OTX2 Homeoprotein Functions in Adult Choroid Plexus
Abstract
:1. Introduction
2. Results and Discussion
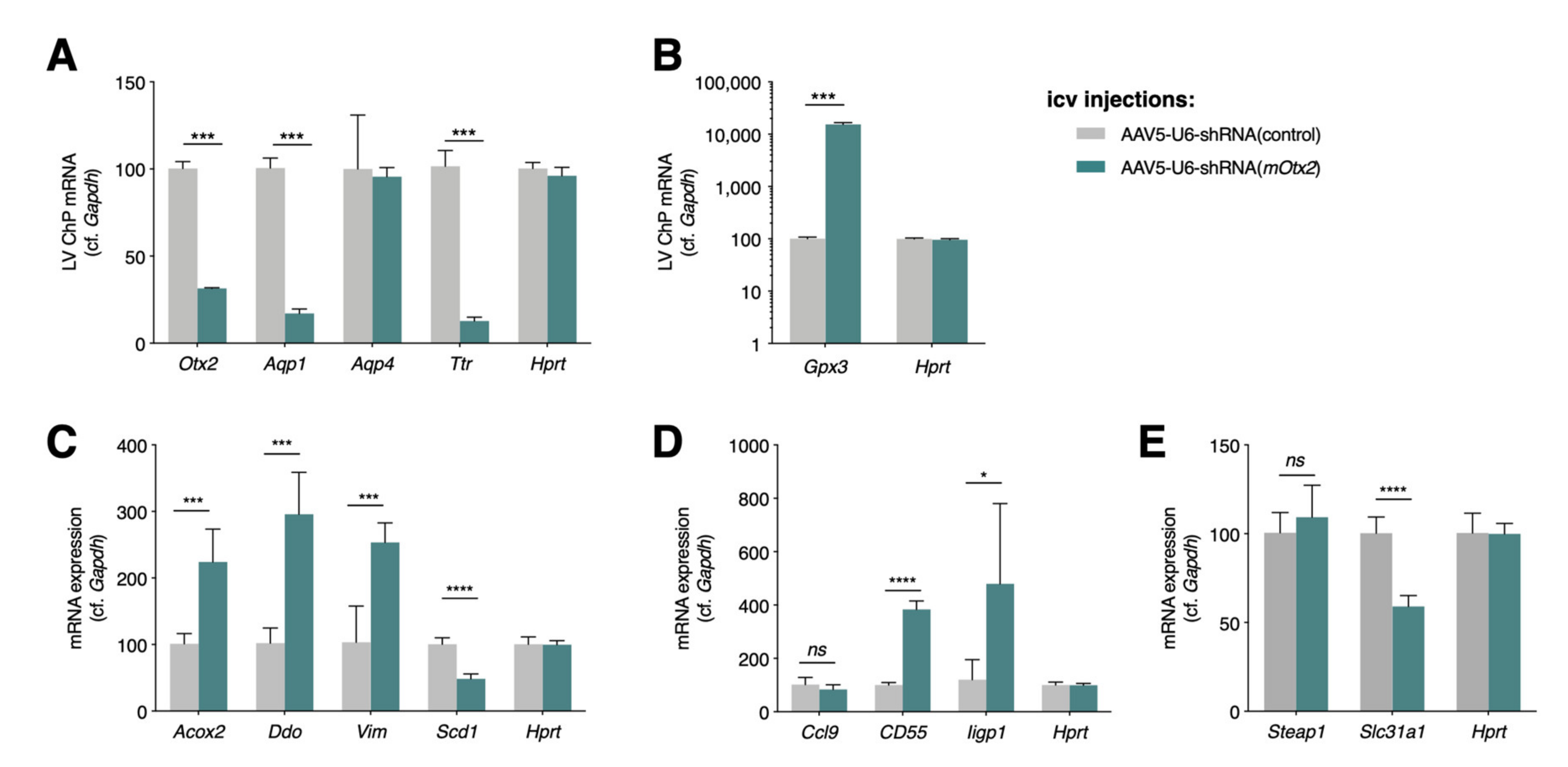
2.1. Conditional and Constitutive Knockdown of Otx2 in Adult ChP
2.2. Altered Expression of ChP Secreted Factors
2.3. Altered Expression of Immune and Stress Factors
2.4. Otx2 Protein Interactions
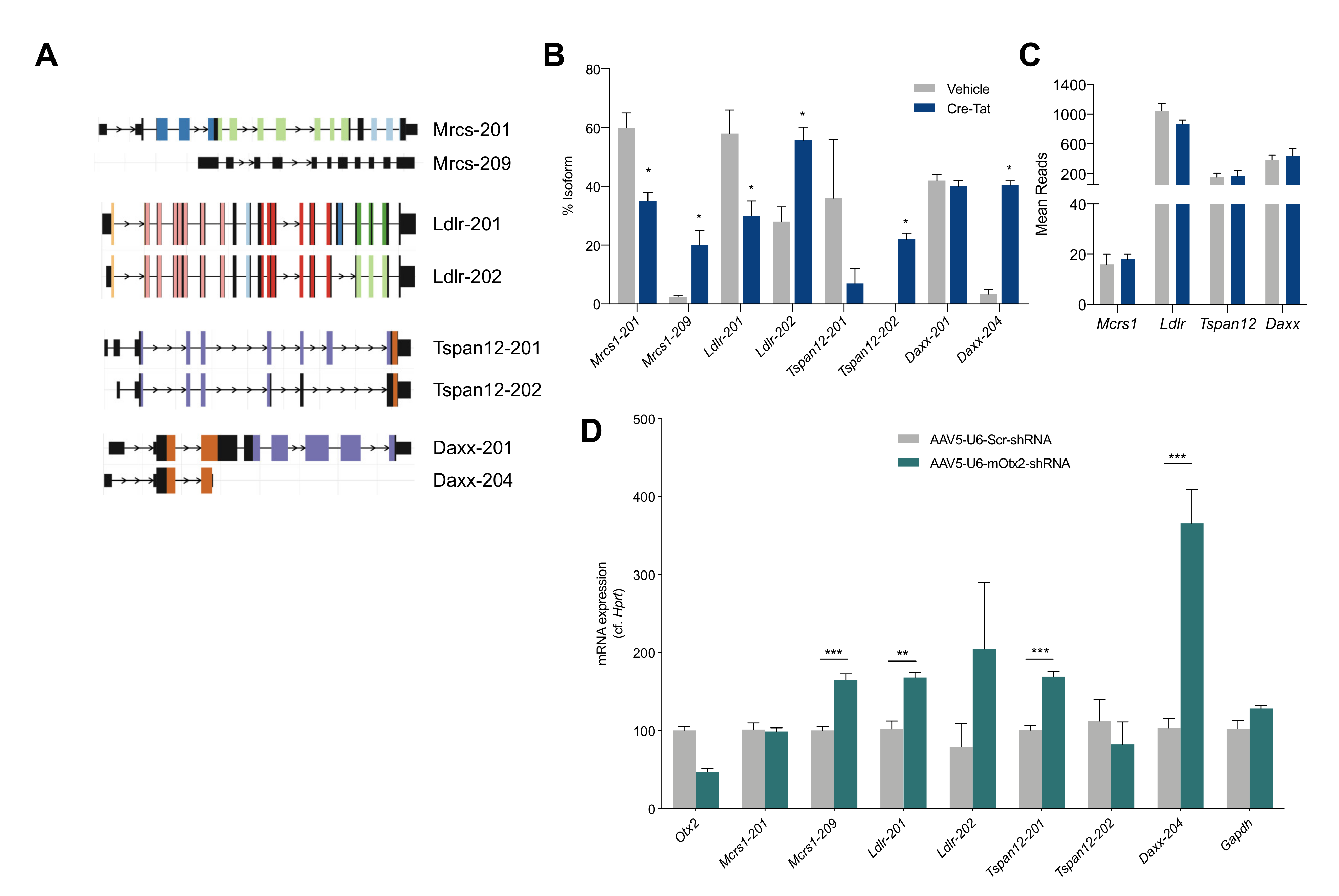
2.5. Splice Variant Analysis
3. Materials and Methods
3.1. Animal Ethics Statement
3.2. Animals and Stereotaxic Surgery
3.3. Quantitative PCR Analysis
3.4. RNA Sequencing Analysis
3.5. Isoform Analysis
3.6. Protein Co-Immunoprecipitation
3.7. Mass Spectrometry Analysis
3.8. Ontology Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fame, R.M.; Lehtinen, M.K. Emergence and Developmental Roles of the Cerebrospinal Fluid System. Dev. Cell 2020, 52, 261–275. [Google Scholar] [CrossRef]
- Johansson, P.A.; Irmler, M.; Acampora, D.; Beckers, J.; Simeone, A.; Götz, M. The transcription factor Otx2 regulates choroid plexus development and function. Development 2013, 140, 1055–1066. [Google Scholar] [CrossRef] [Green Version]
- Lun, M.P.; Johnson, M.B.; Broadbelt, K.G.; Watanabe, M.; Kang, Y.J.; Chau, K.F.; Springel, M.W.; Malesz, A.; Sousa, A.M.M.; Pletikos, M.; et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci. 2015, 35, 4903–4916. [Google Scholar] [CrossRef]
- Dani, N.; Herbst, R.H.; McCabe, C.; Green, G.S.; Kaiser, K.; Head, J.P.; Cui, J.; Shipley, F.B.; Jang, A.; Dionne, D.; et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 2021, 184, 3056–3074.e21. [Google Scholar] [CrossRef]
- Spatazza, J.; Lee, H.H.C.; Di Nardo, A.A.; Tibaldi, L.; Joliot, A.; Hensch, T.K.; Prochiantz, A. Choroid-Plexus-Derived Otx2 Homeoprotein Constrains Adult Cortical Plasticity. Cell Rep. 2013, 3, 1815–1823. [Google Scholar] [CrossRef] [Green Version]
- Rezsohazy, R. Non-transcriptional interactions of Hox proteins: Inventory, facts, and future directions. Dev. Dyn. 2014, 243, 117–131. [Google Scholar] [CrossRef]
- Di Nardo, A.A.; Fuchs, J.; Joshi, R.L.; Moya, K.L.; Prochiantz, A. The physiology of homeoprotein transduction. Physiol. Rev. 2018, 98, 1943–1982. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, A.A.; Joliot, A.; Prochiantz, A. Homeoprotein transduction in neurodevelopment and physiopathology. Sci. Adv. 2020, 6, eabc6374. [Google Scholar] [CrossRef] [PubMed]
- Hoch, R.V.; Lindtner, S.; Price, J.D.; Rubenstein, J.L.R. OTX2 Transcription Factor Controls Regional Patterning within the Medial Ganglionic Eminence and Regional Identity of the Septum. Cell Rep. 2015, 12, 482–494. [Google Scholar] [CrossRef] [Green Version]
- Rohde, K.; Hertz, H.; Rath, M.F. Homeobox genes in melatonin-producing pinealocytes: Otx2 and Crx act to promote hormone synthesis in the mature rat pineal gland. J. Pineal Res. 2019, 66, e12567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, M.; Acampora, D.; Vojtek, M.; Yuan, D.; Simeone, A.; Chambers, I. OTX2 restricts entry to the mouse germline. Nature 2018, 562, 595–599. [Google Scholar] [CrossRef]
- Matsuda, K.; Mikami, T.; Oki, S.; Iida, H.; Andrabi, M.; Boss, J.M.; Yamaguchi, K.; Shigenobu, S.; Kondoh, H. ChIP-seq analysis of genomic binding regions of five major transcription factors in mouse epiblast stem cells that highlights a central role for ZIC2. Development 2017, 144, 1948–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acampora, D.; Omodei, D.; Petrosino, G.; Garofalo, A.; Savarese, M.; Nigro, V.; di Giovannantonio, L.G.; Mercadante, V.; Simeone, A. Loss of the Otx2-Binding Site in the Nanog Promoter Affects the Integrity of Embryonic Stem Cell Subtypes and Specification of Inner Cell Mass-Derived Epiblast. Cell Rep. 2016, 15, 2651–2664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuel, A.; Housset, M.; Fant, B.; Lamonerie, T. Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PLoS ONE 2014, 9, e89110. [Google Scholar] [CrossRef] [Green Version]
