Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives
Abstract
1. Hepatocellular Carcinoma—Epidemiology, Pathophysiology, Risk Factors and Treatments
2. Innate Lymphoid Cells—Hepatic NK Cells and ILC1
3. Diversity within Hepatic NK Cell and ILC1 Subsets
4. Liver Immunity—An Immunotolerant Organ
5. NK Cells in HCC
6. NK Cell Biology
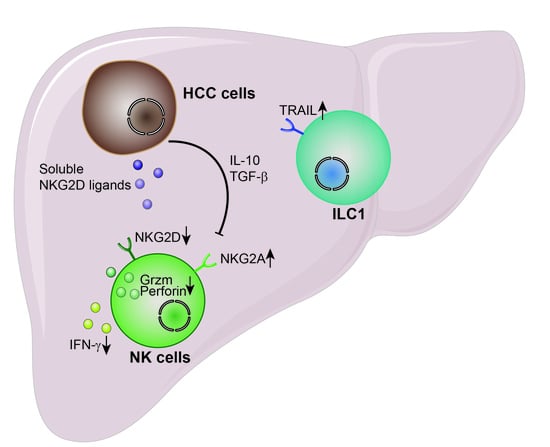
7. NK Cell Dysfunction in HCC
8. Therapeutic Intervention
9. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.J.; von Felden, J.; Garcia-Lezana, T.; Sarcognato, S.; Villanueva, A. Tumour evolution in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Zhai, W.; Lim, T.K.-H.; Zhang, T.; Phang, S.-T.; Tiang, Z.; Guan, P.; Ng, M.-H.; Lim, J.Q.; Yao, F.; Li, Z.; et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 2017, 8, 4565. [Google Scholar] [CrossRef]
- Pok, S.; Barn, V.A.; Wong, H.J.; Blackburn, A.C.; Board, P.; Farrell, G.C.; Teoh, N.C. Testosterone regulation of cyclin E kinase: A key factor in determining gender differences in hepatocarcinogenesis. J. Gastroenterol. Hepatol. 2016, 31, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Simon, T.G.; Duberg, A.-S.; Aleman, S.; Chung, R.T.; Chan, A.T.; Ludvigsson, J.F. Association of Aspirin with Hepatocellular Carcinoma and Liver-Related Mortality. N. Engl. J. Med. 2020, 382, 1018–1028. [Google Scholar] [CrossRef]
- Llovet, J.M.; De Baere, T.; Kulik, L.; Haber, P.K.; Greten, T.F.; Meyer, T.; Lencioni, R. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 293–313. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Bruix, J.; Qin, S.; Merle, P.; Granito, A.; Huang, Y.-H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Breder, V.; et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 389, 56–66. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Meyer, T.; Cheng, A.-L.; El-Khoueiry, A.B.; Rimassa, L.; Ryoo, B.-Y.; Cicin, I.; Merle, P.; Chen, Y.; Park, J.-W.; et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N. Engl. J. Med. 2018, 379, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Kang, Y.-K.; Yen, C.-J.; Finn, R.S.; Galle, P.R.; Llovet, J.M.; Assenat, E.; Brandi, G.; Pracht, M.; Lim, H.Y.; et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019, 20, 282–296. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Hsu, C.; Chan, S.L.; Choo, S.-P.; Kudo, M. Challenges of combination therapy with immune checkpoInt. inhibitors for hepatocellular carcinoma. J. Hepatol. 2020, 72, 307–319. [Google Scholar] [CrossRef]
- Finn, R.S.; Ikeda, M.; Zhu, A.X.; Sung, M.W.; Baron, A.D.; Kudo, M.; Okusaka, T.; Kobayashi, M.; Kumada, H.; Kaneko, S.; et al. Phase Ib Study of Lenvatinib Plus Pembrolizumab in Patients With Unresectable Hepatocellular Carcinoma. J. Clin. Oncol. 2020, 38, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Finn, R.S.; Edeline, J.; Cattan, S.; Ogasawara, S.; Palmer, D.; Verslype, C.; Zagonel, V.; Fartoux, L.; Vogel, A.; et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): A non-randomised, open-label phase 2 trial. Lancet Oncol. 2018, 19, 940–952. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Foerster, F.; Hess, M.; Gerhold-Ay, A.; Marquardt, J.U.; Becker, D.; Galle, P.R.; Schuppan, D.; Binder, H.; Bockamp, E. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci. Rep. 2018, 8, 5351. [Google Scholar] [CrossRef]
- Gabrielson, A.; Wu, Y.; Wang, H.; Jiang, J.; Kallakury, B.; Gatalica, Z.; Reddy, S.; Kleiner, D.; Fishbein, T.; Johnson, L.; et al. Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol. Res. 2016, 4, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, L.; Huang, Q.; Liu, H.; Huang, M.; Wang, J.; Wen, H.; Lin, R.; Qu, K.; Li, K.; et al. Accumulation of Tumor-Infiltrating CD49a+ NK Cells Correlates with Poor Prognosis for Human Hepatocellular Carcinoma. Cancer Immunol. Res. 2019, 7, 1535–1546. [Google Scholar] [CrossRef]
- Nguyen, P.H.D.; Ma, S.; Phua, C.Z.J.; Kaya, N.A.; Lai, H.L.H.; Lim, C.J.; Lim, J.Q.; Wasser, M.; Lai, L.; Tam, W.L.; et al. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nat. Commun. 2021, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Chen, M.-K.; Zhang, Q.-F.; Zhou, Y.-F.; Zhang, M.-Y.; Mai, S.-J.; Zhang, Y.-J.; Chen, M.-S.; Li, X.-X.; Wang, H.-Y. Identification of Immunol.ogical subtypes of hepatocellular carcinoma with expression profiling of immune-modulating genes. Aging 2020, 12, 12187–12205. [Google Scholar] [CrossRef]
- Ma, J.; Zheng, B.; Goswami, S.; Meng, L.; Zhang, D.; Cao, C.; Li, T.; Zhu, F.; Ma, L.; Zhang, Z.; et al. PD1Hi CD8+ T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 331. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Liu, H.; Lan, T.; Xu, L.; Wang, G.; Yuan, K.; Wu, H. Existence of intratumoral tertiary lymphoid structures is associated with immune cells infiltration and predicts better prognosis in early-stage hepatocellular carcinoma. Aging 2020, 12, 3451–3472. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Lin, J.; Weng, Y.; Zeng, D.-N.; Xu, J.; Luo, S.; Xu, L.; Liu, M.; Hua, Q.; Liu, C.-Q.; et al. Myeloid signature reveals immune contexture and predicts the prognosis of hepatocellular carcinoma. J. Clin. Investig. 2020, 130, 4679–4693. [Google Scholar] [CrossRef]
- Cariani, E.; Missale, G. Immune landscape of hepatocellular carcinoma microenvironment: Implications for prognosis and therapeutic applications. Liver Int. 2019, 39, 1608–1621. [Google Scholar] [CrossRef]
- Juengpanich, S.; Shi, L.; Iranmanesh, Y.; Chen, J.; Cheng, Z.; Khoo, A.K.-J.; Pan, L.; Wang, Y.; Cai, X. The role of natural killer cells in hepatocellular carcinoma development and treatment: A narrative review. Transl. Oncol. 2019, 12, 1092–1107. [Google Scholar] [CrossRef]
- Mikulak, J.; Bruni, E.; Oriolo, F.; Di Vito, C.; Mavilio, D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front. Immunol. 2019, 10, 946. [Google Scholar] [CrossRef]
- Vivier, E.; van de Pavert, S.A.; Cooper, M.D.; Belz, G.T. The evolution of innate lymphoid cells. Nat. Immunol. 2016, 17, 790–794. [Google Scholar] [CrossRef]
- Spits, H.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 2013, 13, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Carpentier, S.; Vivier, E.; Belz, G.T. Innate lymphoid cells: Parallel checkpoints and coordinate interactions with T cells. Curr. Opin. Immunol. 2016, 38, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.F.; Jacquelot, N.; Belz, G.T. Deconstructing deployment of the innate immune lymphocyte army for barrier homeostasis and protection. Immunol. Rev. 2018, 286, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; Cornen, S.; Daëron, M.; Morel, Y.; Medzhitov, R.; Vivier, E. Harnessing innate immunity in cancer therapy. Nature 2019, 574, 45–56. [Google Scholar] [CrossRef]
- Ebbo, M.; Crinier, A.; Vély, F.; Vivier, E. Innate lymphoid cells: Major players in inflammatory diseases. Nat. Rev. Immunol. 2017, 17, 665–678. [Google Scholar] [CrossRef]
- Seillet, C.; Jacquelot, N. Sensing of physiological regulators by innate lymphoid cells. Cell Mol. Immunol. 2019, 16, 442–451. [Google Scholar] [CrossRef]
- Guia, S.; Fenis, A.; Vivier, E.; Narni-Mancinelli, E. Activating and inhibitory receptors expressed on innate lymphoid cells. Semin. Immunopathol. 2018, 40, 331–341. [Google Scholar] [CrossRef]
- Mariotti, F.R.; Quatrini, L.; Munari, E.; Vacca, P.; Moretta, L. Innate Lymphoid Cells: Expression of PD-1 and Other Checkpoints in Normal and Pathological Conditions. Front. Immunol. 2019, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Herberman, R.B.; Nunn, M.E.; Holden, H.T.; Lavrin, D.H. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int. J. Cancer 1975, 16, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Herberman, R.B.; Nunn, M.E.; Lavrin, D.H. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int. J. Cancer 1975, 16, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, R.; Klein, E.; Wigzell, H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur. J. Immunol. 1975, 5, 112–117. [Google Scholar] [CrossRef]
- Kiessling, R.; Klein, E.; Pross, H.; Wigzell, H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 1975, 5, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Pross, H.F.; Baines, M.G. Spontaneous human lymphocyte-mediated cytotoxicity againts tumour target cells. I. The effect of malignant disease. Int. J. Cancer 1976, 18, 593–604. [Google Scholar] [CrossRef]
- Lanier, L.L.; Le, A.M.; Civin, C.I.; Loken, M.R.; Phillips, J.H. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J. Immunol. 1986, 136, 4480–4486. [Google Scholar]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Kim, S.; Iizuka, K.; Kang, H.-S.P.; Dokun, A.; French, A.R.; Greco, S.; Yokoyama, W.M. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 2002, 3, 523–528. [Google Scholar] [CrossRef]
- Chiossone, L.; Chaix, J.; Fuseri, N.; Roth, C.; Vivier, E.; Walzer, T. Maturation of mouse NK cells is a 4-stage developmental program. Blood 2009, 113, 5488–5496. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Smyth, M.J. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J. Immunol. 2006, 176, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, C.; Juelke, K.; Falco, M.; Morandi, B.; D’Agostino, A.; Costa, R.; Ratto, G.; Forte, G.; Carrega, P.; Lui, G.; et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J. Immunol. 2007, 178, 4947–4955. [Google Scholar] [CrossRef]
