GhTBL34 Is Associated with Verticillium Wilt Resistance in Cotton
Abstract
:1. Introduction
2. Results
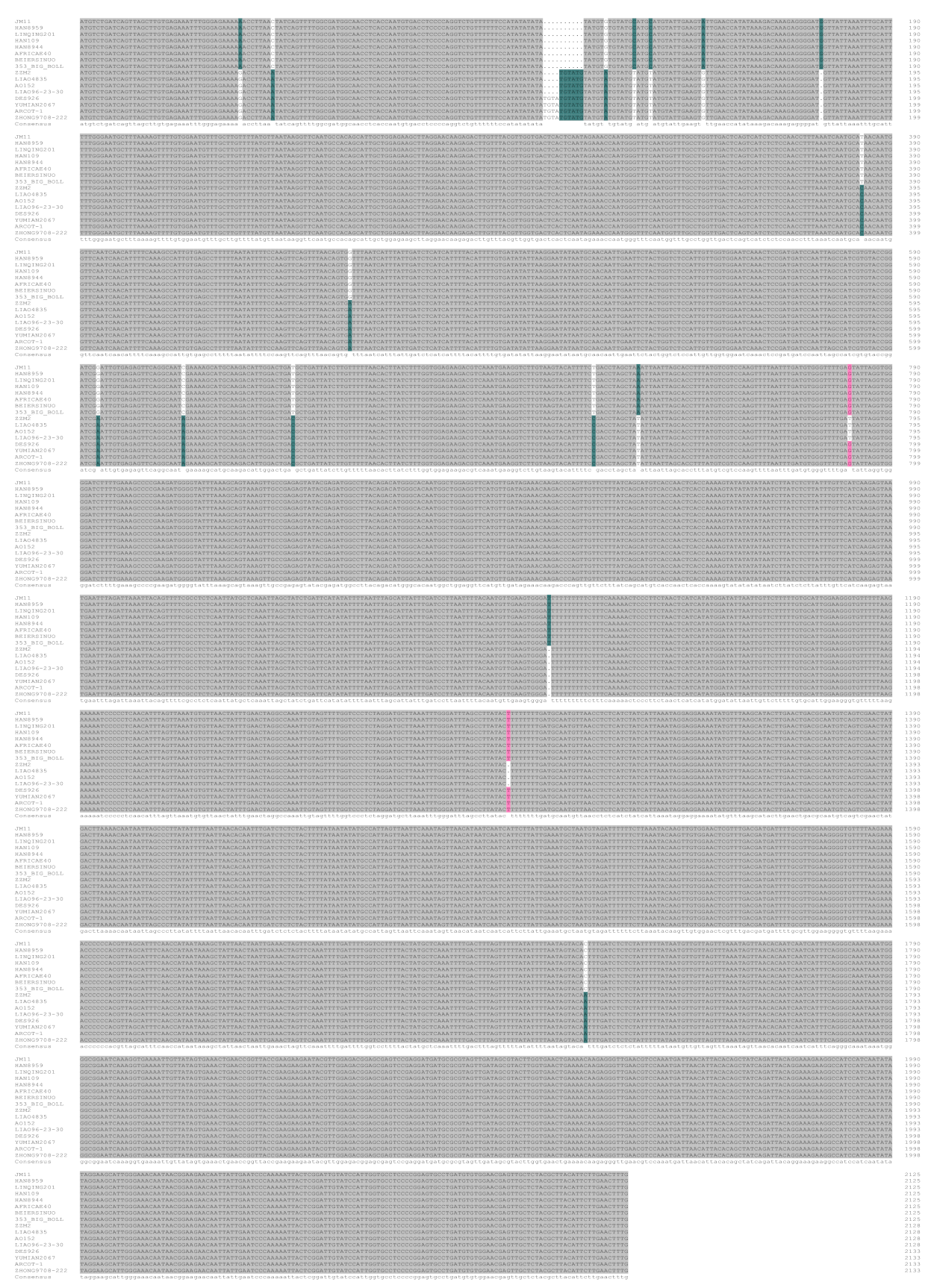
2.1. Analysis of Sequence Variation of GhTBL34 and Detection of Elite Alleles Related to VW Resistance in Cotton
2.2. Expression Pattern of Cotton TBL34
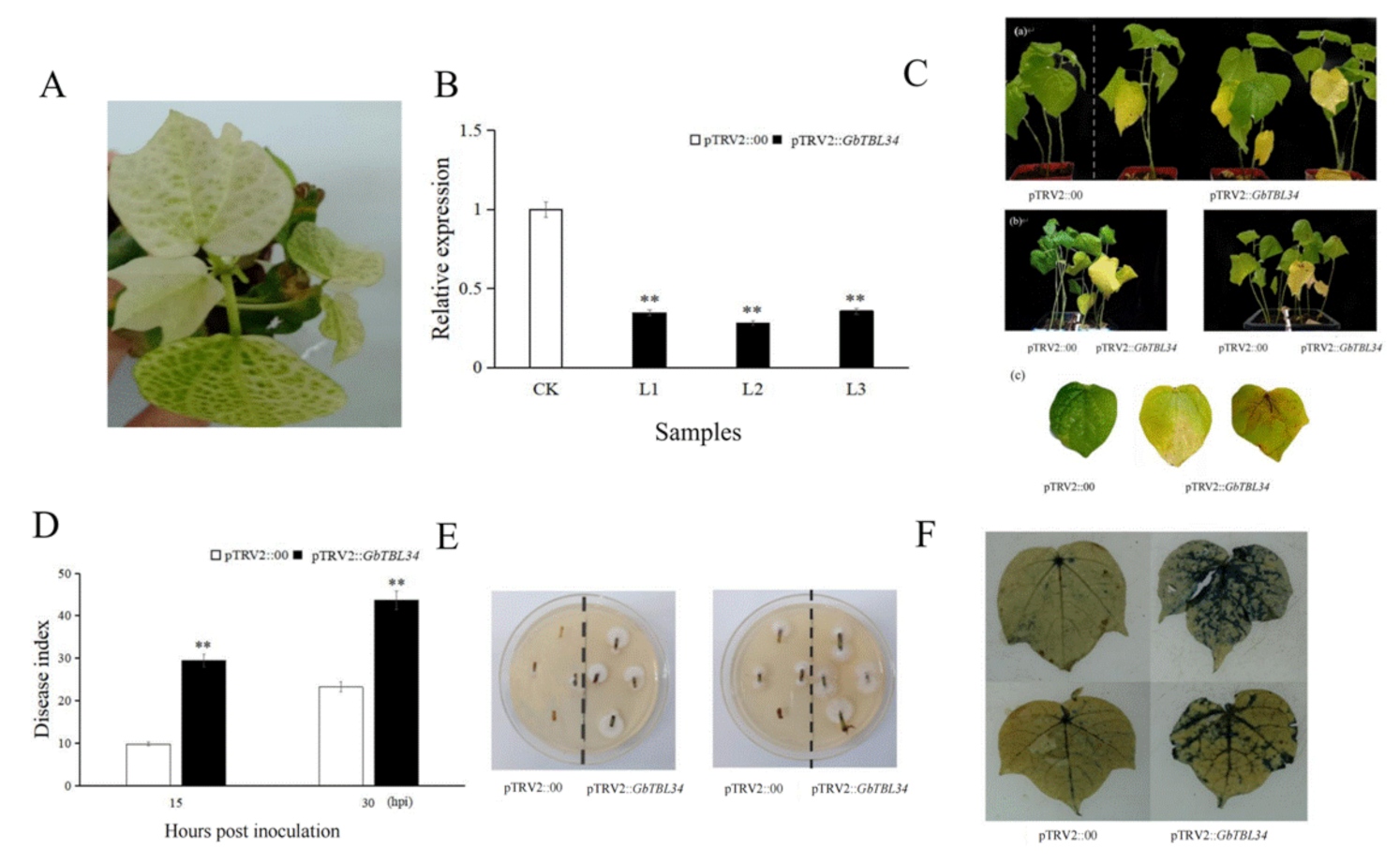
2.3. Silencing of TBL34 Reduced VW Resistance in Cotton
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
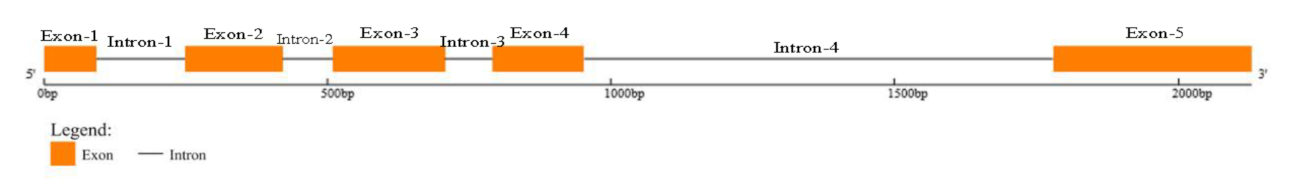
4.2. Cloning the gDNA and cDNA of TBL34 Gene in Cotton
4.3. V. dahliae Materials and Inoculation Methods
4.4. Hormone Processing and Sampling
4.5. Vector Construction for Virus-Induced Gene Silencing (VIGS) in Cotton and VIGS Experiments
4.6. Morbidity Situation Analysis
4.7. Trypan Blue Test
4.8. qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhang, J.F.; Fang, H.; Zhou, H.P.; Sanogo, S.; Ma, Z.Y. Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 2014, 54, 1289–1303. [Google Scholar] [CrossRef]
- Zhou, T.; Zhao, Y.; Guo, H. Secretory proteins are delivered to the septin-organized penetration interface during root infection by Verticillium dahliae. PLoS Pathog. 2017, 13, e1006275. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, W.; Cui, Y.; Sang, X.; Lu, J.; Jing, H.; Wang, W.; Zhao, P.; Wang, H. Detection of candidate genes and development of KASP markers for Verticillium wilt resistance by combining genome-wide association study, QTL-seq and transcriptome sequencing in cotton. Theor. Appl. Genet. 2021, 134, 1063–1081. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Cai, Y.; Gou, J.Y.; Mao, Y.B.; Xu, Y.H.; Jiang, W.H.; Chen, X.Y. Vdnep, an elicitor from verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microbiol. 2004, 70, 4989–4995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huckelhoven, R. Cell wall–associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.F.; Bell, A.A.; Dighe, N.D.; Menz, M.A.; Nichols, R.L.; Stelly, D.M. Introgression of resistance to nematode rotylenchulus reniformis into upland cotton (Gossypium hirsutum) from Gossypium longicalyx. Crop Sci. 2007, 47, 1865–1877. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, L.F.; Tu, L.L.; Liu, L.L.; Yuan, D.J.; Jin, L.; Long, L.; Zhang, X.L. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [Green Version]