- Fant, B.; Samuel, A.; Audebert, S.; Couzon, A.; El Nagar, S.; Billon, N.; Lamonerie, T. Comprehensive interactome of Otx2 in the adult mouse neural retina. Genesis 2015, 53, 685–694. [Google Scholar] [CrossRef]
- Apulei, J.; Kim, N.; Testa, D.; Ribot, J.; Morizet, D.; Bernard, C.; Jourdren, L.; Blugeon, C.; di Nardo, A.A.; Prochiantz, A. Non-cell Autonomous OTX2 Homeoprotein Regulates Visual Cortex Plasticity Through Gadd45b/g. Cereb. Cortex 2019, 29, 2384–2395. [Google Scholar] [CrossRef]
- Sakai, A.; Nakato, R.; Ling, Y.; Hou, X.; Hara, N.; Iijima, T.; Yanagawa, Y.; Kuwano, R.; Okuda, S.; Shirahige, K.; et al. Genome-wide target analyses of Otx2 homeoprotein in postnatal cortex. Front. Neurosci. 2017, 11, 307. [Google Scholar] [CrossRef]
- Peña, C.J.; Kronman, H.G.; Walker, D.M.; Cates, H.M.; Bagot, R.C.; Purushothaman, I.; Issler, O.; Loh, Y.-H.E.; Leong, T.; Kiraly, D.D.; et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 2017, 356, 1185–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planques, A.; Oliveira Moreira, V.; Dubreuil, C.; Prochiantz, A.; Di Nardo, A.A. OTX2 Signals from the Choroid Plexus to Regulate Adult Neurogenesis. eNeuro 2019, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, S.; Di Nardo, A.A.; Aizawa, S.; Matsuo, I.; Volovitch, M.; Prochiantz, A.; Hensch, T.K. Experience-Dependent Transfer of Otx2 Homeoprotein into the Visual Cortex Activates Postnatal Plasticity. Cell 2008, 134, 508–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, C.; Kim, H.T.; Torero Ibad, R.; Lee, E.J.; Simonutti, M.; Picaud, S.; Acampora, D.; Simeone, A.; Di Nardo, A.A.; Prochiantz, A.; et al. Graded Otx2 activities demonstrate dose-sensitive eye and retina phenotypes. Hum. Mol. Genet. 2014, 23, 1742–1753. [Google Scholar] [CrossRef] [Green Version]
- Baruch, K.; Deczkowska, A.; David, E.; Castellano, J.M.; Miller, O.; Kertser, A.; Berkutzki, T.; Barnett-Itzhaki, Z.; Bezalel, D.; Wyss-Coray, T.; et al. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 2014, 346, 89–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Vargas, V.; Maldonado-Soto, A.R.; Mizrak, D.; Codega, P.; Doetsch, F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell 2016, 19, 643–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehtinen, M.K.; Zappaterra, M.W.; Chen, X.; Yang, Y.J.; Hill, A.D.; Lun, M.; Maynard, T.; Gonzalez, D.; Kim, S.; Ye, P.; et al. The Cerebrospinal Fluid Provides a Proliferative Niche for Neural Progenitor Cells. Neuron 2011, 69, 893–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjornsson, C.S.; Apostolopoulou, M.; Tian, Y.; Temple, S. It takes a village: Constructing the neurogenic niche. Dev. Cell 2015, 32, 435–446. [Google Scholar] [CrossRef] [Green Version]
- Nguyen-Ba-Charvet, K.T.; Picard-Riera, N.; Tessier-Lavigne, M.; Baron-Van Evercooren, A.; Sotelo, C.; Chédotal, A. Multiple Roles for Slits in the Control of Cell Migration in the Rostral Migratory Stream. J. Neurosci. 2004, 24, 1497–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawamoto, K.; Wichterle, H.; Gonzalez-Perez, O.; Cholfin, J.A.; Yamada, M.; Spassky, N.; Murcia, N.S.; Garcia-Verdugo, J.M.; Marin, O.; Rubenstein, J.L.R.; et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 2006, 311, 629–632. [Google Scholar] [CrossRef]
- Ziegler, A.N.; Schneider, J.S.; Qin, M.; Tyler, W.A.; Pintar, J.E.; Fraidenraich, D.; Wood, T.L.; Levison, S.W. IGF-II promotes stemness of neural restricted precursors. Stem Cells 2012, 30, 1265–1276. [Google Scholar] [CrossRef] [Green Version]