- Crinier, A.; Milpied, P.; Escalière, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49, 971–986.e5. [Google Scholar] [CrossRef]
- Takeda, K.; Cretney, E.; Hayakawa, Y.; Ota, T.; Akiba, H.; Ogasawara, K.; Yagita, H.; Kinoshita, K.; Okumura, K.; Smyth, M.J. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 2005, 105, 2082–2089. [Google Scholar] [CrossRef]
- Peng, H.; Jiang, X.; Chen, Y.; Sojka, D.K.; Wei, H.; Gao, X.; Sun, R.; Yokoyama, W.M.; Tian, Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 2013, 123, 1444–1456. [Google Scholar] [CrossRef]
- Forkel, M.; Berglin, L.; Kekäläinen, E.; Carlsson, A.; Svedin, E.; Michaëlsson, J.; Nagasawa, M.; Erjefält, J.S.; Mori, M.; Flodström-Tullberg, M.; et al. Composition and functionality of the intrahepatic innate lymphoid cell-compartment in human nonfibrotic and fibrotic livers. Eur. J. Immunol. 2017, 47, 1280–1294. [Google Scholar] [CrossRef]
- Ferlazzo, G.; Thomas, D.; Lin, S.-L.; Goodman, K.; Morandi, B.; Muller, W.A.; Moretta, A.; Münz, C. The Abundant NK Cells in Human Secondary Lymphoid Tissues Require Activation to Express Killer Cell Ig-Like Receptors and Become Cytolytic. J. Immunol. 2004, 172, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Fehniger, T.A.; Cooper, M.A.; Nuovo, G.J.; Cella, M.; Facchetti, F.; Colonna, M.; Caligiuri, M.A. CD56brightnatural killer cells are present in human lymph nodes and are activated by T cell–derived IL-2: A potential new link between adaptive and innate immunity. Blood 2003, 101, 3052–3057. [Google Scholar] [CrossRef]
- Cuff, A.O.; Robertson, F.P.; Stegmann, K.A.; Pallett, L.J.; Maini, M.K.; Davidson, B.R.; Male, V. Eomeshi NK Cells in Human Liver Are Long-Lived and Do Not Recirculate but Can Be Replenished from the Circulation. J. Immunol. 2016, 197, 4283–4291. [Google Scholar] [CrossRef] [PubMed]
- Hudspeth, K.; Donadon, M.; Cimino, M.; Pontarini, E.; Tentorio, P.; Preti, M.; Hong, M.; Bertoletti, A.; Bicciato, S.; Invernizzi, P.; et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J. Autoimmun. 2016, 66, 40–50. [Google Scholar] [CrossRef]
- Stegmann, K.A.; Robertson, F.; Hansi, N.; Gill, U.; Pallant, C.; Christophides, T.; Pallett, L.J.; Peppa, D.; Dunn, C.; Fusai, G.; et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci. Rep. 2016, 6, 26157. [Google Scholar] [CrossRef]
- Harmon, C.; Robinson, M.W.; Fahey, R.; Whelan, S.; Houlihan, D.D.; Geoghegan, J.; O’Farrelly, C. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur. J. Immunol. 2016, 46, 2111–2120. [Google Scholar] [CrossRef]
- Hwang, S.; Han, J.; Baek, J.-S.; Tak, E.; Song, G.-W.; Lee, S.-G.; Jung, D.-H.; Park, G.-C.; Ahn, C.-S.; Kim, N. Cytotoxicity of Human Hepatic Intrasinusoidal CD56bright Natural Killer Cells against Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2019, 20, 1564. [Google Scholar] [CrossRef]
- Harmon, C.; Jameson, G.; Almuaili, D.; Houlihan, D.D.; Hoti, E.; Geoghegan, J.; Robinson, M.W.; O’Farrelly, C. Liver-Derived TGF-β Maintains the EomeshiTbetlo Phenotype of Liver Resident Natural Killer Cells. Front. Immunol. 2019, 10, 1502. [Google Scholar] [CrossRef]
- Marquardt, N.; Béziat, V.; Nyström, S.; Hengst, J.; Ivarsson, M.A.; Kekäläinen, E.; Johansson, H.; Mjösberg, J.; Westgren, M.; Lankisch, T.O.; et al. Cutting Edge: Identification and Characterization of Human Intrahepatic CD49a+ NK Cells. J. Immunol. 2015, 194, 2467–2471. [Google Scholar] [CrossRef] [PubMed]
- Aw Yeang, H.X.; Piersma, S.J.; Lin, Y.; Yang, L.; Malkova, O.N.; Miner, C.; Krupnick, A.S.; Chapman, W.C.; Yokoyama, W.M. Cutting Edge: Human CD49e- NK Cells Are Tissue Resident in the Liver. J. Immunol. 2017, 198, 1417–1422. [Google Scholar] [CrossRef]
- Filipovic, I.; Sönnerborg, I.; Strunz, B.; Friberg, D.; Cornillet, M.; Hertwig, L.; Ivarsson, M.A.; Björkström, N.K. 29-Color Flow Cytometry: Unraveling Human Liver NK Cell Repertoire Diversity. Front. Immunol. 2019, 10, 2692. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Peng, H.; Zhou, J.; Chen, Y.; Wei, H.; Sun, R.; Yokoyama, W.M.; Tian, Z. Differential phenotypic and functional properties of liver-resident NK cells and Mucosal. ILC1s. J. Autoimmun. 2016, 67, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Daussy, C.; Faure, F.; Mayol, K.; Viel, S.; Gasteiger, G.; Charrier, E.; Bienvenu, J.; Henry, T.; Debien, E.; Hasan, U.A.; et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014, 211, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Seillet, C.; Huntington, N.D.; Gangatirkar, P.; Axelsson, E.; Minnich, M.; Brady, H.J.M.; Busslinger, M.; Smyth, M.J.; Belz, G.T.; Carotta, S. Differential Requirement for Nfil3 during NK Cell Development. J. Immunol. 2014, 192, 2667–2676. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.K.; Plougastel-Douglas, B.; Yang, L.; Pak-Wittel, M.A.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B.; Yu, J.; et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife 2014, 3, e01659. [Google Scholar] [CrossRef] [PubMed]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.M.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D.; et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [PubMed]
- Aizarani, N.; Saviano, A.; Mailly, L.; Durand, S.; Herman, J.S.; Pessaux, P.; Saviano, A.; Baumert, T.F.; Grün, D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019, 572, 199–204. [Google Scholar] [CrossRef]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, H.; Wan, L.; Wang, Z.; Wang, H.; Ge, C.; Liu, Y.; Hao, Y.; Zhang, D.; Shi, G.; et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020, 73, 1118–1130. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Liu, Y.; He, X.; Qu, M.; Xu, G.; Wang, H.; Huang, M.; Pan, J.; Liu, Z.; et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Payen, V.L.; Lavergne, A.; Alevra Sarika, N.; Colonval, M.; Karim, L.; Deckers, M.; Najimi, M.; Coppieters, W.; Charloteaux, B.; Sokal, E.M.; et al. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021, 3, 100278. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Toth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labor in the mammalian liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef]
- Chen, T.; Oh, S.; Gregory, S.; Shen, X.; Diehl, A.M. Single-cell omics analysis reveals functional diversification of hepatocytes during liver regeneration. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.S.; Atif, J.; Liu, J.C.; Perciani, C.T.; Ma, X.-Z.; Thoeni, C.; Slyper, M.; Eraslan, G.; Segerstolpe, A.; Manuel, J.; et al. Single Cell, Single Nucleus and Spatial RNA Sequencing of the Human Liver Identifies Hepatic Stellate Cell and Cholangiocyte Heterogeneity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Massalha, H.; Bahar Halpern, K.; Abu-Gazala, S.; Jana, T.; Massasa, E.E.; Moor, A.E.; Buchauer, L.; Rozenberg, M.; Pikarsky, E.; Amit, I.; et al. A single cell atlas of the human liver tumor microenvironment. Mol. Syst. Biol. 2020, 16, e9682. [Google Scholar] [CrossRef]
- Jameson, G.; Robinson, M.W. Insights Into Human Intrahepatic NK Cell Function From Single Cell RNA Sequencing Datasets. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Jiao, Y.; Huntington, N.D.; Belz, G.T.; Seillet, C. Type 1 Innate Lymphoid Cell Biology: Lessons Learnt from Natural Killer Cells. Front. Immunol. 2016, 7, 426. [Google Scholar] [CrossRef]
- Seillet, C.; Brossay, L.; Vivier, E. Natural killers or ILC1s? That is the question. Curr. Opin. Immunol. 2021, 68, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Vienne, M.; Tang, L.; Kerdiles, Y.; Etiennot, M.; Escalière, B.; Galluso, J.; Wei, H.; Sun, R.; Vivier, E.; et al. Liver type 1 innate lymphoid cells develop locally via an interferon-γ-dependent loop. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Constantinides, M.G.; McDonald, B.D.; Verhoef, P.A.; Bendelac, A. A committed hemopoietic precursor to innate lymphoid cells. Nature 2014, 508, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.S.N.; Flach, M.; Möhle, L.; Rogell, L.; Hoyler, T.; Ebert, K.; Fabiunke, C.; Pfeifer, D.; Sexl, V.; Fonseca-Pereira, D.; et al. Differentiation of Type 1 ILCs from a Common Progenitor to All Helper-like Innate Lymphoid Cell Lineages. Cell 2014, 157, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.A.; Wong, P.; Schappe, T.; Berrien-Elliott, M.M.; Cubitt, C.; Jaeger, N.; Lee, M.; Keppel, C.R.; Marin, N.D.; Foltz, J.A.; et al. Stage-Specific Requirement for Eomes in Mature NK Cell Homeostasis and Cytotoxicity. Cell Rep. 2020, 31, 107720. [Google Scholar] [CrossRef] [PubMed]
- Pikovskaya, O.; Chaix, J.; Rothman, N.J.; Collins, A.; Chen, Y.-H.; Scipioni, A.M.; Vivier, E.; Reiner, S.L. Cutting Edge: Eomesodermin Is Sufficient To Direct Type 1 Innate Lymphocyte Development into the Conventional NK Lineage. J. Immunol. 2016, 196, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Seillet, C.; Rankin, L.C.; Groom, J.R.; Mielke, L.A.; Tellier, J.; Chopin, M.; Huntington, N.D.; Belz, G.T.; Carotta, S. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 2014, 211, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, D.M.; Long, E.; Veiga-Fernandes, H.; de Boer, J.; Williams, O.; Seddon, B.; Coles, M.; Kioussis, D.; Brady, H.J.M. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nature Immunol. 2009, 10, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Kamizono, S.; Duncan, G.S.; Seidel, M.G.; Morimoto, A.; Hamada, K.; Grosveld, G.; Akashi, K.; Lind, E.F.; Haight, J.P.; Ohashi, P.S.; et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 2009, 206, 2977–2986. [Google Scholar] [CrossRef] [PubMed]
