Conserved and Distinct Elements of Phagocytosis in Human and C. elegans
Abstract
1. Introduction
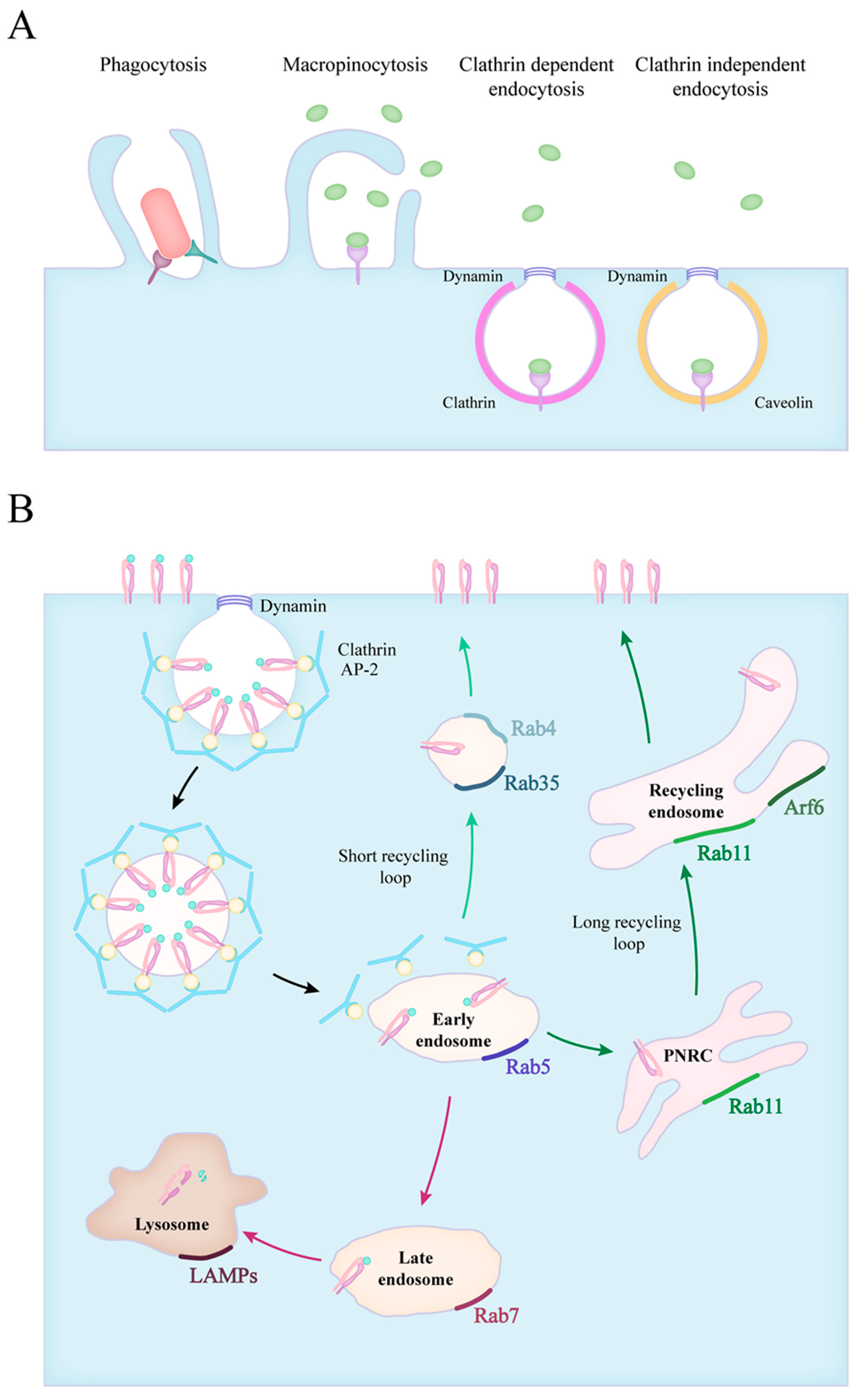
2. Forms of Endocytosis
3. Endocytic Route of Engulfed Particles and Receptor Recyclization
4. Phagocytic Receptors and Pathogen Recognition
4.1. Pattern Recognition and Opsonic Receptors in Mammals
4.2. Innate Immunity and the Lack of Direct Pathogen Recognition in C. elegans
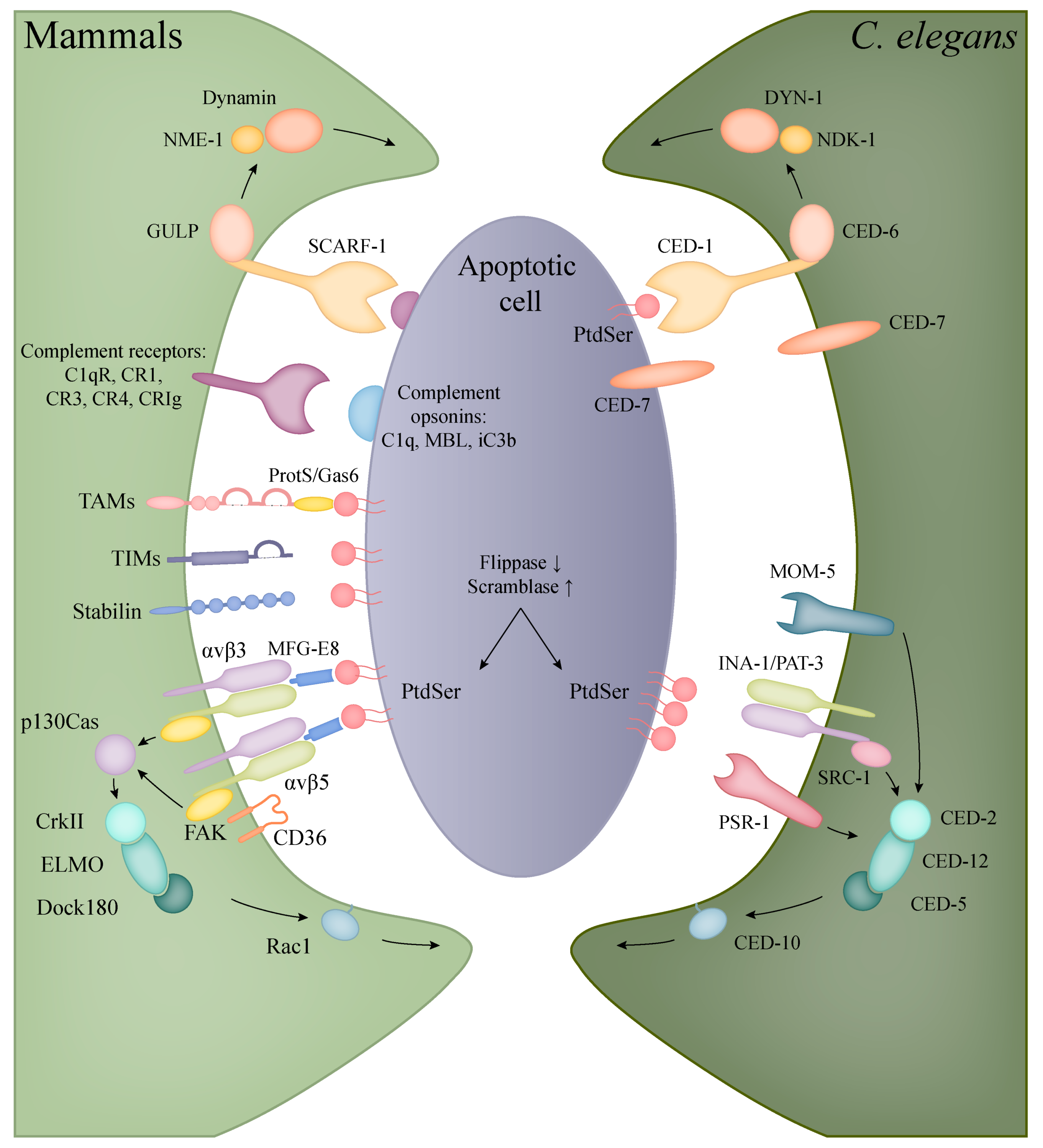
5. Phagocytosis of Apoptotic Cells
5.1. Apoptotic Engulfment in Mammals
5.2. Apoptotic Engulfment and Clearance in C. elegans
5.3. The Interconnection of Innate Immunity with Apoptotic Cell Clearance and the Nervous System
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef]
- Kim, D.H. Signaling in the Innate Immune Response; Wormbook: Pasadena, CA, USA, 2018; pp. 1–35. [Google Scholar] [CrossRef]
- Cummings, R.J.; Barbet, G.; Bongers, G.; Hartmann, B.M.; Gettler, K.; Muniz, L.; Furtado, G.C.; Cho, J.; Lira, S.A.; Blander, J.M. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature 2016, 539, 565–569. [Google Scholar] [CrossRef]
- Hart, S.P.; Dransfield, I.; Rossi, A.G. Phagocytosis of apoptotic cells. Methods 2008, 44, 280–285. [Google Scholar] [CrossRef]
- Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016, 16, 35–50. [Google Scholar] [CrossRef]
- Nathan, C.; Cunningham-Bussel, A. Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 2013, 13, 349–361. [Google Scholar] [CrossRef]
- Segal, B.H.; Grimm, M.J.; Khan, A.N.H.; Han, W.; Blackwell, T.S. Regulation of innate immunity by NADPH oxidase. Free Radic. Biol. Med. 2012, 53, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, H.; Sies, H. Protection against reactive oxygen species by selenoproteins. Biochim. Biophys. Acta 2009, 1790, 1478–1485. [Google Scholar] [CrossRef]
- Cadet, J.; Ravanat, J.-L.; TavernaPorro, M.; Menoni, H.; Angelov, D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 2012, 327, 5–15. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef]
- Lugrin, J.; Rosenblatt-Velin, N.; Parapanov, R.; Liaudet, L. The role of oxidative stress during inflammatory processes. Biol. Chem. 2014, 395, 203–230. [Google Scholar] [CrossRef]
- Gray, M.; Botelho, R.J. Phagocytosis: Hungry, Hungry Cells. Methods Mol. Biol. 2017, 1519, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Conradt, B.; Xue, D. Programmed Cell Death; WormBook: Pasadena, CA, USA, 2005; pp. 1–13. [Google Scholar] [CrossRef]
- Conradt, B.; Wu, Y.-C.; Xue, D. Programmed Cell Death During Caenorhabditis elegans Development. Genetics 2016, 203, 1533–1562. [Google Scholar] [CrossRef] [PubMed]