- Falk, A.; Frisén, J. Amphiregulin is a mitogen for adult neural stem cells. J. Neurosci. Res. 2002, 69, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Sun, Y.; Xie, L.; Batteur, S.; Mao, X.O.; Smelick, C.; Logvinova, A.; Greenberg, D.A. Neurogenesis and aging: FGF-2 and HB-EGF restore neurogenesis in hippocampus and subventricular zone of aged mice. Aging Cell 2003, 2, 175–183. [Google Scholar] [CrossRef]
- Douet, V.; Kerever, A.; Arikawa-Hirasawa, E.; Mercier, F. Fractone-heparan sulphates mediate FGF-2 stimulation of cell proliferation in the adult subventricular zone. Cell Prolif. 2013, 46, 137–145. [Google Scholar] [CrossRef]
- Hayamizu, T.F.; Chan, P.T.; Johanson, C.E. FGF-2 immunoreactivity in adult rat ependyma and choroid plexus: Responses to global forebrain ischemia and intraventricular FGF-2. Neurol. Res. 2001, 23, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Tropepe, V.; Craig, C.G.; Morshead, C.M.; Van Kooy, D. Der Transforming growth factor-α null and senescent mice show decreased neural progenitor cell proliferation in the forebrain subependyma. J. Neurosci. 1997, 17, 7850–7859. [Google Scholar] [CrossRef]
- Wachs, F.P.; Winner, B.; Couillard-Despres, S.; Schiller, T.; Aigner, R.; Winkler, J.; Bogdahn, U.; Aigner, L. Transforming growth factor-β1 is a negative modulator of adult neurogenesis. J. Neuropathol. Exp. Neurol. 2006, 65, 358–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, D.J.; Passini, M.A.; Wolfe, J.H. Transduction of the choroid plexus and ependyma in neonatal mouse brain by vesicular stomatitis virus glycoprotein-pseudotyped lentivirus and adeno-associated virus type 5 vectors. Hum. Gene Ther. 2005, 16, 49–56. [Google Scholar] [CrossRef]
- Arnaud, K.; Moreira, V.O.; Vincent, J.; Dallerac, G.; le Poupon, C.; Richter, M.; Müller, U.; Rondi-Reig, L.; Prochiantz, A.; di Nardo, A. Choroid plexus APP regulates adult brain proliferation and animal behavior. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Khan, S.; Rahman, S.; Singh, L.R. The Extracellular Protein, Transthyretin Is an Oxidative Stress Biomarker. Front. Physiol. 2019, 10, 5. [Google Scholar] [CrossRef]
- Terlecky, S.R. Peroxisomes, oxidative stress, and inflammation. World J. Biol. Chem. 2012, 3, 93. [Google Scholar] [CrossRef]
- De Pablo, Y.; Nilsson, M.; Pekna, M.; Pekny, M. Intermediate filaments are important for astrocyte response to oxidative stress induced by oxygen-glucose deprivation and reperfusion. Histochem. Cell Biol. 2013, 140, 81–91. [Google Scholar] [CrossRef]
- Kim, N.; Acampora, D.; Dingli, F.; Loew, D.; Simeone, A.; Prochiantz, A.; Di Nardo, A.A. Immunoprecipitation and mass spectrometry identify non-cell autonomous Otx2 homeoprotein in the granular and supragranular layers of mouse visual cortex. F1000Research 2014, 3, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, J.Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Nédélec, S.; Foucher, I.; Brunet, I.; Bouillot, C.; Prochiantz, A.; Trembleau, A. Emx2 homeodomain transcription factor interacts with eukaryotic translation initiation factor 4E (eIF4E) in the axons of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 2004, 101, 10815–10820. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Shen, J.; Xie, L.; Wei, Z.; Wong, C.; Li, Y.; Zheng, X.; Li, P.; Song, Y. Mitotic Implantation of the Transcription Factor Prospero via Phase Separation Drives Terminal Neuronal Differentiation. Dev. Cell 2020, 52, 277–293.e8. [Google Scholar] [CrossRef] [PubMed]