- Firth, M.A.; Madera, S.; Beaulieu, A.M.; Gasteiger, G.; Castillo, E.F.; Schluns, K.S.; Kubo, M.; Rothman, P.B.; Vivier, E.; Sun, J.C. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med. 2013, 210, 2981–2990. [Google Scholar] [CrossRef]
- Cortez, V.S.; Fuchs, A.; Cella, M.; Gilfillan, S.; Colonna, M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 2014, 192, 4487–4491. [Google Scholar] [CrossRef]
- Fuchs, A.; Vermi, W.; Lee, J.S.; Lonardi, S.; Gilfillan, S.; Newberry, R.D.; Cella, M.; Colonna, M. Intraepithelial Type 1 Innate Lymphoid Cells Are a Unique Subset of IL-12- and IL-15-Responsive IFN-γ-Producing Cells. Immunity 2013, 38, 769–781. [Google Scholar] [CrossRef]
- Gasteiger, G.; Fan, X.; Dikiy, S.; Lee, S.Y.; Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and non-lymphoid organs. Science 2015, 350, 981–985. [Google Scholar] [CrossRef]
- Huang, Q.; Seillet, C.; Belz, G.T. Shaping Innate Lymphoid Cell Diversity. Front. Immunol. 2017, 8, 1569. [Google Scholar] [CrossRef]
- Almeida, F.F.; Belz, G.T. Innate lymphoid cells: Models of plasticity for immune homeostasis and rapid responsiveness in protection. Mucosal. Immunol. 2016, 9, 1103–1112. [Google Scholar] [CrossRef]
- Cortez, V.S.; Ulland, T.K.; Cervantes-Barragan, L.; Bando, J.K.; Robinette, M.L.; Wang, Q.; White, A.J.; Gilfillan, S.; Cella, M.; Colonna, M. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 2017, 18, 995–1003. [Google Scholar] [CrossRef]
- Gao, Y.; Souza-Fonseca-Guimaraes, F.; Bald, T.; Ng, S.S.; Young, A.; Ngiow, S.F.; Rautela, J.; Straube, J.; Waddell, N.; Blake, S.J.; et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017, 18, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Gressner, A.M.; Weiskirchen, R.; Breitkopf, K.; Dooley, S. Roles of TGF-beta in hepatic fibrosis. Front. Biosci. 2002, 7, d793–807. [Google Scholar] [CrossRef]
- Schon, H.-T.; Weiskirchen, R. Immunomodulatory effects of transforming growth factor-β in the liver. Hepatobiliary Surg. Nutr. 2014, 3, 386–406. [Google Scholar] [CrossRef]
- El-Bassiouni, N.E.; Nosseir, M.M.F.; Madkour, M.E.; Zoheiry, M.M.K.; Bekheit, I.W.; Ibrahim, R.A.; Ibrahim, I.M.; El Bassiouny, A.E. Role of fibrogenic markers in chronic hepatitis C and associated hepatocellular carcinoma. Mol. Biol. Rep. 2012, 39, 6843–6850. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xu, J.; Huang, Q.; Huang, M.; Li, K.; Qu, K.; Wen, H.; Lin, R.; Zheng, M.; Wei, H.; et al. Reduced CD160 Expression Contributes to Impaired NK-cell Function and Poor Clinical Outcomes in Patients with HCC. Cancer Res. 2018, 78, 6581–6593. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, K.; Hattori, E.; Tamura, K.; Kiso, S.; Watanabe, H.; Saito, K.; Saito, T.; Togashi, H.; Kawata, S. Possible contribution of circulating transforming growth factor-beta1 to immunity and prognosis in unresectable hepatocellular carcinoma. Liver Int. 2004, 24, 21–28. [Google Scholar] [CrossRef]
- Cortez, V.S.; Cervantes-Barragan, L.; Robinette, M.L.; Bando, J.K.; Wang, Y.; Geiger, T.L.; Gilfillan, S.; Fuchs, A.; Vivier, E.; Sun, J.C.; et al. Transforming Growth Factor-β Signaling Guides the Differentiation of Innate Lymphoid Cells in Salivary Glands. Immunity 2016, 44, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Rautela, J.; Dagley, L.F.; de Oliveira, C.C.; Schuster, I.S.; Hediyeh-Zadeh, S.; Delconte, R.B.; Cursons, J.; Hennessy, R.; Hutchinson, D.S.; Harrison, C.; et al. Therapeutic blockade of activin-A improves NK cell function and antitumor immunity. Sci. Signal. 2019, 12, eaat7527. [Google Scholar] [CrossRef]
- Bald, T.; Wagner, M.; Gao, Y.; Koyasu, S.; Smyth, M.J. Hide and seek: Plasticity of innate lymphoid cells in cancer. Semin. Immunol. 2019, 41, 101273. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Jeong, W.-I.; Tian, Z. Liver: An organ with predominant innate immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Chiossone, L.; Dumas, P.-Y.; Vienne, M.; Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018, 18, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Stamataki, Z.; Swadling, L. The liver as an Immunol.ogical barrier redefined by single-cell analysis. Immunology 2020, 160, 157–170. [Google Scholar] [CrossRef]
- Mackay, I.R. HepatoImmunol.ogy: A perspective. Immunol. Cell Biol. 2002, 80, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Tian, Z. Liver-Mediated Adaptive Immune Tolerance. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Dou, L.; Ono, Y.; Chen, Y.; Thomson, A.W.; Chen, X. Hepatic Dendritic Cells, the Tolerogenic Liver Environment, and Liver Disease. Semin. Liver Dis. 2018, 38, 170–180. [Google Scholar] [CrossRef]
- Orci, L.A.; Kreutzfeldt, M.; Goossens, N.; Rubbia-Brandt, L.; Slits, F.; Hammad, K.; Delaune, V.; Oldani, G.; Negro, F.; Clément, S.; et al. Tolerogenic properties of liver macrophages in non-alcoholic steatohepatitis. Liver Int. 2020, 40, 609–621. [Google Scholar] [CrossRef]
- Horst, A.K.; Neumann, K.; Diehl, L.; Tiegs, G. Modulation of liver tolerance by conventional and nonconventional antigen-presenting cells and regulatory immune cells. Cell Mol. Immunol. 2016, 13, 277–292. [Google Scholar] [CrossRef]
- You, Q.; Cheng, L.; Kedl, R.M.; Ju, C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatol. 2008, 48, 978–990. [Google Scholar] [CrossRef]
- Shouval, D.; Rager-Zisman, B.; Quan, P.; Shafritz, D.A.; Bloom, B.R.; Reid, L.M. Role in nude mice of interferon and natural killer cells in inhibiting the tumorigenicity of human hepatocellular carcinoma cells infected with hepatitis B virus. J. Clin. Investig. 1983, 72, 707–717. [Google Scholar] [CrossRef]
- Gorelik, E.; Wiltrout, R.H.; Okumura, K.; Habu, S.; Herberman, R.B. Role of NK cells in the control of metastatic spread and growth of tumor cells in mice. Int. J. Cancer 1982, 30, 107–112. [Google Scholar] [CrossRef]
- Talmadge, J.E.; Meyers, K.M.; Prieur, D.J.; Starkey, J.R. Role of natural killer cells in tumor growth and metastasis: C57BL/6 normal and beige mice. J. Natl. Cancer Inst. 1980, 65, 929–935. [Google Scholar]
- Sathe, P.; Delconte, R.B.; Souza-Fonseca-Guimaraes, F.; Seillet, C.; Chopin, M.; Vandenberg, C.J.; Rankin, L.C.; Mielke, L.A.; Vikstrom, I.; Kolesnik, T.B.; et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat. Commun. 2014, 5, 4539. [Google Scholar] [CrossRef]
- Dupaul-Chicoine, J.; Arabzadeh, A.; Dagenais, M.; Douglas, T.; Champagne, C.; Morizot, A.; Rodrigue-Gervais, I.G.; Breton, V.; Colpitts, S.L.; Beauchemin, N.; et al. The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity. Immunity 2015, 43, 751–763. [Google Scholar] [CrossRef]
- Imai, K.; Matsuyama, S.; Miyake, S.; Suga, K.; Nakachi, K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet 2000, 356, 1795–1799. [Google Scholar] [CrossRef]
- Hersey, P.; Edwards, A.; Honeyman, M.; McCarthy, W.H. Low natural-killer-cell activity in familial melanoma patients and their relatives. Br. J. Cancer 1979, 40, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Strayer, D.R.; Carter, W.A.; Mayberry, S.D.; Pequignot, E.; Brodsky, I. Low Natural Cytotoxicity of Peripheral Blood Mononuclear Cells in Individuals with High Familial Incidences of Cancer. Cancer Res. 1984, 44, 370–374. [Google Scholar] [PubMed]
- Hoechst, B.; Voigtlaender, T.; Ormandy, L.; Gamrekelashvili, J.; Zhao, F.; Wedemeyer, H.; Lehner, F.; Manns, M.P.; Greten, T.F.; Korangy, F. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology 2009, 50, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Chew, V.; Chen, J.; Lee, D.; Loh, E.; Lee, J.; Lim, K.H.; Weber, A.; Slankamenac, K.; Poon, R.T.P.; Yang, H.; et al. Chemokine-driven lymphocyte infiltration: An early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut 2012, 61, 427–438. [Google Scholar] [CrossRef]
- Wu, M.; Mei, F.; Liu, W.; Jiang, J. Comprehensive characterization of tumor infiltrating natural killer cells and clinical significance in hepatocellular carcinoma based on gene expression profiles. Biomed. Pharmacother. 2020, 121, 109637. [Google Scholar] [CrossRef]
- Dunk, A.A.; Novick, D.; Thomas, H.C. Natural killer cell activity in hepatocellular carcinoma. In vitro and in vivo responses to interferon. Scand. J. Gastroenterol. 1987, 22, 1245–1250. [Google Scholar] [CrossRef]
- Hirofuji, H.; Kakumu, S.; Fuji, A.; Ohtani, Y.; Murase, K.; Tahara, H. Natural killer and activated killer activities in chronic liver disease and hepatocellular carcinoma: Evidence for a decreased lymphokine-induced activity of effector cells. Clin. Exp. Immunol. 1987, 68, 348–356. [Google Scholar]
- Guo, C.-L.; Yang, H.-C.; Yang, X.-H.; Cheng, W.; Dong, T.-X.; Zhu, W.-J.; Xu, Z.; Zhao, L. Associations between infiltrating lymphocyte subsets and hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 5909–5913. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kuang, D.-M.; Pan, W.-D.; Wan, Y.-L.; Lao, X.-M.; Wang, D.; Li, X.-F.; Zheng, L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology 2013, 57, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Mizushima, N.; Kanai, K. Relationship between natural killer activity and development of hepatocellular carcinoma in patients with cirrhosis of the liver. Jpn. J. Clin. Oncol. 1987, 17, 327–332. [Google Scholar]