- Cantu, D.; Vicente, A.R.; Greve, L.C.; Dewey, F.M.; Bennett, A.B.; Labavitch, J.M.; Powell, A.L.T. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 2008, 105, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Sun, Z.X.; Zhu, J.; Xu, T.; Gary, E.H.; Matteo, L.; Sheri, W. Rice transformation with cell wall degrading enzyme genes from Trichoderma atroviride and its effect on plant growth and resistance to fungal pathogens. J. Zhejiang Univ. 2004, 30, 447. [Google Scholar]
- Toshihisa, K.; Tsutomu, A.; Ko, H.; Ami, S.; Yasuko, K.; Yoichi, T.; Hiroshi, T.; Shinji, K. Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 2011, 62, 2053–2062. [Google Scholar]
- Ono, E.; Wong, H.L.; Kawasaki, T. Essential role of the small gtpase rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef]
- Akamatsu, A.; Wong, H.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y. An oscebip/oscerk1-osracgef1-osrac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Gille, S.; Pauly, M. O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 2012, 12, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Manabe, Y.; Nafisi, M.; Verhertbruggen, Y.; Orfila, C.; Gille, S.; Rautengarten, C.; Cherk, C.; Marcus, S.E.; Somerville, S.; Pauly, M.; et al. Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011, 155, 1068–1078. [Google Scholar]
- Bischoff, V.; Joachim, S.; Wolf-Rudiger, S. Involvement of TBL/DUF231 proteins into cell wall biology. Plant Signal. Behav. 2010, 5, 1057–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultink, A.; Naylor, D.; Dama, M.; Pauly, M. The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol. 2015, 167, 1271–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.; Morrison, S.; McInerney, P.; Hadi, M.Z.; et al. Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.X.; Teng, Q.; Zhong, R.Q.; Ye, Z.H. The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O-and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol. 2013, 54, 1186–1199. [Google Scholar] [CrossRef]