- Blaudin de Thé, F.; Rekaik, H.; Peze-Heidsieck, E.; Massiani-Beaudoin, O.; Joshi, R.L.; Fuchs, J.; Prochiantz, A. Engrailed homeoprotein blocks degeneration in adult dopaminergic neurons through LINE-1 repression. EMBO J. 2018, 37, e97374. [Google Scholar] [CrossRef]
- Agoston, Z.; Schulte, D. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development 2009, 136, 3311–3322. [Google Scholar] [CrossRef] [Green Version]
- Heimbucher, T.; Murko, C.; Bajoghli, B.; Aghaallaei, N.; Huber, A.; Stebegg, R.; Eberhard, D.; Fink, M.; Simeone, A.; Czerny, T. Gbx2 and Otx2 Interact with the WD40 Domain of Groucho/Tle Corepressors. Mol. Cell. Biol. 2007, 27, 340–351. [Google Scholar] [CrossRef] [Green Version]
- McCartney, A.J.; Zhang, Y.; Weisman, L.S. Phosphatidylinositol 3,5-bisphosphate: Low abundance, high significance. BioEssays 2014, 36, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topisirovic, I.; Kentsis, A.; Perez, J.M.; Guzman, M.L.; Jordan, C.T.; Borden, K.L.B. Eukaryotic Translation Initiation Factor 4E Activity Is Modulated by HOXA9 at Multiple Levels. Mol. Cell. Biol. 2005, 25, 1100–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Holst, A.; Egbers, U.; Prochiantz, A.; Faissner, A. Neural stem/progenitor cells express 20 tenascin C isoforms that are differentially regulated by Pax6. J. Biol. Chem. 2007, 282, 9172–9181. [Google Scholar] [CrossRef] [Green Version]
- Balbinot, C.; Vanier, M.; Armant, O.; Nair, A.; Penichon, J.; Soret, C.; Martin, E.; Saandi, T.; Reimund, J.M.; Deschamps, J.; et al. Fine-tuning and autoregulation of the intestinal determinant and tumor suppressor homeobox gene CDX2 by alternative splicing. Cell Death Differ. 2017, 24, 2173–2186. [Google Scholar] [CrossRef] [Green Version]
- Fossat, N.; Chatelain, G.; Brun, G.; Lamonerie, T. Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout. EMBO Rep. 2006, 7, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Acampora, D.; di Giovannantonio, L.G.; di Salvio, M.; Mancuso, P.; Simeone, A. Selective inactivation of Otx2 mRNA isoforms reveals isoform-specific requirement for visceral endoderm anteriorization and head morphogenesis and highlights cell diversity in the visceral endoderm. Mech. Dev. 2009, 126, 882–897. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitting-Seerup, K.; Sandelin, A. IsoformSwitchAnalyzeR: Analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A. The landscape of isoform switches in human cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.S.; Jespersen, M.C.; Nielsen, H.; Jensen, K.K.; Jurtz, V.I.; Sønderby, C.K.; Sommer, M.O.A.; Winther, O.; Nielsen, M.; Petersen, B.; et al. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Prot. Struct. Funct. Bioinform. 2019, 87, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poullet, P.; Carpentier, S.; Barillot, E. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 2007, 7, 2553–2556. [Google Scholar] [CrossRef]
- Spivak, M.; Weston, J.; Bottou, L.; Käll, L.; Noble, W.S. Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J. Proteome Res. 2009, 8, 3737–3745. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Vincent, C.; Gilabert-Juan, J.; Gibel-Russo, R.; Alvarez-Fischer, D.; Krebs, M.-O.; Le Pen, G.; Prochiantz, A.; Di Nardo, A.A. Non-cell-autonomous OTX2 transcription factor regulates anxiety-related behavior in the mouse. Mol. Psychiatry 2021, 1–12. [Google Scholar] [CrossRef]


| Gene Symbol | Function | Mean Reads Combined | Mean Reads 4V ChP | Mean Reads LV ChP | Fold Change LV vs. 4V | p-adj |
|---|---|---|---|---|---|---|
| Ttr | T4 and retinol transport | 2,623,541 | 2,348,304 | 2,898,779 | 1.2 | 0.4700 |
| Enpp2 | Extracellular signaling | 593,932 | 612,769 | 575,094 | 0.94 | 1 |
| Malat1 | RNA processing | 112,726 | 157,395 | 68,058 | 0.43 | 0.1482 |