- Klöditz, K.; Chen, Y.Z.; Xue, D.; Fadeel, B. Programmed cell clearance: From nematodes to humans. Biochem. Biophys. Res. Commun. 2017, 482, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Yuan, L.; Jing, H.; Zheng, Q.; Xiao, H. Defective apoptotic cell clearance activates innate immune response to protect Caenorhabditis elegans against pathogenic bacteria. Virulence 2021, 12, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Walpole, G.F.W.; Grinstein, S. Endocytosis and the internalization of pathogenic organisms: Focus on phosphoinositides. F1000Research 2020, 9, 1–17. [Google Scholar] [CrossRef]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Schlam, D.; Breuer, C.; Gütschow, M.; Glogauer, M.; Grinstein, S. Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat. Commun. 2016, 7, 11284. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.L.; Frolov, V.A. Dynamin: Functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 2011, 27, 79–105. [Google Scholar] [CrossRef] [PubMed]
- Marks, B.; Stowell, M.H.; Vallis, Y.; Mills, I.G.; Gibson, A.; Hopkins, C.R.; McMahon, H.T. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 2001, 410, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Mayor, S.; Riezman, H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004, 5, 110–120. [Google Scholar] [CrossRef]
- Howes, M.T.; Mayor, S.; Parton, R.G. Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Curr. Opin. Cell Biol. 2010, 22, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. Phagocytosis: Phenotypically Simple Yet a Mechanistically Complex Process. Int. Rev. Immunol. 2020, 39, 118–150. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 1996, 12, 575–625. [Google Scholar] [CrossRef]
- Blander, J.M.; Medzhitov, R. On regulation of phagosome maturation and antigen presentation. Nat. Immunol. 2006, 7, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Gruenberg, J.; van der Goot, F.G. Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 2006, 7, 495–504. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Christoforidis, S.; McBride, H.M.; Burgoyne, R.D.; Zerial, M. The Rab5 effector EEA1 is a core component of endosome docking. Nature 1999, 397, 621–625. [Google Scholar] [CrossRef]
- Callaghan, J.; Nixon, S.; Bucci, C.; Toh, B.H.; Stenmark, H. Direct interaction of EEA1 with Rab5b. Eur. J. Biochem. 1999, 265, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Daro, E.; van der Sluijs, P.; Galli, T.; Mellman, I. Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA 1996, 93, 9559–9564. [Google Scholar] [CrossRef]
- van der Sluijs, P.; Hull, M.; Webster, P.; Mâle, P.; Goud, B.; Mellman, I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 1992, 70, 729–740. [Google Scholar] [CrossRef]