- Cai, L.; Zhang, Z.; Zhou, L.; Wang, H.; Fu, J.; Zhang, S.; Shi, M.; Zhang, H.; Yang, Y.; Wu, H.; et al. Functional impairment in circulating and intrahepatic NK cells and relative mechanism in hepatocellular carcinoma patients. Clin. Immunol. 2008, 129, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Fathy, A.; Eldin, M.M.; Metwally, L.; Eida, M.; Abdel-Rehim, M. Diminished absolute counts of CD56dim and CD56bright natural killer cells in peripheral blood from Egyptian patients with hepatocellular carcinoma. Egypt. J. Immunol. 2009, 16, 17–25. [Google Scholar] [PubMed]
- Zecca, A.; Barili, V.; Rizzo, D.; Olivani, A.; Biasini, E.; Laccabue, D.; Dalla Valle, R.; Ferrari, C.; Cariani, E.; Missale, G. Intratumor Regulatory Noncytotoxic NK Cells in Patients with Hepatocellular Carcinoma. Cells 2021, 10, 614. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Huang, Q.; Huang, M.; Wen, H.; Lin, R.; Zheng, M.; Qu, K.; Li, K.; Wei, H.; Xiao, W.; et al. Human CD96 Correlates to Natural Killer Cell Exhaustion and Predicts the Prognosis of Human Hepatocellular Carcinoma. Hepatology 2019, 70, 168–183. [Google Scholar] [CrossRef]
- Sun, C.; Sun, H.; Xiao, W.; Zhang, C.; Tian, Z. Natural killer cell dysfunction in hepatocellular carcinoma and NK cell-based Immunother.apy. Acta Pharmacol. Sin. 2015, 36, 1191–1199. [Google Scholar] [CrossRef]
- Thielens, A.; Vivier, E.; Romagné, F. NK cell MHC class I specific receptors (KIR): From biology to clinical intervention. Curr. Opin. Immunol. 2012, 24, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Long, E.O.; Kim, H.S.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef]
- Lanier, L.L.; Le, A.M.; Phillips, J.H.; Warner, N.L.; Babcock, G.F. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J. Immunol. 1983, 131, 1789–1796. [Google Scholar]
- Chang, C.; Rodríguez, A.; Carretero, M.; López-Botet, M.; Phillips, J.H.; Lanier, L.L. Molecular characterization of human CD94: A type II membrane glycoprotein related to the C-type lectin superfamily. Eur. J. Immunol. 1995, 25, 2433–2437. [Google Scholar] [CrossRef] [PubMed]
- Lazetic, S.; Chang, C.; Houchins, J.P.; Lanier, L.L.; Phillips, J.H. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 1996, 157, 4741–4745. [Google Scholar] [PubMed]
- Wu, J.; Song, Y.; Bakker, A.B.; Bauer, S.; Spies, T.; Lanier, L.L.; Phillips, J.H. An activating immunoreceptor complex formed by NKG2D and DAP10. Science 1999, 285, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Groh, V.; Wu, J.; Steinle, A.; Phillips, J.H.; Lanier, L.L.; Spies, T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999, 285, 727–729. [Google Scholar] [CrossRef]
- Sivori, S.; Vitale, M.; Morelli, L.; Sanseverino, L.; Augugliaro, R.; Bottino, C.; Moretta, L.; Moretta, A. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J. Exp. Med. 1997, 186, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Pessino, A.; Sivori, S.; Bottino, C.; Malaspina, A.; Morelli, L.; Moretta, L.; Biassoni, R.; Moretta, A. Molecular cloning of NKp46: A novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998, 188, 953–960. [Google Scholar] [CrossRef]
- Vitale, M.; Bottino, C.; Sivori, S.; Sanseverino, L.; Castriconi, R.; Marcenaro, E.; Augugliaro, R.; Moretta, L.; Moretta, A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998, 187, 2065–2072. [Google Scholar] [CrossRef]
- Pende, D.; Parolini, S.; Pessino, A.; Sivori, S.; Augugliaro, R.; Morelli, L.; Marcenaro, E.; Accame, L.; Malaspina, A.; Biassoni, R.; et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999, 190, 1505–1516. [Google Scholar] [CrossRef]
- Shibuya, A.; Campbell, D.; Hannum, C.; Yssel, H.; Franz-Bacon, K.; McClanahan, T.; Kitamura, T.; Nicholl, J.; Sutherland, G.R.; Lanier, L.L.; et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996, 4, 573–581. [Google Scholar] [CrossRef]
- Johnson, L.A.; Vaidya, S.V.; Goldfarb, R.H.; Mathew, P.A. 2B4(CD244)-mediated activation of NK cells reduces metastases of B16F10 melanoma in mice. Anticancer. Res. 2003, 23, 3651–3655. [Google Scholar]
- Garni-Wagner, B.A.; Purohit, A.; Mathew, P.A.; Bennett, M.; Kumar, V. A novel function-associated molecule related to non-MHC-restricted cytotoxicity mediated by activated natural killer cells and T cells. J. Immunol. 1993, 151, 60–70. [Google Scholar]
- Turman, M.A.; Yabe, T.; McSherry, C.; Bach, F.H.; Houchins, J.P. Characterization of a novel gene (NKG7) on human chromosome 19 that is expressed in natural killer cells and T cells. Hum. Immunol. 1993, 36, 34–40. [Google Scholar] [CrossRef]
- Lanier, L.L.; Yu, G.; Phillips, J.H. Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature 1989, 342, 803–805. [Google Scholar] [CrossRef]
- Lanier, L.L.; Ruitenberg, J.J.; Phillips, J.H. Functional and biochemical analysis of CD16 antigen on natural killer cells and granulocytes. J. Immunol. 1988, 141, 3478–3485. [Google Scholar]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Vance, R.E.; Jamieson, A.M.; Raulet, D.H. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 1999, 190, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- López-Botet, M.; Muntasell, A.; Vilches, C. The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin. Immunol. 2014, 26, 145–151. [Google Scholar] [CrossRef]
- Lanier, L.L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015, 3, 575–582. [Google Scholar] [CrossRef]
- Diefenbach, A.; Jensen, E.R.; Jamieson, A.M.; Raulet, D.H. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature 2001, 413, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, D.E.; Roberts, S.J.; Clarke, S.L.; Filler, R.; Lewis, J.M.; Tigelaar, R.E.; Girardi, M.; Hayday, A.C. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat. Immunol. 2005, 6, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The Natural Cytotoxicity Receptors in Health and Disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef]
- Narni-Mancinelli, E.; Gauthier, L.; Baratin, M.; Guia, S.; Fenis, A.; Deghmane, A.-E.; Rossi, B.; Fourquet, P.; Escalière, B.; Kerdiles, Y.M.; et al. Complement factor P is a ligand for the natural killer cell-activating receptor NKp46. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Edeling, M.A.; Trifonov, V.; Luo, J.; Goyal, P.; Bohl, B.; Bando, J.K.; Kim, A.H.; Walker, J.; Andahazy, M.; et al. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell 2018, 172, 534–548.e19. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef]
- Dougall, W.C.; Kurtulus, S.; Smyth, M.J.; Anderson, A.C. TIGIT and CD96: New checkpoInt. receptor targets for cancer Immunother.apy. Immunol. Rev. 2017, 276, 112–120. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Madore, J.; Li, X.-Y.; Smyth, M.J. Tumor intrinsic and extrinsic immune functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Pahima, H.; Puzzovio, P.G.; Levi-Schaffer, F. 2B4 and CD48: A powerful couple of the immune system. Clin. Immunol. 2019, 204, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.S.; De Labastida Rivera, F.; Yan, J.; Corvino, D.; Das, I.; Zhang, P.; Kuns, R.; Chauhan, S.B.; Hou, J.; Li, X.-Y.; et al. The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation. Nat. Immunol. 2020, 21, 1205–1218. [Google Scholar] [CrossRef]
- Morikawa, Y.; Murakami, M.; Kondo, H.; Nemoto, N.; Iwabuchi, K.; Eshima, K. Natural Killer Cell Group 7 Sequence in Cytotoxic Cells Optimizes Exocytosis of Lytic Granules Essential for the Perforin-Dependent, but Not Fas Ligand–Dependent, Cytolytic Pathway. ImmunoHorizons 2021, 5, 234–245. [Google Scholar] [CrossRef]
- Brooks, A.G.; Posch, P.E.; Scorzelli, C.J.; Borrego, F.; Coligan, J.E. NKG2A complexed with CD94 defines a novel inhibitory natural killer cell receptor. J. Exp. Med. 1997, 185, 795–800. [Google Scholar] [CrossRef]
- Carretero, M.; Cantoni, C.; Bellón, T.; Bottino, C.; Biassoni, R.; Rodríguez, A.; Pérez-Villar, J.J.; Moretta, L.; Moretta, A.; López-Botet, M. The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur. J. Immunol. 1997, 27, 563–567. [Google Scholar] [CrossRef]
- Pérez-Villar, J.J.; Melero, I.; Navarro, F.; Carretero, M.; Bellón, T.; Llano, M.; Colonna, M.; Geraghty, D.E.; López-Botet, M. The CD94/NKG2-A inhibitory receptor complex is involved in natural killer cell-mediated recognition of cells expressing HLA-G1. J. Immunol. 1997, 158, 5736–5743. [Google Scholar] [PubMed]
- Triebel, F.; Jitsukawa, S.; Baixeras, E.; Roman-Roman, S.; Genevee, C.; Viegas-Pequignot, E.; Hercend, T. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 1990, 171, 1393–1405. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Dierich, A.; Benoist, C.; Mathis, D. Independent modes of natural killing distinguished in mice lacking Lag3. Science 1996, 272, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Orr, M.T.; Lanier, L.L. Inhibitory Ly49 receptors on mouse natural killer cells. Curr. Top. MicroBiol. Immunol. 2011, 350, 67–87. [Google Scholar] [CrossRef]
- Vance, R.E.; Kraft, J.R.; Altman, J.D.; Jensen, P.E.; Raulet, D.H. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b). J. Exp. Med. 1998, 188, 1841–1848. [Google Scholar] [CrossRef]