| Apoe | Lipid transport | 64,482 | 54,600 | 74,365 | 1.4 | 0.7231 |
| Trpm3 | Cation channel | 39,627 | 46,150 | 33,105 | 0.72 | 0.1606 |
| Bsg | Cell adhesion | 49,354 | 41,701 | 57,007 | 1.4 | 0.5023 |
| Kl | Cell signaling | 45,044 | 40,667 | 49,422 | 1.2 | 0.4792 |
| Abhd2 | Lipid metabolism | 41,094 | 36,405 | 45,783 | 1.3 | 0.2413 |
| AY036118 | Hemopoiesis | 25,992 | 33,903 | 18,081 | 0.53 | 1 |
| Slc4a10 | Solute transport | 30,242 | 30,563 | 29,920 | 0.98 | 1 |
| Psap | Trophic, metabolism | 33,695 | 28,768 | 38,623 | 1.3 | 0.0646 |
| Igfbp2 | IGF-binding | 25,336 | 28,556 | 22,117 | 0.77 | 0.4457 |
| Hspa5 | ER chaperone | 25,426 | 24,960 | 25,891 | 1.0 | 1 |
| F5 | Hemostasis | 25,758 | 24,672 | 26,843 | 1.1 | 0.9813 |
| Slc12a2 | Solute transport | 23,869 | 24,279 | 23,459 | 0.97 | 1 |
| Ctsd | APP processing | 28,200 | 23,672 | 32,728 | 1.4 | 0.0423 |
| Prlr | Hormone receptor | 27,452 | 22,728 | 32,176 | 1.4 | 0.7565 |
| Atp1a1 | Ion transport | 22,896 | 22,067 | 23,724 | 1.1 | 1 |
| Clu | Extracellular chaperone | 26,747 | 21,750 | 31,744 | 1.5 | 0.0323 |
| App | Cell adhesion, signaling | 19,290 | 21,197 | 17,383 | 0.82 | 0.4130 |
| Cntn1 | Cell adhesion | 25,888 | 21,017 | 30,759 | 1.5 | 0.0175 |
| Atp2b3 | Ion transport | 20,580 | 19,745 | 21,415 | 1.1 | 1 |
| Ahcyl2 | Solute transport | 20,726 | 19,427 | 22,025 | 1.1 | 0.8161 |
| Igf2 | Growth hormone | 24,978 | 18,973 | 30,984 | 1.6 | 0.0123 |
| Hsp90b1 | Chaperone | 18,816 | 18,514 | 19,117 | 1.0 | 1 |
| Sptbn1 | Cytoskeleton | 18,883 | 18,276 | 19,490 | 1.1 | 1 |
| Cpe | Prohormone processing | 17,182 | 17,449 | 16,916 | 0.97 | 1 |
| Car12 | Metabolism | 18,529 | 17,277 | 19,782 | 1.1 | 0.7979 |
| Clic6 | Ion channel | 16,953 | 16,432 | 17,473 | 1.1 | 1 |
| Strip2 | Cytoskeleton | 14,776 | 16,187 | 13,366 | 0.83 | 0.4745 |
| Timp3 | Collagenase inhibitor | 18,530 | 15,562 | 21,499 | 1.4 | 0.0292 |
| Itpr1 | ER Ca2+ release | 16,252 | 15,546 | 16,957 | 1.1 | 0.9741 |
| Kcne2 | Potassium channel | 13,861 | 15,416 | 12,305 | 0.8 | 0.8581 |
| Cgnl1 | Cell adhesion | 15,183 | 15,292 | 15,073 | 0.99 | 1 |
| Gpm6a | Membrane structure | 22,456 | 15,181 | 29,732 | 2.0 | 0.0000 |
| Slc4a2 | Solute carrier | 15,501 | 14,912 | 16,090 | 1.1 | 1 |
| Atp5a1 | Metabolism | 15,922 | 14,766 | 17,078 | 1.2 | 0.7456 |
| Nsg2 | Vesicle trafficking | 15,587 | 14,669 | 16,506 | 1.1 | 0.8949 |
| Zbtb20 | Transcription factor | 12,600 | 14,603 | 10,597 | 0.73 | 0.5636 |
| Stk39 | Stress response | 14,417 | 14,507 | 14,326 | 0.99 | 1 |
| Tmem72 | 14,821 | 14,343 | 15,298 | 1.1 | 1 | |
| Cab39l | Cell polarity | 15,438 | 14,311 | 16,565 | 1.2 | 0.8214 |
| Nedd4 | Ubiquitination | 15,837 | 14,272 | 17,402 | 1.2 | 0.4738 |
| Macf1 | Cytoskeleton | 12,704 | 14,027 | 11,382 | 0.81 | 0.5999 |
| Vat1l | 14,589 | 13,983 | 15,196 | 1.1 | 1 | |
| Hsp90ab1 | Chaperone | 14,368 | 13,756 | 14,979 | 1.1 | 1 |
| Calr | Chaperone | 13,889 | 13,606 | 14,171 | 1.0 | 1 |
| Htr2c | Serotonin receptor | 13,453 | 13,343 | 13,564 | 1.0 | 1 |
| Slc5a3 | Solute transport | 11,648 | 13,281 | 10,015 | 0.75 | 0.0914 |
| Sptan1 | Cytoskeleton, Secretion | 11,913 | 13,161 | 10,665 | 0.81 | 0.3292 |
| Gene Symbol | Mean Reads Vehicle | Mean Reads Cre-Tat | Fold Change | p-adj |
|---|---|---|---|---|
| Upregulated in lateral ventricle ChP | ||||
| Igkv1-135 | 0.6 | 33 | 61 | 0.0000 |
| Slc1a6 | 2.1 | 82 | 39 | 0.0000 |
| Mup5 | 123 | 4119 | 34 | 0.0000 |
| Gpx3 | 295 | 7229 | 25 | 0.0000 |
| Ighv1-67 | 1.6 | 36 | 22 | 0.0000 |
| Saa3 | 2.2 | 47 | 22 | 0.0000 |
| Tnfrsf11b | 6.3 | 116 | 18 | 0.0000 |
| Cacnb3 | 24 | 325 | 13 | 0.0000 |
| Ndnf | 89 | 1098 | 12 | 0.0000 |
| Gm4841 | 3.8 | 44 | 12 | 0.0000 |