- Perrin, L.; Laura, P.; Lacas-Gervais, S.; Sandra, L.-G.; Gilleron, J.; Jérôme, G.; Ceppo, F.; Franck, C.; Prodon, F.; François, P.; et al. Rab4b controls an early endosome sorting event by interacting with the γ-subunit of the clathrin adaptor complex 1. J. Cell Sci. 2013, 126, 4950–4962. [Google Scholar] [CrossRef]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Vieira, O.V.; Bucci, C.; Harrison, R.E.; Trimble, W.S.; Lanzetti, L.; Gruenberg, J.; Schreiber, A.D.; Stahl, P.D.; Grinstein, S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol. Cell. Biol. 2003, 23, 2501–2514. [Google Scholar] [CrossRef]
- Beyenbach, K.W.; Wieczorek, H. The V-type H+ ATPase: Molecular structure and function, physiological roles and regulation. J. Exp. Biol. 2006, 209, 577–589. [Google Scholar] [CrossRef]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef]
- Harding, C.V.; Geuze, H.J. Class II MHC molecules are present in macrophage lysosomes and phagolysosomes that function in the phagocytic processing of Listeria monocytogenes for presentation to T cells. J. Cell Biol. 1992, 119, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Minakami, R.; Sumimotoa, H. Phagocytosis-coupled activation of the superoxide-producing phagocyte oxidase, a member of the NADPH oxidase (nox) family. Int. J. Hematol. 2006, 84, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Balce, D.R.; Li, B.; Allan, E.R.O.; Rybicka, J.M.; Krohn, R.M.; Yates, R.M. Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood 2011, 118, 4199–4208. [Google Scholar] [CrossRef] [PubMed]
- Yates, R.M.; Hermetter, A.; Taylor, G.A.; Russell, D.G. Macrophage activation downregulates the degradative capacity of the phagosome. Traffic 2007, 8, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Delamarre, L.; Pack, M.; Chang, H.; Mellman, I.; Trombetta, E.S. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 2005, 307, 1630–1634. [Google Scholar] [CrossRef]
- Delamarre, L.; Couture, R.; Mellman, I.; Trombetta, E.S. Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. J. Exp. Med. 2006, 203, 2049–2055. [Google Scholar] [CrossRef]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef]
- Lobert, V.H.; Brech, A.; Pedersen, N.M.; Wesche, J.; Oppelt, A.; Malerød, L.; Stenmark, H. Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell 2010, 19, 148–159. [Google Scholar] [CrossRef]
- Roberts, M.; Barry, S.; Woods, A.; van der Sluijs, P.; Norman, J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 2001, 11, 1392–1402. [Google Scholar] [CrossRef]