- Wu, N.; Veillette, A. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 2016, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Sun, H.; Zhang, C.; Tian, Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol. Immunol. 2015, 12, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Cózar, B.; Greppi, M.; Carpentier, S.; Narni-Mancinelli, E.; Chiossone, L.; Vivier, E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021, 11, 34–44. [Google Scholar] [CrossRef]
- Sun, C.; Xu, J.; Huang, Q.; Huang, M.; Wen, H.; Zhang, C.; Wang, J.; Song, J.; Zheng, M.; Sun, H.; et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. OncoImmunology 2017, 6, e1264562. [Google Scholar] [CrossRef]
- Tao, L.; Wang, S.; Yang, L.; Jiang, L.; Li, J.; Wang, X. Reduced Siglec-7 expression on NK cells predicts NK cell dysfunction in primary hepatocellular carcinoma. Clin. Exp. Immunol. 2020, 201, 161–170. [Google Scholar] [CrossRef]
- Lassen, M.G.; Lukens, J.R.; Dolina, J.S.; Brown, M.G.; Hahn, Y.S. Intrahepatic IL-10 Maintains NKG2A+Ly49− Liver NK Cells in a Functionally Hyporesponsive State. J. Immunol. 2010, 184, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Molgora, M.; Bonavita, E.; Ponzetta, A.; Riva, F.; Barbagallo, M.; Jaillon, S.; Popović, B.; Bernardini, G.; Magrini, E.; Gianni, F.; et al. IL-1R8 is a checkpoInt. in NK cells regulating anti-tumour and anti-viral activity. Nature 2017, 551, 110–114. [Google Scholar] [CrossRef]
- Mantovani, S.; Oliviero, B.; Lombardi, A.; Varchetta, S.; Mele, D.; Sangiovanni, A.; Rossi, G.; Donadon, M.; Torzilli, G.; Soldani, C.; et al. Deficient Natural Killer Cell NKp30-Mediated Function and Altered NCR3 Splice Variants in Hepatocellular Carcinoma. Hepatology 2019, 69, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Delahaye, N.F.; Rusakiewicz, S.; Martins, I.; Ménard, C.; Roux, S.; Lyonnet, L.; Paul, P.; Sarabi, M.; Chaput, N.; Semeraro, M.; et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat. Med. 2011, 17, 700–707. [Google Scholar] [CrossRef]
- Fend, L.; Rusakiewicz, S.; Adam, J.; Bastien, B.; Caignard, A.; Messaoudene, M.; Iribarren, C.; Cremer, I.; Marabelle, A.; Borg, C.; et al. Prognostic impact of the expression of NCR1 and NCR3 NK cell receptors and PD-L1 on advanced non-small cell lung cancer. OncoImmunology 2017, 6, e1163456. [Google Scholar] [CrossRef] [PubMed]
- Messaoudene, M.; Fregni, G.; Enot, D.; Jacquelot, N.; Neves, E.; Germaud, N.; Garchon, H.J.; Boukouaci, W.; Tamouza, R.; Chanal, J.; et al. NKp30 isoforms and NKp46 transcripts in metastatic melanoma patients: Unique NKp30 pattern in rare melanoma patients with favorable evolution. OncoImmunology 2016, 5, e1154251. [Google Scholar] [CrossRef]
- Semeraro, M.; Rusakiewicz, S.; Minard-Colin, V.; Delahaye, N.F.; Enot, D.; Vély, F.; Marabelle, A.; Papoular, B.; Piperoglou, C.; Ponzoni, M.; et al. Clinical impact of the NKp30/B7-H6 axis in high-risk neuroblastoma patients. Sci. Transl. Med. 2015, 7, 283ra55. [Google Scholar] [CrossRef]
- Rusakiewicz, S.; Perier, A.; Semeraro, M.; Pitt, J.M.; Pogge von Strandmann, E.; Reiners, K.S.; Aspeslagh, S.; Pipéroglou, C.; Vély, F.; Ivagnes, A.; et al. NKp30 isoforms and NKp30 ligands are predictive biomarkers of response to imatinib mesylate in metastatic GIST patients. OncoImmunology 2017, 6, e1137418. [Google Scholar] [CrossRef]
- Rusakiewicz, S.; Nocturne, G.; Lazure, T.; Semeraro, M.; Flament, C.; Caillat-Zucman, S.; Sène, D.; Delahaye, N.; Vivier, E.; Chaba, K.; et al. NCR3/NKp30 contributes to pathogenesis in primary Sjogren’s syndrome. Sci. Transl. Med. 2013, 5, 195ra96. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Qian, Y.; Fu, B.; Jiao, D.; Jiang, Y.; Chen, P.; Shen, Y.; Zhang, H.; Sun, R.; Tian, Z.; et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat. Immunol. 2019, 20, 1656–1667. [Google Scholar] [CrossRef]
- Weston, C.J.; Zimmermann, H.W.; Adams, D.H. The Role of Myeloid-Derived Cells in the Progression of Liver Disease. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Baglieri, J.; Brenner, D.A.; Kisseleva, T. The Role of Fibrosis and Liver-Associated Fibroblasts in the Pathogenesis of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 1723. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Bozward, A.G.; Warricker, F.; Oo, Y.H.; Khakoo, S.I. Natural Killer Cells and Regulatory T Cells Cross Talk in Hepatocellular Carcinoma: Exploring Therapeutic Options for the Next Decade. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Han, S.; Toker, A.; Liu, Z.Q.; Ohashi, P.S. Turning the Tide Against Regulatory T Cells. Front. Oncol. 2019, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, Y.; Hua, X.; Wang, G.; Liu, W.; Jia, C.; Tai, Y.; Zhang, Q.; Chen, G. Hepatocellular carcinoma-associated fibroblasts trigger NK cell dysfunction via PGE2 and IDO. Cancer Lett. 2012, 318, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Ma, C.; Heinrich, B.; Brown, Z.J.; Sandhu, M.; Zhang, Q.; Fu, Q.; Agdashian, D.; Rosato, U.; Korangy, F.; et al. Targeting the crosstalk between cytokine-induced killer cells and myeloid-derived suppressor cells in hepatocellular carcinoma. J. Hepatol. 2019, 70, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Chew, V.; Lai, L.; Pan, L.; Lim, C.J.; Li, J.; Ong, R.; Chua, C.; Leong, J.Y.; Lim, K.H.; Toh, H.C.; et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. PNAS 2017, 114, E5900–E5909. [Google Scholar] [CrossRef]
- Tu, Z.; Bozorgzadeh, A.; Pierce, R.H.; Kurtis, J.; Crispe, I.N.; Orloff, M.S. TLR-dependent cross talk between human Kupffer cells and NK cells. J. Exp. Med. 2008, 205, 233–244. [Google Scholar] [CrossRef]
- Langhans, B.; Nischalke, H.D.; Krämer, B.; Dold, L.; Lutz, P.; Mohr, R.; Vogt, A.; Toma, M.; Eis-Hübinger, A.M.; Nattermann, J.; et al. Role of regulatory T cells and checkpoInt. inhibition in hepatocellular carcinoma. Cancer Immunol. Immunother. 2019, 68, 2055–2066. [Google Scholar] [CrossRef]
- Langhans, B.; Alwan, A.W.; Krämer, B.; Glässner, A.; Lutz, P.; Strassburg, C.P.; Nattermann, J.; Spengler, U. Regulatory CD4+ T cells modulate the interaction between NK cells and hepatic stellate cells by acting on either cell type. J. Hepatol. 2015, 62, 398–404. [Google Scholar] [CrossRef]
- Kohga, K.; Tatsumi, T.; Tsunematsu, H.; Aono, S.; Shimizu, S.; Kodama, T.; Hikita, H.; Yamamoto, M.; Oze, T.; Aketa, H.; et al. Interleukin-1β enhances the production of soluble MICA in human hepatocellular carcinoma. Cancer Immunol. Immunother. 2012, 61, 1425–1432. [Google Scholar] [CrossRef]
- Goto, K.; Arai, J.; Stephanou, A.; Kato, N. Novel therapeutic features of disulfiram against hepatocellular carcinoma cells with inhibitory effects on a disintegrin and metalloproteinase 10. Oncotarget 2018, 9, 18821–18831. [Google Scholar] [CrossRef]
- Zingoni, A.; Vulpis, E.; Loconte, L.; Santoni, A. NKG2D Ligand Shedding in Response to Stress: Role of ADAM10. Front. Immunol. 2020, 11, 447. [Google Scholar] [CrossRef]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Ishida, H.; Miyagi, T.; Hosui, A.; Hayashi, N. Sorafenib inhibits the shedding of major histocompatibility complex class I-related chain A on hepatocellular carcinoma cells by down-regulating a disintegrin and metalloproteinase 9. Hepatology 2010, 51, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Arai, J.; Goto, K.; Tanoue, Y.; Ito, S.; Muroyama, R.; Matsubara, Y.; Nakagawa, R.; Kaise, Y.; Lim, L.A.; Yoshida, H.; et al. Enzymatic inhibition of MICA sheddase ADAM17 by lomofungin in hepatocellular carcinoma cells. Int. J. Cancer 2018, 143, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Z.; Zhou, X.; Zhang, H.; Yang, N.; Wu, Y.; Chen, Y.; Yang, G.; Ren, T. Loss of expression of MHC class I-related chain A (MICA) is a frequent event and predicts poor survival in patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3123–3131. [Google Scholar]
- Fang, L.; Gong, J.; Wang, Y.; Liu, R.; Li, Z.; Wang, Z.; Zhang, Y.; Zhang, C.; Song, C.; Yang, A.; et al. MICA/B expression is inhibited by unfolded protein response and associated with poor prognosis in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2014, 33, 76. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-J.; Pan, K.; Gu, M.-F.; Chen, M.-S.; Zhao, J.-J.; Wang, H.; Liang, X.-T.; Sun, J.-C.; Xia, J.-C. Prognostic value of soluble MICA levels in the serum of patients with advanced hepatocellular carcinoma. Chin. J. Cancer 2013, 32, 141–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, V.; Yi Lo, P.H.; Sawai, H.; Kato, N.; Takahashi, A.; Deng, Z.; Urabe, Y.; Mbarek, H.; Tokunaga, K.; Tanaka, Y.; et al. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV-induced hepatocellular carcinoma. PLoS ONE 2012, 7, e44743. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Luo, W.; Zhu, Q.; Huang, H.; Peng, H.; Liu, R.; Xie, M.; Li, S.; Li, M.; Hu, X.; et al. Tumor-Derived Soluble MICA Obstructs the NKG2D Pathway to Restrain NK Cytotoxicity. Aging Dis. 2020, 11, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Groh, V.; Wu, J.; Yee, C.; Spies, T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 2002, 419, 734–738. [Google Scholar] [CrossRef]
- Jinushi, M.; Takehara, T.; Tatsumi, T.; Hiramatsu, N.; Sakamori, R.; Yamaguchi, S.; Hayashi, N. Impairment of natural killer cell and dendritic cell functions by the soluble form of MHC class I-related chain A in advanced human hepatocellular carcinomas. J. Hepatol. 2005, 43, 1013–1020. [Google Scholar] [CrossRef]