| Downregulated in lateral ventricle ChP | ||||
| Ngfr | 444 | 83 | −5.3 | 0.0000 |
| Nrn1 | 1924 | 496 | −3.9 | 0.0000 |
| B3galt2 | 109 | 31 | −3.6 | 0.0007 |
| Dazl | 207 | 60 | −3.4 | 0.0000 |
| Itga10 | 184 | 54 | −3.4 | 0.0000 |
| Slc26a7 | 1670 | 533 | −3.1 | 0.0000 |
| Steap1 | 2224 | 735 | −3.0 | 0.0000 |
| Defb11 | 473 | 169 | −2.8 | 0.0000 |
| Entpd3 | 240 | 87 | −2.8 | 0.0001 |
| Ccl9 | 2037 | 774 | −2.6 | 0.0000 |
| Upregulated in 4th ventricle ChP | ||||
| 9030619P08Rik | 0.00 | 28.6 | infinite | 0.0000 |
| Tmigd1 | 0.00 | 33.1 | infinite | 0.0000 |
| A730020M07Rik | 1.3 | 76 | 61 | 0.0000 |
| Gpx3 | 181 | 10,780 | 59 | 0.0000 |
| Cacnb3 | 18 | 778 | 43 | 0.0000 |
| Mup5 | 26 | 1053 | 41 | 0.0000 |
| Fmod * | 286 | 8688 | 30 | 0.0000 |
| Slitrk6 | 4.7 | 125 | 27 | 0.0000 |
| Ndnf | 84 | 2070 | 25 | 0.0000 |
| Adcy8 | 10 | 233 | 23 | 0.0000 |
| Downregulated in 4th ventricle ChP | ||||
| Ngfr | 684 | 159 | −4.3 | 0.0000 |
| Steap1 * | 1225 | 326 | −3.8 | 0.0000 |
| Elfn1 | 104 | 28 | −3.7 | 0.0000 |
| Gnmt | 122 | 33 | −3.7 | 0.0000 |
| Gm22650 | 141 | 40 | −3.5 | 0.0000 |
| Mir448 | 95 | 28 | −3.4 | 0.0002 |
| Igf2os | 89 | 26 | −3.4 | 0.0005 |
| B3galt2 | 141 | 44 | −3.2 | 0.0000 |
| Slc26a7 | 843 | 271 | −3.1 | 0.0000 |
| Crhr2 | 504 | 164 | −3.1 | 0.0000 |
| Up-Regulated | Function | Downregulated | Function |
|---|---|---|---|
| Adora1 | Adenosine receptor | Aqp1 * | Osmotic gradient |
| Arrb1 | Receptor signaling | Atp2b4 | Ion transport |
| Atp1a2 * | Ion transport | B3galt2 | Glycosylation |
| Cadm1 | Cell adhesion | Elfn1 | Signaling cascade |
| Cd55 | Complement cascade | Entpd3 | |
| Cfap46 | Fam132a | Glucose uptake | |
| Chn2 | Signaling cascade | Igf2 | Growth factor |
| Col11a1 | Collagen II fibrils | Ins2 | Glucose uptake |
| Col1a2 | Collagen I fibrils | Kalrn | Signaling cascade |
| Edn3 | Vasoconstriction | Klhl36 | Ubiquitination |
| Eva1a | Cell death | Mapk9 | Cell signaling |
| Fam211b | Myo5b * | Cell trafficking | |
| Fgf1 | Growth factor | Myrip * | Cell trafficking |
| Flrt1 | FGF signaling | Nav3 | Immune response |
| Fmod * | Collagen I and II fibrils | Otx2 | Transcription factor |
| Gda | Metabolism | Pcnx * | |
| Gpx3 | Oxidative stress | Pitpnm1 * | Cytoskeleton |
| Hopx * | Chromatin structure | Pomgnt1 | Metabolism |
| Layn | Hyaluronan receptor | Rcn1 | ER regulation |
| Lrrc18 | Spermatogenesis | Scg5 | Cell secretion |
| Mapk10 | Cell signaling | Sfrp1 | Wnt signaling |
| Matn2 | Extracellular matrix | Slc29a4 | Cation transport |
| Megf11 * | Cell adhesion | Slc2a12 * | Glucose transport |
| Mlc1 | Osmotic gradient | Slc35f1 | Solute transport |
| Mup5 | Pheromone activity | Slc41a2 | Magnesium transport |
| Ndnf | Cell adhesion, growth | Stra6 | Retinol transport |
| Ndrg3 | Tbc1d2 | Cell adhesion | |
| Pi15 | Protease inhibitor | Tbcd | Cytoskeleton |
| Plin4 | Adipocyte formation | Thumpd3 | |
| Rufy4 | Autophagy | Tmem255b | |
| Sel1l3 | Tmprss11a | Cellular senescence | |
| Sema5a * | Cell adhesion | Tspan33 * | Notch signaling |
| Shisal1 | Ttr | Retinol and T4 transport | |
| Smrp1 | Cilia function | Wdr17 * | |
| Sncg * | Neurofilament network | ||
| Sned1 * | |||
| Sorcs2 | Signaling cascade | ||
| Sorl1 | Cell trafficking | ||
| Sulf2 | Extracellular matrix | ||
| Tm4sf1 | |||
| Vim | Cell filaments | ||
| Vwa5b1 |
| Gene | Choroid Plexus | Mean Reads, Control (Either Veh or Wildtype) | Mean Reads, Knockdown (Either Cre-Tat or Otx2+/GFP) | Fold Change | p-adj |
|---|---|---|---|---|---|
| Bmp7 | Otx2lox/lox LV | 5125 | 7090 | 1.4 | 0.1845 |
| Otx2lox/lox 4V | 2547 | 4036 | 1.6 | 0.0014 | |
| Otx2+/GFP 4V | 2215 | 2261 | 1.0 | 1 | |
| Wnt2b | Otx2lox/lox LV | 10 | 66 | 6.6 | 0.0002 |