- Arjonen, A.; Alanko, J.; Veltel, S.; Ivaska, J. Distinct recycling of active and inactive β1 integrins. Traffic 2012, 13, 610–625. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, O.; Molecular, E. Rab11 Regulates Recycling through the Pericentriolar Recycling Endosome. J. Cell Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Powelka, A.M.; Sun, J.; Li, J.; Gao, M.; Shaw, L.M.; Sonnenberg, A.; Hsu, V.W. Stimulation-dependent recycling of integrin beta1 regulated by ARF6 and Rab11. Traffic 2004, 5, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, N.; Scott, D.W.; Castle, J.D.; Casanova, J.E.; Schwartz, M.A. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat. Cell Biol. 2007, 9, 1381–1391. [Google Scholar] [CrossRef]
- Kouranti, I.; Sachse, M.; Arouche, N.; Goud, B.; Echard, A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 2006, 16, 1719–1725. [Google Scholar] [CrossRef]
- Fares, H.; Grant, B. Deciphering endocytosis in Caenorhabditis elegans. Traffic 2002, 3, 11–19. [Google Scholar] [CrossRef]
- Balklava, Z.; Pant, S.; Fares, H.; Grant, B.D. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 2007, 9, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Fares, H.; Greenwald, I. Genetic analysis of endocytosis in Caenorhabditis elegans: Coelomocyte uptake defective mutants. Genetics 2001, 159, 133–145. [Google Scholar] [CrossRef]
- Stuart, L.M.; Ezekowitz, R.A.B. Phagocytosis: Elegant complexity. Immunity 2005, 22, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Xu, X.; Hereld, D. Chemotaxis, chemokine receptors and human disease. Cytokine 2008, 44, 1–8. [Google Scholar] [CrossRef]
- Sozzani, S.; Sallusto, F.; Luini, W.; Zhou, D.; Piemonti, L.; Allavena, P.; Van Damme, J.; Valitutti, S.; Lanzavecchia, A.; Mantovani, A. Migration of dendritic cells in response to formyl peptides, C5a, and a distinct set of chemokines. J. Immunol. 1995, 155, 3292–3295. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, K.; Yoshimura, T.; Liu, Y.; Gong, W.; Wang, A.; Gao, J.-L.; Murphy, P.M.; Wang, J.M. Formylpeptide receptors are critical for rapid neutrophil mobilization in host defense against Listeria monocytogenes. Sci. Rep. 2012, 2, 1–7. [Google Scholar] [CrossRef]
- Capucetti, A.; Albano, F.; Bonecchi, R. Multiple Roles for Chemokines in Neutrophil Biology. Front. Immunol. 2020, 11, 1259. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Platnich, J.M.; Muruve, D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Olagnier, D.; Lin, R. Host and Viral Modulation of RIG-I-Mediated Antiviral Immunity. Front. Immunol. 2016, 7, 662. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Gordon, S. Immune recognition. A new receptor for beta-glucans. Nature 2001, 413, 36–37. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Tsoni, S.V.; Willment, J.A.; Dennehy, K.M.; Rosas, M.; Findon, H.; Haynes, K.; Steele, C.; Botto, M.; Gordon, S.; et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 2007, 8, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Lima-Junior, D.S.; Mineo, T.W.P.; Calich, V.L.G.; Zamboni, D.S. Dectin-1 Activation during Leishmania amazonensis Phagocytosis Prompts Syk-Dependent Reactive Oxygen Species Production to Trigger Inflammasome Assembly and Restriction of Parasite Replication. J. Immunol. 2017, 199, 2055–2068. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Schorey, J.S. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood 2006, 108, 3168–3175. [Google Scholar] [CrossRef]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- McGreal, E.P.; Rosas, M.; Brown, G.D.; Zamze, S.; Wong, S.Y.C.; Gordon, S.; Martinez-Pomares, L.; Taylor, P.R. The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 2006, 16, 422–430. [Google Scholar] [CrossRef]
- Sato, K.; Yang, X.; Yudate, T.; Chung, J.-S.; Wu, J.; Luby-Phelps, K.; Kimberly, R.P.; Underhill, D.; Cruz, P.D.; Ariizumi, K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem. 2006, 281, 38854–38866. [Google Scholar] [CrossRef] [PubMed]
- Yonekawa, A.; Saijo, S.; Hoshino, Y.; Miyake, Y.; Ishikawa, E.; Suzukawa, M.; Inoue, H.; Tanaka, M.; Yoneyama, M.; Oh-Hora, M.; et al. Dectin-2 is a direct receptor for mannose-capped lipoarabinomannan of mycobacteria. Immunity 2014, 41, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Saijo, S.; Ikeda, S.; Yamabe, K.; Kakuta, S.; Ishigame, H.; Akitsu, A.; Fujikado, N.; Kusaka, T.; Kubo, S.; Chung, S.; et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 2010, 32, 681–691. [Google Scholar] [CrossRef]
- Ishikawa, E.; Ishikawa, T.; Morita, Y.S.; Toyonaga, K.; Yamada, H.; Takeuchi, O.; Kinoshita, T.; Akira, S.; Yoshikai, Y.; Yamasaki, S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 2009, 206, 2879–2888. [Google Scholar] [CrossRef]
- Lu, X.; Nagata, M.; Yamasaki, S. Mincle: 20 years of a versatile sensor of insults. Int. Immunol. 2018, 30, 233–239. [Google Scholar] [CrossRef]
- Yamasaki, S.; Matsumoto, M.; Takeuchi, O.; Matsuzawa, T.; Ishikawa, E.; Sakuma, M.; Tateno, H.; Uno, J.; Hirabayashi, J.; Mikami, Y.; et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc. Natl. Acad. Sci. USA 2009, 106, 1897–1902. [Google Scholar] [CrossRef]
- Behler-Janbeck, F.; Takano, T.; Maus, R.; Stolper, J.; Jonigk, D.; Tort Tarrés, M.; Fuehner, T.; Prasse, A.; Welte, T.; Timmer, M.S.M.; et al. C-type Lectin Mincle Recognizes Glucosyl-diacylglycerol of Streptococcus pneumoniae and Plays a Protective Role in Pneumococcal Pneumonia. PLoS Pathog. 2016, 12, e1006038. [Google Scholar] [CrossRef]