- Easom, N.J.W.; Marks, M.; Jobe, D.; Gillmore, R.; Meyer, T.; Maini, M.K.; Njie, R. ULBP1 Is Elevated in Human Hepatocellular Carcinoma and Predicts Outcome. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Cursons, J.; Huntington, N.D. The Emergence of Natural Killer Cells as a Major Target in Cancer Immunother.apy. Trends Immunol. 2019, 40, 142–158. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, Y.; Liu, L.L.; Lundqvist, A. Strategies to Augment Natural Killer (NK) Cell Activity against Solid Tumors. Cancers 2019, 11, 1040. [Google Scholar] [CrossRef]
- Li, J.; Tao, L.; Wang, X. Cytotoxic immune cell-based Immunother.apy for hepatocellular carcinoma. Hepatoma Res. 2020, 6. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Gobius, I.; Souza-Fonseca-Guimaraes, F. Natural killer cell engineering—A new hope for cancer Immunotherapy. Semin. Hematol. 2020, 57, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.S.; Jang, J.W. Natural Killer Cell Dysfunction in Hepatocellular Carcinoma: Pathogenesis and Clinical Implications. Int. J. Mol. Sci. 2018, 19, 3648. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, L.; Capanni, M.; Urbani, E.; Perruccio, K.; Shlomchik, W.D.; Tosti, A.; Posati, S.; Rogaia, D.; Frassoni, F.; Aversa, F.; et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002, 295, 2097–2100. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Lotze, M.T.; Muul, L.M.; Leitman, S.; Chang, A.E.; Ettinghausen, S.E.; Matory, Y.L.; Skibber, J.M.; Shiloni, E.; Vetto, J.T. Observations on the systemic administration of autologous lymphokine-activated killer cells and recombinant interleukin-2 to patients with metastatic cancer. N. Engl. J. Med. 1985, 313, 1485–1492. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, H.; Liu, L.; Cao, S.; Ren, B.; Zhang, N.; An, X.; Yu, J.; Li, H.; Ren, X. A randomized phase II study of autologous cytokine-induced killer cells in treatment of hepatocellular carcinoma. J. Clin. Immunol. 2014, 34, 194–203. [Google Scholar] [CrossRef]
- Jia, C.-C.; Chen, Y.-H.; Cai, X.-R.; Li, Y.; Zheng, X.-F.; Yao, Z.-C.; Zhao, L.-Y.; Qiu, D.-B.; Xie, S.-J.; Chen, W.-J.; et al. Efficacy of cytokine-induced killer cell-based Immunother.apy for hepatocellular carcinoma. Am. J. Cancer Res. 2019, 9, 1254–1265. [Google Scholar]
- Lee, J.H.; Lee, J.-H.; Lim, Y.-S.; Yeon, J.E.; Song, T.-J.; Yu, S.J.; Gwak, G.-Y.; Kim, K.M.; Kim, Y.J.; Lee, J.W.; et al. Adjuvant Immunother.apy with autologous cytokine-induced killer cells for hepatocellular carcinoma. Gastroenterology 2015, 148, 1383–1391.e6. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, J.H.; Lim, Y.-S.; Yeon, J.E.; Song, T.-J.; Yu, S.J.; Gwak, G.-Y.; Kim, K.M.; Kim, Y.J.; Lee, J.W.; et al. Sustained efficacy of adjuvant Immunother.apy with cytokine-induced killer cells for hepatocellular carcinoma: An extended 5-year follow-up. Cancer Immunol. Immunother. 2019, 68, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.-Z.; Liu, Q.; Zhou, Y.-Q.; Zhao, J.-J.; Wang, Q.-J.; Li, Y.-Q.; Tang, Y.; Gu, J.-M.; He, J.; Chen, S.-P.; et al. CIK cell cytotoxicity is a predictive biomarker for CIK cell Immunother.apy in postoperative patients with hepatocellular carcinoma. Cancer Immunol. Immunother. 2020, 69, 825–834. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Z.; Liu, Z.; Wei, D.; Wu, X.; Bian, H.; Chen, Z. Adoptive cell transfer therapy for hepatocellular carcinoma. Front. Med. 2019, 13, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Parkhurst, M.R.; Riley, J.P.; Dudley, M.E.; Rosenberg, S.A. Adoptive transfer of autologous natural killer cells leads to high levels of circulating natural killer cells but does not mediate tumor regression. Clin. Cancer Res. 2011, 17, 6287–6297. [Google Scholar] [CrossRef]
- Chandran, S.S.; Somerville, R.P.T.; Yang, J.C.; Sherry, R.M.; Klebanoff, C.A.; Goff, S.L.; Wunderlich, J.R.; Danforth, D.N.; Zlott, D.; Paria, B.C.; et al. Treatment of metastatic uveal melanoma with adoptive transfer of tumour-infiltrating lymphocytes: A single-centre, two-stage, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 792–802. [Google Scholar] [CrossRef]
- Mehta, G.U.; Malekzadeh, P.; Shelton, T.; White, D.E.; Butman, J.A.; Yang, J.C.; Kammula, U.S.; Goff, S.L.; Rosenberg, S.A.; Sherry, R.M. Outcomes of Adoptive Cell Transfer With Tumor-infiltrating Lymphocytes for Metastatic Melanoma Patients With and Without Brain Metastases. J. Immunother. 2018, 41, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Saibil, S.D.; Sotov, V.; Le, M.X.; Khoja, L.; Ghazarian, D.; Bonilla, L.; Majeed, H.; Hogg, D.; Joshua, A.M.; et al. Phase II clinical trial of adoptive cell therapy for patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and low-dose interleukin-2. Cancer Immunol. Immunother. 2019, 68, 773–785. [Google Scholar] [CrossRef]
- Dafni, U.; Michielin, O.; Lluesma, S.M.; Tsourti, Z.; Polydoropoulou, V.; Karlis, D.; Besser, M.J.; Haanen, J.; Svane, I.-M.; Ohashi, P.S.; et al. Efficacy of adoptive therapy with tumor-infiltrating lymphocytes and recombinant interleukin-2 in advanced cutaneous melanoma: A systematic review and meta-analysis. Ann. Oncol. 2019, 30, 1902–1913. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized Immunother.apy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef]
- Geukes Foppen, M.H.; Donia, M.; Svane, I.M.; Haanen, J.B. Tumor-infiltrating lymphocytes for the treatment of metastatic cancer. Mol. Oncol. 2015, 9, 1918–1935. [Google Scholar] [CrossRef]
- Krishna, S.; Lowery, F.J.; Copeland, A.R.; Bahadiroglu, E.; Mukherjee, R.; Jia, L.; Anibal, J.T.; Sachs, A.; Adebola, S.O.; Gurusamy, D.; et al. Stem-like CD8 T cells mediate response of adoptive cell Immunother.apy against human cancer. Science 2020, 370, 1328–1334. [Google Scholar] [CrossRef]
- Kremer, V.; Ligtenberg, M.A.; Zendehdel, R.; Seitz, C.; Duivenvoorden, A.; Wennerberg, E.; Colón, E.; Scherman-Plogell, A.-H.; Lundqvist, A. Genetic engineering of human NK cells to express CXCR2 improves migration to renal cell carcinoma. J. Immunother. Cancer 2017, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Liang, S.; Wang, X.; Liang, Y.; Zhang, M.; Chen, J.; Niu, L.; Xu, K. Cryoablation combined with allogenic natural killer cell Immunother.apy improves the curative effect in patients with advanced hepatocellular cancer. Oncotarget 2017, 8, 81967–81977. [Google Scholar] [CrossRef]
- Alnaggar, M.; Lin, M.; Mesmar, A.; Liang, S.; Qaid, A.; Xu, K.; Chen, J.; Niu, L.; Yin, Z. Allogenic Natural Killer Cell Immunother.apy Combined with Irreversible Electroporation for Stage IV Hepatocellular Carcinoma: Survival Outcome. CPB 2018, 48, 1882–1893. [Google Scholar] [CrossRef]
- Marofi, F.; Al-Awad, A.S.; Sulaiman Rahman, H.; Markov, A.; Abdelbasset, W.K.; Ivanovna Enina, Y.; Mahmoodi, M.; Hassanzadeh, A.; Yazdanifar, M.; Stanley Chartrand, M.; et al. CAR-NK Cell: A New Paradigm in Tumor Immunother.apy. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-J.; Qi, C.-Y.; Xiao, W.-K.; Li, S.-Q.; Peng, B.-G.; Liang, L.-J. Glypican-3 is a potential prognostic biomarker for hepatocellular carcinoma after curative resection. Surgery 2013, 154, 536–544. [Google Scholar] [CrossRef]
- Shih, T.-C.; Wang, L.; Wang, H.-C.; Wan, Y.-J.Y. Glypican-3: A molecular marker for the detection and treatment of hepatocellular carcinoma. Liver Res. 2020, 4, 168–172. [Google Scholar] [CrossRef]
- Yu, M.; Luo, H.; Fan, M.; Wu, X.; Shi, B.; Di, S.; Liu, Y.; Pan, Z.; Jiang, H.; Li, Z. Development of GPC3-Specific Chimeric Antigen Receptor-Engineered Natural Killer Cells for the Treatment of Hepatocellular Carcinoma. Mol. Ther. 2018, 26, 366–378. [Google Scholar] [CrossRef]
- Tseng, H.; Xiong, W.; Badeti, S.; Yang, Y.; Ma, M.; Liu, T.; Ramos, C.A.; Dotti, G.; Fritzky, L.; Jiang, J.; et al. Efficacy of anti-CD147 chimeric antigen receptors targeting hepatocellular carcinoma. Nat. Commun. 2020, 11, 4810. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Z.-Z.; Zhou, M.-L.; Lin, J.-W.; Chen, X.-M.; Li, Z.; Gao, W.-B.; Yu, Z.-D.; Liu, T. Development of c-MET-specific chimeric antigen receptor-engineered natural killer cells with cytotoxic effects on human liver cancer HepG2 cells. Mol. Med. Rep. 2019, 20, 2823–2831. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rao, B.; Lou, J.; Li, J.; Liu, Z.; Li, A.; Cui, G.; Ren, Z.; Yu, Z. The Function of the HGF/c-Met Axis in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2020, 8, 55. [Google Scholar] [CrossRef]
- Xiong, L.; Edwards, C.K.; Zhou, L. The biological function and clinical utilization of CD147 in human diseases: A review of the current Sci.entific literature. Int. J. Mol. Sci. 2014, 15, 17411–17441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guo, L.; Song, Y.; Zhang, Y.; Lin, D.; Hu, B.; Mei, Y.; Sandikin, D.; Liu, H. Augmented anti-tumor activity of NK-92 cells expressing chimeric receptors of TGF-βR II and NKG2D. Cancer Immunol. Immunother. 2017, 66, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Sivori, S.; Della Chiesa, M.; Carlomagno, S.; Quatrini, L.; Munari, E.; Vacca, P.; Tumino, N.; Mariotti, F.R.; Mingari, M.C.; Pende, D.; et al. Inhibitory Receptors and Checkpoints in Human NK Cells, Implications for the Immunotherapy of Cancer. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Sanchez-Correa, B.; Lopez-Sejas, N.; Duran, E.; Labella, F.; Alonso, C.; Solana, R.; Tarazona, R. Modulation of NK cells with checkpoint inhibitors in the context of cancer Immunotherapy. Cancer Immunol. Immunother. 2019, 68, 861–870. [Google Scholar] [CrossRef]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a CheckpoInt. Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Seow, S.V.; Wong, D.; Robinson, M.; Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest. 2019, 129, 2094–2106. [Google Scholar] [CrossRef]