| Otx2lox/lox 4V | 12 | 66 | 5.4 | 0.0001 | |
| Otx2+/GFP 4V | 29 | 35 | 1.2 | 1 | |
| Tgm2 | Otx2lox/lox LV | 276 | 422 | 1.5 | 0.2496 |
| Otx2lox/lox 4V | 138 | 265 | 1.9 | 0.0040 | |
| Otx2+/GFP 4V | 388 | 404 | 1.0 | 1 | |
| Sfrp1 | Otx2lox/lox LV | 8536 | 4511 | 0.53 | 0.0000 |
| Otx2lox/lox 4V | 5121 | 2726 | 0.53 | 0.0000 | |
| Otx2+/GFP 4V | 4087 | 2001 | 0.49 | 0.0392 | |
| Sostdc1 | Otx2lox/lox LV | 8627 | 4429 | 0.51 | 0.0000 |
| Otx2lox/lox 4V | 5412 | 2793 | 0.52 | 0.0076 | |
| Otx2+/GFP 4V | 8932 | 8458 | 1.1 | 1 | |
| Shh | Otx2lox/lox LV | 0 | 1.4 | infinite | 1 |
| Otx2lox/lox 4V | 0.7 | 4.6 | 6.6 | 1 | |
| Otx2+/GFP 4V | 0 | 64 | infinite | 0.0000 | |
| Slit2 | Otx2lox/lox LV | 2302 | 2674 | 1.2 | 1 |
| Otx2lox/lox 4V | 3471 | 4803 | 1.4 | 0.7159 | |
| Otx2+/GFP 4V | 4323 | 6099 | 1.4 | 0.6487 | |
| Fgf2 | Otx2lox/lox LV | 140 | 123 | 0.88 | 1 |
| Otx2lox/lox 4V | 64 | 84 | 1.3 | 0.9878 | |
| Otx2+/GFP 4V | 67 | 66 | 1.0 | 1 | |
| Areg | Otx2lox/lox LV | 0 | 0.9 | infinite | 1 |
| Otx2lox/lox 4V | NA | NA | NA | NA | |
| Otx2+/GFP 4V | 0 | 0 | NA | NA | |
| Tgf-α | Otx2lox/lox LV | 2140 | 1425 | 0.67 | 0.0620 |
| Otx2lox/lox 4V | 1357 | 999 | 0.74 | 0.1611 | |
| Otx2+/GFP 4V | 1040 | 1673 | 1.6 | 0.2163 | |
| Tgf-β2 | Otx2lox/lox LV | 9233 | 4747 | 0.51 | 0.0000 |
| Otx2lox/lox 4V | 3755 | 2169 | 0.58 | 0.0000 | |
| Otx2+/GFP 4V | 3324 | 2563 | 0.77 | 0.3602 | |
| Igf2 | Otx2lox/lox LV | 42,336 | 19,144 | 0.45 | 0.0008 |
| Otx2lox/lox 4V | 18,717 | 8087 | 0.43 | 0.0000 | |
| Otx2+/GFP 4V | 42,542 | 17,197 | 0.40 | 0.0000 | |
| Igfbp2 | Otx2lox/lox LV | 30,195 | 20,992 | 0.70 | 0.3255 |
| Otx2lox/lox 4V | 28,166 | 13,235 | 0.47 | 0.0003 | |
| Otx2+/GFP 4V | 47,529 | 21,660 | 0.46 | 0.0000 |
| List Name | Choroid Plexus | SVZ | RMS | Visual Cortex |
|---|---|---|---|---|
| Total proteins OTX2 | 4814 | 1138 | 2425 | 2644 |
| Total proteins IgG | 3602 | 1776 | 2274 | 2667 |
| Unique proteins OTX2 (≥3 peptides) | 392 | 17 | 40 | 29 |
| Unique proteins IgG (≥3 peptides) | 59 | 139 | 22 | 25 |
| Selected proteins OTX2 (≥50% rel. ∆) | 653 | 6 | 75 | 37 |
| Unique small proteins Otx2 (≤25 kDa) | 182 | 31 | 68 | 48 |
| Total OTX2 partners | 1195 of 4814 | 52 of 1138 | 180 of 2425 | 109 of 2644 |
| Protein | Function | Protein | Function |
|---|---|---|---|
| ABCF1 * | Translation | MAP4 | Cytoskeleton |
| ACOT11 | Lipid metabolism | MCM3AP | RNA export |
| AGO1 * | RNA silencing | MECP2 | Transcription, epigenetics |
| APC | Cell adhesion | MLYCD | Metabolism |
| ARHGEF6 | Trafficking | MOV10 * | RNA and LINE-1 silencing |
| ARHGEF7 | Trafficking, cell adhesion | MSI2 * | Translation |
| ARVCF | Cell adhesion | MYCBP2 | Transcription |
| CDH2 | Cell adhesion | NFATC2 | Signaling |
| CDH3 | Cell adhesion | PIKFYVE | Trafficking |
| CHD4 | Cell adhesion | PITPNM2 | Trafficking |
| CTNNA1 | Cell adhesion | POLDIP3 | Translation |
| CTNNA2 | Cell adhesion | PRRC2A * | RNA splicing, stress granule |
| CTNNB1 | Cell adhesion | RBM39 | RNA splicing |
| DDX41 | RNA splicing | RHOT1 | Mitochondrial trafficking |
| EDC4 * | RNA processing | RPL19 | |
| EPB41L5 | Cell adhesion | RPL21 | Translation |
| ERBIN | Signaling | RPL22 | |
| FIG4 | Trafficking | RPL29 | Translation |
| FMNL3 | Cytoskeleton | RPL35 | Translation |
| GIT1 | Trafficking, cell adhesion | RPL36A | |
| GIT2 | Trafficking | SRRM2 | RNA splicing |
| GJA1 | Gap junction | STRAP * | Stress response |
| GPAM | Metabolism | TJP2 | Cell adhesion |
| GTPBP1 | RNA processing | TMPO | Nuclear membrane |
| HNRPLL * | RNA splicing | TNS2 | Signaling |
| ILF2 | Transcription | TRPV4 | Osmotic sensitivity |
| KIFAP3 | Chromosome structure | VAC14 | Trafficking |