- Gao, Y.P.; He, C.W.; Zhang, D.M.; Liu, X.L.; Xu, Z.P.; Tian, Y.B.; Liu, X.H.; Zang, S.S.; Pauly, M.; Zhou, Y.H.; et al. Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 2017, 173, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Xin, Z.G.; Browse, J. eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 1998, 95, 7799–7804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.F.; Sun, Y.; Zhang, B.C.; Mansoori, N.; Wan, J.X.; Liu, Y.; Wang, Z.W.; Shi, Y.Z.M.; Zhou, Y.H.; Zheng, S.J. TRICHOME BIREFRINGENCE-LIKE27 affects aluminum sensitivity by modulating the O-Acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol. 2014, 166, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.X.; Teng, Q.; Zhong, R.Q.; Ye, Z.H. Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci. 2016, 243, 120–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, B.X.; Zhao, Y.L.; Chen, W.; Wang, H.M.; Guo, Z.J.; Gong, H.Y.; Sang, X.H.; Cui, Y.L.; Wang, C.H. Analysis of upland cotton (Gossypium hirsutum) response to Verticillium dahliae inoculation by transcriptome sequencing. Genet. Mol. Res. 2015, 14, 13120–13130. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Zuo, K.; Wang, J.; Wu, W.; Chai, Y.; Sun, X.; Tang, K. Identification and characterization of differentially expressed ESTs of Gossypium barbadense infected by Verticillium dahliae with suppression subtractive hybridization. Mol. Biol. 2005, 39, 191–199. [Google Scholar] [CrossRef]
- Zuo, K.J.; Qin, J.; Zhao, J.Y.; Ling, H.; Zhang, L.D.; Cao, Y.F.; Tang, K.X. Overexpression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene 2007, 391, 80–90. [Google Scholar] [CrossRef]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Bioch. 2018, 125, 193–204. [Google Scholar] [CrossRef]
- Mo, H.J.; Sun, Y.X.; Zhu, X.L.; Wang, X.F.; Zhang, Y.; Yang, J.; Yan, G.J.; Ma, Z.Y. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 2016, 243, 1023–1039. [Google Scholar] [CrossRef]
- Ding, L.N.; Xu, H.B.; Yi, H.Y.; Yang, L.M.; Kong, Z.X.; Zhang, L.X.; Xue, S.L.; Jia, H.Y.; Ma, Z.Q. Resistance to hemi-biotrophic F-graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS ONE 2011, 6, e19008. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhu, L.; Zhang, X. Research on resistance mechanism of cotton to Verticillium wilt. Acta Agron. Sin. 2012, 38, 1553–1560. [Google Scholar] [CrossRef]