- Vey, N.; Karlin, L.; Sadot-Lebouvier, S.; Broussais, F.; Berton-Rigaud, D.; Rey, J.; Charbonnier, A.; Marie, D.; André, P.; Paturel, C.; et al. A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 2018, 9, 17675–17688. [Google Scholar] [CrossRef] [PubMed]
- Rettman, P.; Blunt, M.D.; Fulton, R.J.; Vallejo, A.F.; Bastidas-Legarda, L.Y.; España-Serrano, L.; Polak, M.E.; Al-Shamkhani, A.; Retiere, C.; Khakoo, S.I. Peptide: MHC-based DNA vaccination strategy to activate natural killer cells by targeting killer cell immunoglobulin-like receptors. J. Immunother. Cancer 2021, 9, e001912. [Google Scholar] [CrossRef]
- Romagné, F.; André, P.; Spee, P.; Zahn, S.; Anfossi, N.; Gauthier, L.; Capanni, M.; Ruggeri, L.; Benson, D.M.; Blaser, B.W.; et al. Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood 2009, 114, 2667–2677. [Google Scholar] [CrossRef]
- Vey, N.; Bourhis, J.-H.; Boissel, N.; Bordessoule, D.; Prebet, T.; Charbonnier, A.; Etienne, A.; Andre, P.; Romagne, F.; Benson, D.; et al. A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood 2012, 120, 4317–4323. [Google Scholar] [CrossRef]
- Benson, D.M.; Cohen, A.D.; Jagannath, S.; Munshi, N.C.; Spitzer, G.; Hofmeister, C.C.; Efebera, Y.A.; Andre, P.; Zerbib, R.; Caligiuri, M.A. A Phase I Trial of the Anti-KIR Antibody IPH2101 and Lenalidomide in Patients with Relapsed/Refractory Multiple Myeloma. Clin. Cancer Res. 2015, 21, 4055–4061. [Google Scholar] [CrossRef]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoInt. receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Démoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Burova, E.; Hermann, A.; Dai, J.; Ullman, E.; Halasz, G.; Potocky, T.; Hong, S.; Liu, M.; Allbritton, O.; Woodruff, A.; et al. Preclinical Development of the Anti-LAG-3 Antibody REGN3767: Characterization and Activity in Combination with the Anti-PD-1 Antibody Cemiplimab in Human PD-1xLAG-3-Knockin Mice. Mol. Cancer Ther. 2019, 18, 2051–2062. [Google Scholar] [CrossRef]
- Yu, X.; Huang, X.; Chen, X.; Liu, J.; Wu, C.; Pu, Q.; Wang, Y.; Kang, X.; Zhou, L. Characterization of a novel anti-human lymphocyte activation gene 3 (LAG-3) antibody for cancer Immunotherapy. mAbs 2019, 11, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; Gauthier, L.; Debroas, G.; Vivier, E. Natural killer cell engagers in cancer Immunotherapy: Next generation of immuno-Oncol.ogy treatments. Eur. J. Immunol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, L.; Morel, A.; Anceriz, N.; Rossi, B.; Blanchard-Alvarez, A.; Grondin, G.; Trichard, S.; Cesari, C.; Sapet, M.; Bosco, F.; et al. Multifunctional Natural Killer Cell Engagers Targeting NKp46 Trigger Protective Tumor Immunity. Cell 2019, 177, 1701–1713.e16. [Google Scholar] [CrossRef] [PubMed]
- Wiernik, A.; Foley, B.; Zhang, B.; Verneris, M.R.; Warlick, E.; Gleason, M.K.; Ross, J.A.; Luo, X.; Weisdorf, D.J.; Walcheck, B.; et al. Targeting natural killer cells to acute myeloid leukemia in vitro with a CD16 x 33 bispecific killer cell engager and ADAM17 inhibition. Clin. Cancer Res. 2013, 19, 3844–3855. [Google Scholar] [CrossRef] [PubMed]
- von Strandmann, E.P.; Hansen, H.P.; Reiners, K.S.; Schnell, R.; Borchmann, P.; Merkert, S.; Simhadri, V.R.; Draube, A.; Reiser, M.; Purr, I.; et al. A novel bispecific protein (ULBP2-BB4) targeting the NKG2D receptor on natural killer (NK) cells and CD138 activates NK cells and has potent antitumor activity against human multiple myeloma in vitro and in vivo. Blood 2006, 107, 1955–1962. [Google Scholar] [CrossRef]
- Rothe, A.; Sasse, S.; Topp, M.S.; Eichenauer, D.A.; Hummel, H.; Reiners, K.S.; Dietlein, M.; Kuhnert, G.; Kessler, J.; Buerkle, C.; et al. A phase 1 study of the bispecific anti-CD30/CD16A antibody construct AFM13 in patients with relapsed or refractory Hodgkin lymphoma. Blood 2015, 125, 4024–4031. [Google Scholar] [CrossRef]
- Bartlett, N.L.; Herrera, A.F.; Domingo-Domenech, E.; Mehta, A.; Forero-Torres, A.; Garcia-Sanz, R.; Armand, P.; Devata, S.; Izquierdo, A.R.; Lossos, I.S.; et al. A phase 1b study of AFM13 in combination with pembrolizumab in patients with relapsed or refractory Hodgkin lymphoma. Blood 2020, 136, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Easom, N.J.W.; Stegmann, K.A.; Swadling, L.; Pallett, L.J.; Burton, A.R.; Odera, D.; Schmidt, N.; Huang, W.-C.; Fusai, G.; Davidson, B.; et al. IL-15 Overcomes Hepatocellular Carcinoma-Induced NK Cell Dysfunction. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Rautela, J.; Huntington, N.D. IL-15 signaling in NK cell cancer Immunother.apy. Curr. Opin. Immunol. 2017, 44, 1–6. [Google Scholar] [CrossRef]
- Conlon, K.C.; Lugli, E.; Welles, H.C.; Rosenberg, S.A.; Fojo, A.T.; Morris, J.C.; Fleisher, T.A.; Dubois, S.P.; Perera, L.P.; Stewart, D.M.; et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J. Clin. Oncol. 2015, 33, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, C.; Bear, J.; Rosati, M.; Beach, R.K.; Alicea, C.; Sowder, R.; Chertova, E.; Rosenberg, S.A.; Felber, B.K.; Pavlakis, G.N. Circulating IL-15 exists as heterodimeric complex with soluble IL-15Rα in human and mouse serum. Blood 2012, 120, e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Burkett, P.R.; Koka, R.; Chien, M.; Chai, S.; Boone, D.L.; Ma, A. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J. Exp. Med. 2004, 200, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Schluns, K.S.; Nowak, E.C.; Cabrera-Hernandez, A.; Puddington, L.; Lefrançois, L.; Aguila, H.L. Distinct cell types control lymphoid subset development by means of IL-15 and IL-15 receptor alpha expression. Proc. Natl. Acad Sci. USA 2004, 101, 5616–5621. [Google Scholar] [CrossRef]
- Guo, Y.; Luan, L.; Rabacal, W.; Bohannon, J.K.; Fensterheim, B.A.; Hernandez, A.; Sherwood, E.R. IL-15 Superagonist-Mediated Immunotoxicity: Role of NK Cells and IFN-γ. J. Immunol. 2015, 195, 2353–2364. [Google Scholar] [CrossRef]
- Desbois, M.; Béal, C.; Charrier, M.; Besse, B.; Meurice, G.; Cagnard, N.; Jacques, Y.; Béchard, D.; Cassard, L.; Chaput, N. IL-15 superagonist RLI has potent immunostimulatory properties on NK cells: Implications for antimetastatic treatment. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Huntington, N.D.; Legrand, N.; Alves, N.L.; Jaron, B.; Weijer, K.; Plet, A.; Corcuff, E.; Mortier, E.; Jacques, Y.; Spits, H.; et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009, 206, 25–34. [Google Scholar] [CrossRef]
- Bergamaschi, C.; Pandit, H.; Nagy, B.A.; Stellas, D.; Jensen, S.M.; Bear, J.; Cam, M.; Valentin, A.; Fox, B.A.; Felber, B.K.; et al. Heterodimeric IL-15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN-γ, CXCL9 and CXCL10. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Liu, B.; Kong, L.; Han, K.; Hong, H.; Marcus, W.D.; Chen, X.; Jeng, E.K.; Alter, S.; Zhu, X.; Rubinstein, M.P.; et al. A Novel Fusion of ALT-803 (Interleukin (IL)-15 Superagonist) with an Antibody Demonstrates Antigen-specific Antitumor Responses. J. Biol. Chem. 2016, 291, 23869–23881. [Google Scholar] [CrossRef] [PubMed]
- Rosario, M.; Liu, B.; Kong, L.; Collins, L.I.; Schneider, S.E.; Chen, X.; Han, K.; Jeng, E.K.; Rhode, P.R.; Leong, J.W.; et al. The IL-15-Based ALT-803 Complex Enhances FcγRIIIa-Triggered NK Cell Responses and In Vivo Clearance of B Cell Lymphomas. Clin. Cancer Res. 2016, 22, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Mathios, D.; Park, C.-K.; Marcus, W.D.; Alter, S.; Rhode, P.R.; Jeng, E.K.; Wong, H.C.; Pardoll, D.M.; Lim, M. Therapeutic administration of IL-15 superagonist complex ALT-803 leads to long-term survival and durable antitumor immune response in a murine glioblastoma model. Int. J. Cancer 2016, 138, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.M.R.; Maas, R.J.A.; Guldevall, K.; Klarenaar, K.; de Jonge, P.K.J.D.; Evert, J.S.H.; van der Waart, A.B.; Cany, J.; Safrit, J.T.; Lee, J.H.; et al. IL-15 superagonist N-803 improves IFNγ production and killing of leukemia and ovarian cancer cells by CD34+ progenitor-derived NK cells. Cancer Immunol. Immunother. 2021, 70, 1305–1321. [Google Scholar] [CrossRef] [PubMed]
- Pinette, A.; McMichael, E.; Courtney, N.B.; Duggan, M.; Benner, B.N.; Choueiry, F.; Yu, L.; Abood, D.; Mace, T.A.; Carson, W.E. An IL-15-based superagonist ALT-803 enhances the NK cell response to cetuximab-treated squamous cell carcinoma of the head and neck. Cancer Immunol. Immunother. 2019, 68, 1379–1389. [Google Scholar] [CrossRef]
- Foltz, J.A.; Hess, B.T.; Bachanova, V.; Bartlett, N.L.; Berrien-Elliott, M.M.; McClain, E.; Becker-Hapak, M.; Foster, M.; Schappe, T.; Kahl, B.; et al. Phase I Trial of N-803, an IL15 Receptor Agonist, with Rituximab in Patients with Indolent Non-Hodgkin Lymphoma. Clin. Cancer Res. 2021, 27, 3339–3350. [Google Scholar] [CrossRef]