- Sharma, A.; Steichen, A.L.; Jondle, C.N.; Mishra, B.B.; Sharma, J. Protective role of Mincle in bacterial pneumonia by regulation of neutrophil mediated phagocytosis and extracellular trap formation. J. Infect. Dis. 2014, 209, 1837–1846. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Toyonaga, K.; Mori, D.; Kakuta, S.; Hoshino, Y.; Oyamada, A.; Yamada, H.; Ono, K.-I.; Suyama, M.; Iwakura, Y.; et al. C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity 2013, 38, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.J.; Marakalala, M.J.; Hoving, J.C.; van Laarhoven, A.; Drummond, R.A.; Kerscher, B.; Keeton, R.; van de Vosse, E.; Ottenhoff, T.H.M.; Plantinga, T.S.; et al. The C-type lectin receptor CLECSF8/CLEC4D is a key component of anti-mycobacterial immunity. Cell Host Microbe 2015, 17, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Pascual, A.; Saether, P.C.; Fossum, S.; Dissen, E.; Daws, M.R. Mincle, the receptor for mycobacterial cord factor, forms a functional receptor complex with MCL and FcεRI-γ. Eur. J. Immunol. 2013, 43, 3167–3174. [Google Scholar] [CrossRef] [PubMed]
- van Liempt, E.; Bank, C.M.C.; Mehta, P.; Garciá-Vallejo, J.J.; Kawar, Z.S.; Geyer, R.; Alvarez, R.A.; Cummings, R.D.; Kooyk, Y.; van Die, I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006, 580, 6123–6131. [Google Scholar] [CrossRef]
- Appelmelk, B.J.; van Die, I.; van Vliet, S.J.; Vandenbroucke-Grauls, C.M.J.E.; Geijtenbeek, T.B.H.; van Kooyk, Y. Cutting edge: Carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 2003, 170, 1635–1639. [Google Scholar] [CrossRef] [PubMed]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.A.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.L.; et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.P.; Lasala, F.; Carrillo, J.; Muñiz, O.; Corbí, A.L.; Delgado, R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002, 76, 6841–6844. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Gramberg, T.; Simmons, G.; Möller, P.; Rennekamp, A.J.; Krumbiegel, M.; Geier, M.; Eisemann, J.; Turza, N.; Saunier, B.; et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 12090–12095. [Google Scholar] [CrossRef]
- Cormier, E.G.; Durso, R.J.; Tsamis, F.; Boussemart, L.; Manix, C.; Olson, W.C.; Gardner, J.P.; Dragic, T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA 2004, 101, 14067–14072. [Google Scholar] [CrossRef]
- Halary, F.; Amara, A.; Lortat-Jacob, H.; Messerle, M.; Delaunay, T.; Houlès, C.; Fieschi, F.; Arenzana-Seisdedos, F.; Moreau, J.F.; Déchanet-Merville, J. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 2002, 17, 653–664. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Middel, J.; Cornelissen, I.L.M.H.A.; Nottet, H.S.L.M.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef]