- Guo, R.Y.; Yu, F.F.; Gao, Z.; An, H.L.; Cao, X.C.; Guo, X.Q. GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol. Biol. Rep. 2011, 38, 49–58. [Google Scholar] [CrossRef]
- Luo, X.; Mao, H.; Wei, Y.; Cai, J.; Xie, C.; Sui, A.; Yang, X.; Dong, J. The fungal specific transcription factor Vdpf influences conidia production, melanized microsclerotia formation and pathogenicity in Verticillium dahliae. Mol. Plant Pathol. 2016, 17, 1364–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, D.; Rajendran, S.R.C.K.; Gaur, M.; Sajeesh, P.K.; Kumar, A. Plant defense signaling and responses against necrotrophic fungal pathogens. J. Plant Growth Regul. 2016, 35, 1159–1174. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Ye, Z.H. Regiospecific Acetylation of Xylan is Mediated by a Group of DUF231-Containing O-Acetyltransferases. Plant Cell Physiol. 2017, 58, 2126–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, H.; Chen, W.; Li, Y. Genetic structure, linkage disequilibrium and association mapping of Verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PLoS ONE 2014, 9, e86308. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Jing, H.; Zhao, P.; Chen, W.; Li, X.; Sang, X.; Lu, J.; Wang, H. GhTBL34 Is Associated with Verticillium Wilt Resistance in Cotton. Int. J. Mol. Sci. 2021, 22, 9115. https://doi.org/10.3390/ijms22179115
Zhao Y, Jing H, Zhao P, Chen W, Li X, Sang X, Lu J, Wang H. GhTBL34 Is Associated with Verticillium Wilt Resistance in Cotton. International Journal of Molecular Sciences. 2021; 22(17):9115. https://doi.org/10.3390/ijms22179115
Chicago/Turabian StyleZhao, Yunlei, Huijuan Jing, Pei Zhao, Wei Chen, Xuelin Li, Xiaohui Sang, Jianhua Lu, and Hongmei Wang. 2021. "GhTBL34 Is Associated with Verticillium Wilt Resistance in Cotton" International Journal of Molecular Sciences 22, no. 17: 9115. https://doi.org/10.3390/ijms22179115
APA StyleZhao, Y., Jing, H., Zhao, P., Chen, W., Li, X., Sang, X., Lu, J., & Wang, H. (2021). GhTBL34 Is Associated with Verticillium Wilt Resistance in Cotton. International Journal of Molecular Sciences, 22(17), 9115. https://doi.org/10.3390/ijms22179115