| LBR | Metabolism | VRK3 | Signaling |
| LIG3 | DNA repair | WDR70 | |
| MAP1A | Cytoskeleton | ZFR | RNA export |
| Protein | Function | SVZ | RMS | VCx | ChP | ||||
|---|---|---|---|---|---|---|---|---|---|
| Otx2 co-IP | IgG co-IP | Otx2 co-IP | IgG co-IP | Otx2 co-IP | IgG co-IP | Otx2 co-IP | IgG co-IP | ||
| ACIN1 | mRNA splicing | 3 | 0 | 11 | 1 | 16 | 4 | ||
| ACOT11 | Lipid metabolism | 4 | 0 | 5 | 0 | 8 | 1 | 34 | 7 |
| ARCN1 | Protein transport | 3 | 1 | 7 | 2 | 4 | 1 | ||
| DDX46 | mRNA splicing | 30 | 0 | 45 | 1 | 34 | 3 | ||
| EIF4A3 | mRNA translation | 4 | 0 | 10 | 2 | 11 | 5 | ||
| FIG4 | PI(3,5)P2 regulation, MVB | 20 | 0 | 26 | 0 | 27 | 0 | 43 | 0 |
| KCND3 | Potassium channel | 6 | 0 | 5 | 0 | 7 | 0 | 6 | 0 |
| PIKFYVE | PI(3,5)P2 regulation, MVB | 36 | 0 | 42 | 0 | 59 | 0 | 122 | 1 |
| RBM25 | mRNA splicing | 5 | 0 | 11 | 1 | 12 | 3 | ||
| SF3A1 | mRNA splicing | 3 | 0 | 11 | 3 | 13 | 6 | ||
| SF3B1 | mRNA splicing | 3 | 0 | 22 | 0 | 25 | 9 | ||
| SNRNP200 | mRNA splicing | 5 | 0 | 14 | 1 | 31 | 4 | ||
| THOC2 | mRNA export | 3 | 0 | 5 | 0 | 3 | 0 | ||
| VAC14 | PI(3,5)P2 regulation, MVB | 39 | 2 | 49 | 3 | 56 | 0 | 75 | 0 |
| KEGG Pathway | Size | p-adj |
|---|---|---|
| Choroid plexus (1326 proteins) | ||
| Tight junction | 27 of 137 | 0.0000 |
| Metabolic pathways | 77 of 1184 | 0.0000 |
| Protein processing in ER | 26 of 169 | 0.0000 |
| RNA transport | 24 of 168 | 0.0000 |
| Ribosome biogenesis | 18 or 86 | 0.0000 |
| Regulation of actin cytoskeleton | 27 of 216 | 0.0000 |
| Ribosome | 19 of 119 | 0.0000 |
| Spliceosome | 20 of 138 | 0.0000 |
| Oxidative phosphorylation | 20 of 147 | 0.0000 |
| Axon guidance | 18 of 131 | 0.0000 |
| SVZ (79 proteins) | ||
| Ribosome | 15 of 119 | 0.0000 |
| Spliceosome | 15 of 138 | 0.0000 |
| RNA transport | 4 of 168 | 0.0004 |
| mRNA surveillance | 2 of 93 | 0.0219 |
| Neurotrophin signaling | 2 of 131 | 0.0297 |
| RMS (219 proteins) | ||
| Spliceosome | 28 of 138 | 0.0000 |
| Oxidative phosphorylation | 11 of 147 | 0.0000 |
| Metabolic pathways | 24 of 1184 | 0.0000 |
| Alzheimer disease | 10 of 188 | 0.0000 |
| mRNA surveillance | 8 of 93 | 0.0000 |
| Visual cortex (192 proteins) | ||
| Spliceosome | 17 of 138 | 0.0000 |
| RNA transport | 7 of 168 | 0.0000 |
| Metabolic pathways | 14 of 1184 | 0.0005 |
| Insulin signaling | 5 of 137 | 0.0007 |
| Oxidative phosphorylation | 5 of 147 | 0.0007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Planques, A.; Oliveira Moreira, V.; Benacom, D.; Bernard, C.; Jourdren, L.; Blugeon, C.; Dingli, F.; Masson, V.; Loew, D.; Prochiantz, A.; et al. OTX2 Homeoprotein Functions in Adult Choroid Plexus. Int. J. Mol. Sci. 2021, 22, 8951. https://doi.org/10.3390/ijms22168951
Planques A, Oliveira Moreira V, Benacom D, Bernard C, Jourdren L, Blugeon C, Dingli F, Masson V, Loew D, Prochiantz A, et al. OTX2 Homeoprotein Functions in Adult Choroid Plexus. International Journal of Molecular Sciences. 2021; 22(16):8951. https://doi.org/10.3390/ijms22168951
Chicago/Turabian StylePlanques, Anabelle, Vanessa Oliveira Moreira, David Benacom, Clémence Bernard, Laurent Jourdren, Corinne Blugeon, Florent Dingli, Vanessa Masson, Damarys Loew, Alain Prochiantz, and et al. 2021. "OTX2 Homeoprotein Functions in Adult Choroid Plexus" International Journal of Molecular Sciences 22, no. 16: 8951. https://doi.org/10.3390/ijms22168951
APA StylePlanques, A., Oliveira Moreira, V., Benacom, D., Bernard, C., Jourdren, L., Blugeon, C., Dingli, F., Masson, V., Loew, D., Prochiantz, A., & Di Nardo, A. A. (2021). OTX2 Homeoprotein Functions in Adult Choroid Plexus. International Journal of Molecular Sciences, 22(16), 8951. https://doi.org/10.3390/ijms22168951