- Trumpfheller, C.; Park, C.G.; Finke, J.; Steinman, R.M.; Granelli-Piperno, A. Cell type-dependent retention and transmission of HIV-1 by DC-SIGN. Int. Immunol. 2003, 15, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.A.; Gilbert, C.; Richard, M.; Beaulieu, A.D.; Tremblay, M.J. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood 2008, 112, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Bloem, K.; Vuist, I.M.; van der Plas, A.-J.; Knippels, L.M.J.; Garssen, J.; García-Vallejo, J.J.; van Vliet, S.J.; van Kooyk, Y. Ligand binding and signaling of dendritic cell immunoreceptor (DCIR) is modulated by the glycosylation of the carbohydrate recognition domain. PLoS ONE 2013, 8, e66266. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; de Wet, B.J.M.; DeWet, B.J.M.; Martinez-Pomares, L.; Radcliffe, C.M.; Dwek, R.A.; Rudd, P.M.; Gordon, S. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008, 4, e17. [Google Scholar] [CrossRef]
- Nguyen, D.G.; Hildreth, J.E.K. Involvement of macrophage mannose receptor in the binding and transmission of HIV by macrophages. Eur. J. Immunol. 2003, 33, 483–493. [Google Scholar] [CrossRef]
- Taylor, M.E.; Drickamer, K. Structural requirements for high affinity binding of complex ligands by the macrophage mannose receptor. J. Biol. Chem. 1993, 268, 399–404. [Google Scholar] [CrossRef]
- Gazi, U.; Martinez-Pomares, L. Influence of the mannose receptor in host immune responses. Immunobiology 2009, 214, 554–561. [Google Scholar] [CrossRef]
- Schlesinger, L.S. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J. Immunol. 1993, 150, 2920–2930. [Google Scholar]
- Schulert, G.S.; Allen, L.-A.H. Differential infection of mononuclear phagocytes by Francisella tularensis: Role of the macrophage mannose receptor. J. Leukoc. Biol. 2006, 80, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, H.; Taylor, M.E.; Razi, N.; McBride, R.; Knirel, Y.A.; Graham, S.A.; Drickamer, K.; Weis, W.I. Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J. Mol. Biol. 2011, 405, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Tateno, H.; Ohnishi, K.; Yabe, R.; Hayatsu, N.; Sato, T.; Takeya, M.; Narimatsu, H.; Hirabayashi, J. Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J. Biol. Chem. 2010, 285, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.A.W.P.; Vriend, L.E.M.; Theelen, B.; Taylor, M.E.; Fluitsma, D.; Boekhout, T.; Geijtenbeek, T.B.H. C-type lectin Langerin is a beta-glucan receptor on human Langerhans cells that recognizes opportunistic and pathogenic fungi. Mol. Immunol. 2010, 47, 1216–1225. [Google Scholar] [CrossRef]
- de Witte, L.; Nabatov, A.; Pion, M.; Fluitsma, D.; de Jong, M.A.W.P.; de Gruijl, T.; Piguet, V.; van Kooyk, Y.; Geijtenbeek, T.B.H. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 2007, 13, 367–371. [Google Scholar] [CrossRef]
