google Scholar] [CrossRef]
- Zhang, F.; Qi, L.; Feng, Q.; Zhang, B.; Li, X.; Liu, C.; Li, W.; Liu, Q.; Yang, D.; Yin, Y.; et al. HIPK2 phosphorylates HDAC3 for NF-κB acetylation to ameliorate colitis-associated colorectal carcinoma and sepsis. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Thevenet, L.; Méjean, C.; Moniot, B.; Bonneaud, N.; Galéotti, N.; Aldrian-Herrada, G.; Poulat, F.; Berta, P.; Benkirane, M.; Boizet-Bonhoure, B. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO J. 2004, 23, 3336–3345. [Google Scholar] [CrossRef]
- Takami, Y.; Nakayama, T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J. Biol. Chem. 2000, 275, 16191–16201. [Google Scholar] [CrossRef]
- Yang, W.M.; Tsai, S.C.; Wen, Y.D.; Fejer, G.; Seto, E. Functional domains of histone deacetylase-3. J. Biol. Chem. 2002, 277, 9447–9454. [Google Scholar] [CrossRef]
- Gao, Z.; He, Q.; Peng, B.; Chiao, P.J.; Ye, J. Regulation of nuclear translocation of HDAC3 by IkappaBalpha is required for tumor necrosis factor inhibition of peroxisome proliferator-activated receptor gamma function. J. Biol. Chem. 2006, 281, 4540–4547. [Google Scholar] [CrossRef]
- Bacon, T.; Seiler, C.; Wolny, M.; Hughes, R.; Watson, P.; Schwabe, J.; Grigg, R.; Peckham, M. Histone deacetylase 3 indirectly modulates tubulin acetylation. Biochem. J. 2015, 472, 367–377. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishii, S. The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action. Int. J. Mol. Sci. 2021, 22, 9138. https://doi.org/10.3390/ijms22179138
Ishii S. The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action. International Journal of Molecular Sciences. 2021; 22(17):9138. https://doi.org/10.3390/ijms22179138
Chicago/Turabian StyleIshii, Sumiyasu. 2021. "The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action" International Journal of Molecular Sciences 22, no. 17: 9138. https://doi.org/10.3390/ijms22179138
APA StyleIshii, S. (2021). The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action. International Journal of Molecular Sciences, 22(17), 9138. https://doi.org/10.3390/ijms22179138