- Faivre, S.; Santoro, A.; Kelley, R.K.; Gane, E.; Costentin, C.E.; Gueorguieva, I.; Smith, C.; Cleverly, A.; Lahn, M.M.; Raymond, E.; et al. Novel transforming growth factor beta receptor I kinase inhibitor galunisertib (LY2157299) in advanced hepatocellular carcinoma. Liver Int. 2019, 39, 1468–1477. [Google Scholar] [CrossRef]
- Kelley, R.K.; Gane, E.; Assenat, E.; Siebler, J.; Galle, P.R.; Merle, P.; Hourmand, I.O.; Cleverly, A.; Zhao, Y.; Gueorguieva, I.; et al. A Phase 2 Study of Galunisertib (TGF-β1 Receptor Type I Inhibitor) and Sorafenib in Patients With Advanced Hepatocellular Carcinoma. Clin. Transl. Gastroenterol. 2019, 10, e00056. [Google Scholar] [CrossRef]
- Serova, M.; Tijeras-Raballand, A.; Dos Santos, C.; Albuquerque, M.; Paradis, V.; Neuzillet, C.; Benhadji, K.A.; Raymond, E.; Faivre, S.; de Gramont, A. Effects of TGF-beta signalling inhibition with galunisertib (LY2157299) in hepatocellular carcinoma models and in ex vivo whole tumor tissue samples from patients. Oncotarget 2015, 6, 21614–21627. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.F.; Reisinger, F.; Puschnik, A.; Ringelhan, M.; Ackermann, K.; Hartmann, D.; Schiemann, M.; Weinmann, A.; Galle, P.R.; Schuchmann, M.; et al. Sorafenib perpetuates cellular anticancer effector functions by modulating the crosstalk between macrophages and natural killer cells. Hepatology 2013, 57, 2358–2368. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, F.; Verdi, J.; Ai, J.; Hajighasemlou, S.; Seyhoun, I.; Parvizpour, F.; Hosseinzadeh, F.; Iranikhah, A.; Shirian, S. Combinational immune-cell therapy of natural killer cells and sorafenib for advanced hepatocellular carcinoma: A review. Cancer Cell Int. 2018, 18, 133. [Google Scholar] [CrossRef]
- Otoyama, Y.; Arai, J.; Goto, K.; Nozawa, H.; Nakagawa, R.; Muroyama, R.; Sugiura, I.; Nakajima, Y.; Kajiwara, A.; Tojo, M.; et al. Retinoids Decrease Soluble MICA Concentration by Inhibiting the Enzymatic Activity of ADAM9 and ADAM10. Anticancer. Res. 2021, 41, 2307–2320. [Google Scholar] [CrossRef]
- Ferrari de Andrade, L.; Tay, R.E.; Pan, D.; Luoma, A.M.; Ito, Y.; Badrinath, S.; Tsoucas, D.; Franz, B.; May, K.F.; Harvey, C.J.; et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science 2018, 359, 1537–1542. [Google Scholar] [CrossRef] [PubMed]
- Cording, S.; Medvedovic, J.; Lécuyer, E.; Aychek, T.; Déjardin, F.; Eberl, G. Mouse models for the study of fate and function of innate lymphoid cells. Eur. J. Immunol. 2018, 48, 1271–1280. [Google Scholar] [CrossRef]
- Narni-Mancinelli, E.; Chaix, J.; Fenis, A.; Kerdiles, Y.M.; Yessaad, N.; Reynders, A.; Gregoire, C.; Luche, H.; Ugolini, S.; Tomasello, E.; et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad Sci. USA 2011, 108, 18324–18329. [Google Scholar] [CrossRef]
- Eckelhart, E.; Warsch, W.; Zebedin, E.; Simma, O.; Stoiber, D.; Kolbe, T.; Rülicke, T.; Mueller, M.; Casanova, E.; Sexl, V. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood 2011, 117, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Merzoug, L.B.; Marie, S.; Satoh-Takayama, N.; Lesjean, S.; Albanesi, M.; Luche, H.; Fehling, H.J.; Di Santo, J.P.; Vosshenrich, C.A.J. Conditional ablation of NKp46+ cells using a novel Ncr1(greenCre) mouse strain: NK cells are essential for protection against pulmonary B16 metastases. Eur. J. Immunol. 2014, 44, 3380–3391. [Google Scholar] [CrossRef]
- Kwong, B.; Rua, R.; Gao, Y.; Flickinger, J.; Wang, Y.; Kruhlak, M.J.; Zhu, J.; Vivier, E.; McGavern, D.B.; Lazarevic, V. T-bet-dependent NKp46+ innate lymphoid cells regulate the onset of TH17-induced neuroinflammation. Nat. Immunol. 2017, 18, 1117–1127. [Google Scholar] [CrossRef]
- Ducimetière, L.; Lucchiari, G.; Litscher, G.; Nater, M.; Heeb, L.; Nuñez, N.G.; Wyss, L.; Burri, D.; Vermeer, M.; Gschwend, J.; et al. Conventional NK cells and tissue-resident ILC1s join forces to control liver metastasis. Proc. Natl. Acad Sci. USA 2021, 118, e2026271118. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.B.; Vallera, D.A.; Miller, J.S.; Felices, M. Natural killer cells unleashed: CheckpoInt. receptor blockade and BiKE/TriKE utilization in NK-mediated anti-tumor Immunother.apy. Semin. Immunol. 2017, 31, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Vallera, D.A.; Felices, M.; McElmurry, R.; McCullar, V.; Zhou, X.; Schmohl, J.U.; Zhang, B.; Lenvik, A.J.; Panoskaltsis-Mortari, A.; Verneris, M.R.; et al. IL15 Trispecific Killer Engagers (TriKE) Make Natural Killer Cells Specific to CD33+ Targets While Also Inducing Persistence, In Vivo Expansion, and Enhanced Function. Clin. Cancer Res. 2016, 22, 3440–3450. [Google Scholar] [CrossRef] [PubMed]
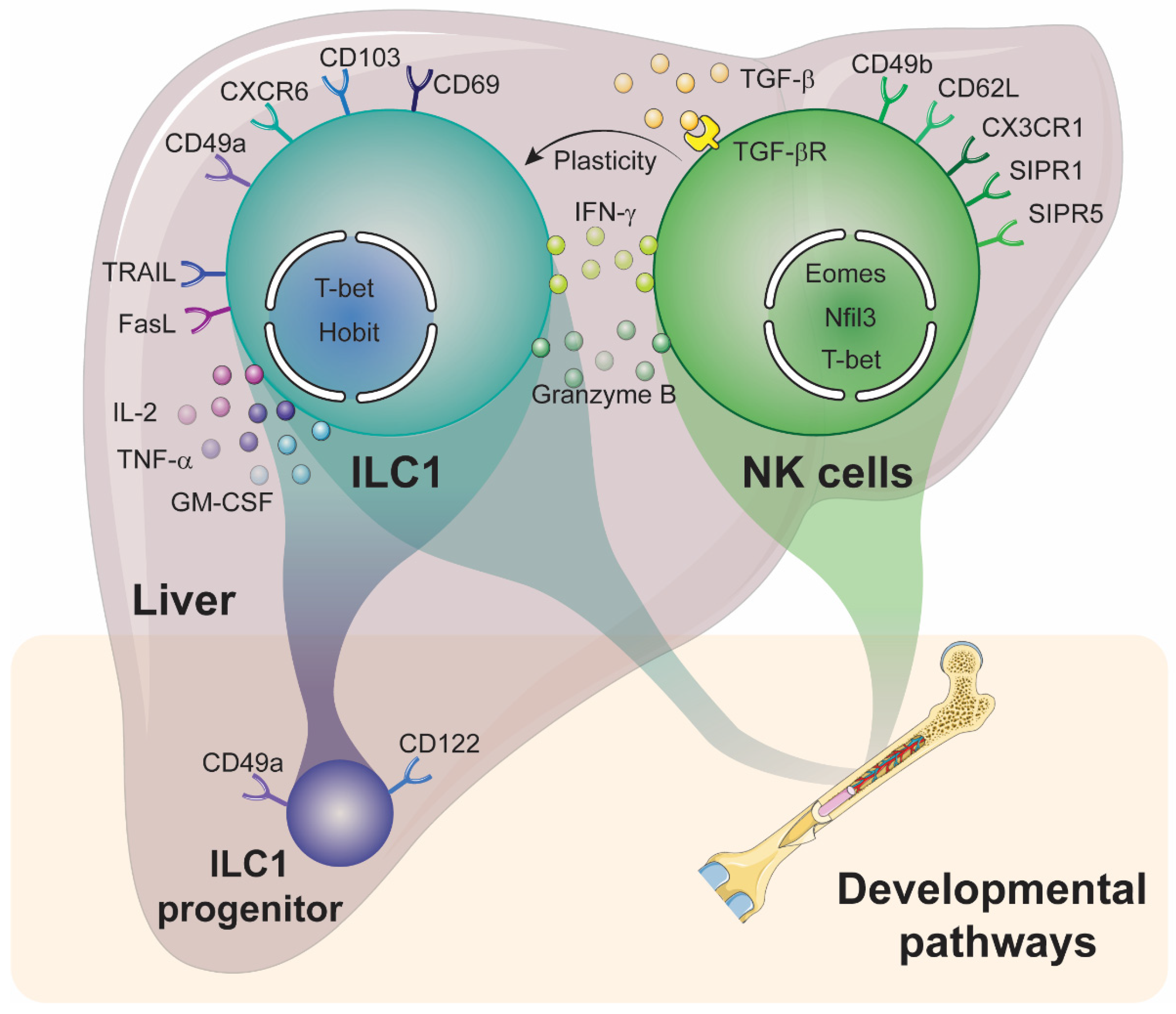

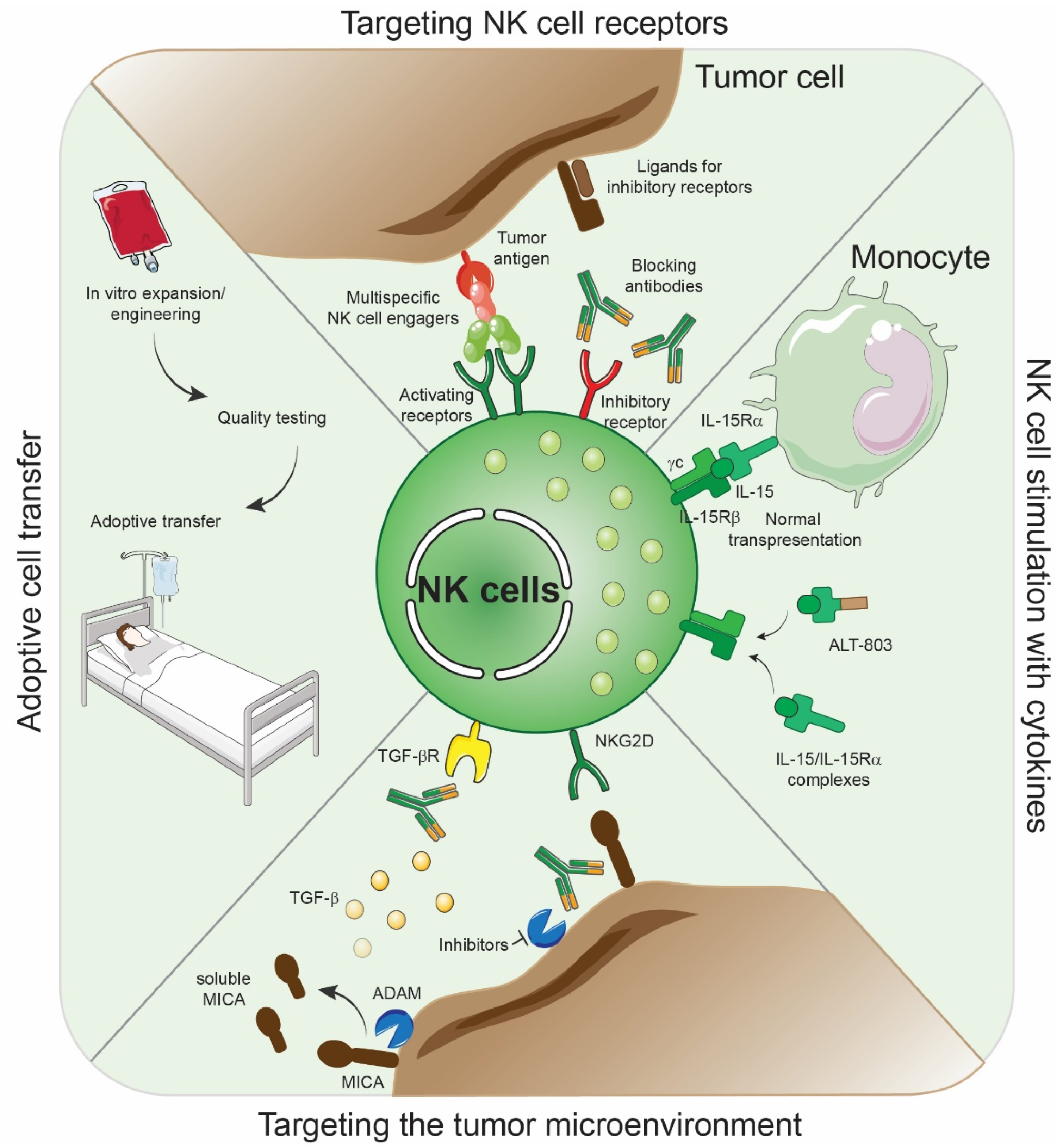
| Phenotypic Markers | Effector Molecules | References | |||
|---|---|---|---|---|---|
| Mouse | Human | ||||
| cNK Cells | lrNK Cells | ||||
| NK cells | NK1.1+, NKp46+, CD49a−, Eomes+, T-bet+, CD49b+, CD200R−, Ly49+/−, CXCR6−, Killer cell lectin-like receptor G1(KLRG1)+/−, CD62L+/− | CD56dim/bright, CD16bright/low, NKG2D+/−, NKp46+, CD49b+, CD49e+, CD200R−, Eomes+, T-bet+ | CD56+, CD16−, NKG2D+, NKG2C+, NKG2A−, NKp46+, CD49e−, CXCR6+, CD69+, Eomeshi, T-bet− | Granzyme A and B, Perforin, IFN-γ | [55,60,61,62,66,67,68,69,70,71,72] |
| ILC1 | NK1.1+, NKp46+, CD49a+, T-bet+, TRAIL+, CD49b−, CD200R+, CD62L−, KLRG1−, CXCR6+, CD127+ | Lineage−CD127+CD16−NKG2A−NKp44−CD117− | Granzyme B+, interleukin (IL)-2, IFN-γ, Tumor necrosis factor (TNF)-α, granulocyte-macrophage colony-stimulating factor (GM-CSF) | [55,57,69,70,71,72,73] | |
| Targeted Gene | Liver NK Cells | Liver ILC1 | References |
|---|---|---|---|
| Tbx21 | Strongly reduced | Absent | [69,70,87] |
| Eomes | Absent | No impact | [90,91] |
| Nfil3 | Absent | Reduced or no impact | [69,71,93,94] |
| Hobit | No impact | Absent | [73] |
| Treatment Strategy | Allocation | Outcomes Measured | Clinical Trials.gov Identifier |
|---|---|---|---|
| FT500, an allogeneic NK cell product, as monotherapy and in combination with IL-2 and/or immune checkpoint inhibitors (anti-PD_1 and anti-PD-L1) | Non-Randomized | Primary outcome measures: Dose Limiting toxicities Secondary outcome measures: Objective-response rates (ORR) and pharmacokinetics of FT500 in blood | NCT03841110 |
| A Phase II/III clinical trial withe ex vivo expanded autologous immune killer cells to treat liver cancer patients as an adjunct therapy | Randomized Autologous NK cells + transcatheter arterial chemoembolization (TACE) vs. TACE alone | Primary outcome measures: Change of tumor size and progression-free survival (PFS) Secondary outcome measures: Promotion of anti-cancer immune responses | NCT03592706 |
| Safety and efficacy of allogeneic NK cell therapy in patients with advanced hepatocellular carcinoma | Non-Randomized | Primary outcome measures: Incidence of Adverse events and Overall survival (OS) Secondary outcome measures: Disease control rate | NCT04162158 |
| FATE-NK100 as monotherapy and in combination with monoclonal antibody in subjects with advanced solid tumor | Non-Randomized | Primary outcome measures: Dose Limiting toxicities Secondary outcome measures: ORR and pharmacokinetics of FATE-NK100 in blood | NCT03319459 |
| By using adoptive transfer of autologous NK cells to prevent recurrence of hepatocellular carcinoma after curative therapy | Randomized NK cell infusion + curative therapy (stage I and II patients only) vs. curative therapy only | Primary outcome measures: Recurrence free survival and OS | NCT02725996 |
| Study of SO-C101 (IL-15 superagonist) and SO-C101 in combination with pembrolizumab in adult patients with advanced/metastatic solid tumors | Non-Randomized | Primary outcome measures: Dose Limiting toxicities and adverse events Secondary outcome measures: Pharmacokinetics, ORR, best overall response, duration of response (DOR), clinical benefit rate, PFS and immunogenicity | NCT04234113 |
| QUILT-3.055: A study of combination immunotherapies (N-803, an IL-15 superagonist + ICB) in patients who have previously received treatment with immune checkpoint inhibitors | Non-Randomized | Primary outcome measures: ORR Secondary outcome measures: Disease-specific survival, OS, time to response, DOR, incidence of adverse events, quality of life and PFS | NCT03228667 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacquelot, N.; Seillet, C.; Souza-Fonseca-Guimaraes, F.; Sacher, A.G.; Belz, G.T.; Ohashi, P.S. Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 9044. https://doi.org/10.3390/ijms22169044
Jacquelot N, Seillet C, Souza-Fonseca-Guimaraes F, Sacher AG, Belz GT, Ohashi PS. Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives. International Journal of Molecular Sciences. 2021; 22(16):9044. https://doi.org/10.3390/ijms22169044
Chicago/Turabian StyleJacquelot, Nicolas, Cyril Seillet, Fernando Souza-Fonseca-Guimaraes, Adrian G. Sacher, Gabrielle T. Belz, and Pamela S. Ohashi. 2021. "Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives" International Journal of Molecular Sciences 22, no. 16: 9044. https://doi.org/10.3390/ijms22169044
APA StyleJacquelot, N., Seillet, C., Souza-Fonseca-Guimaraes, F., Sacher, A. G., Belz, G. T., & Ohashi, P. S. (2021). Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives. International Journal of Molecular Sciences, 22(16), 9044. https://doi.org/10.3390/ijms22169044