- van der Vlist, M.; de Witte, L.; de Vries, R.D.; Litjens, M.; de Jong, M.A.W.P.; Fluitsma, D.; de Swart, R.L.; Geijtenbeek, T.B.H. Human Langerhans cells capture measles virus through Langerin and present viral antigens to CD4+ T cells but are incapable of cross-presentation. Eur. J. Immunol. 2011, 41, 2619–2631. [Google Scholar] [CrossRef]
- Stambach, N.S.; Taylor, M.E. Characterization of carbohydrate recognition by langerin, a C-type lectin of Langerhans cells. Glycobiology 2003, 13, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Penberthy, K.K.; Ravichandran, K.S. Apoptotic cell recognition receptors and scavenger receptors. Immunol. Rev. 2016, 269, 44–59. [Google Scholar] [CrossRef] [PubMed]
- PrabhuDas, M.R.; Baldwin, C.L.; Bollyky, P.L.; Bowdish, D.M.E.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; et al. A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J. Immunol. 2017, 198, 3775–3789. [Google Scholar] [CrossRef] [PubMed]
- Kodama, T.; Freeman, M.; Rohrer, L.; Zabrecky, J.; Matsudaira, P.; Krieger, M. Type I macrophage scavenger receptor contains alpha-helical and collagen-like coiled coils. Nature 1990, 343, 531–535. [Google Scholar] [CrossRef]
- Rohrer, L.; Freeman, M.; Kodama, T.; Penman, M.; Krieger, M. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature 1990, 343, 570–572. [Google Scholar] [CrossRef]
- Plüddemann, A.; Neyen, C.; Gordon, S. Macrophage scavenger receptors and host-derived ligands. Methods 2007, 43, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Dunne, D.W.; Resnick, D.; Greenberg, J.; Krieger, M.; Joiner, K.A. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc. Natl. Acad. Sci. USA 1994, 91, 1863–1867. [Google Scholar] [CrossRef]
- Hampton, R.Y.; Golenbock, D.T.; Penman, M.; Krieger, M.; Raetz, C.R.H. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature 1991, 352, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Palecanda, A.; Paulauskis, J.; Al-Mutairi, E.; Imrich, A.; Qin, G.; Suzuki, H.; Kodama, T.; Tryggvason, K.; Koziel, H.; Kobzik, L. Role of the scavenger receptor MARCO in alveolar macrophage binding of unopsonized environmental particles. J. Exp. Med. 1999, 189, 1497–1506. [Google Scholar] [CrossRef]
- van der Laan, L.J.; Döpp, E.A.; Haworth, R.; Pikkarainen, T.; Kangas, M.; Elomaa, O.; Dijkstra, C.D.; Gordon, S.; Tryggvason, K.; Kraal, G. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 1999, 162, 939–947. [Google Scholar]
- Greenwalt, D.E.; Lipsky, R.H.; Ockenhouse, C.F.; Ikeda, H.; Tandon, N.N.; Jamieson, G.A. Membrane glycoprotein CD36: A review of its roles in adherence, signal transduction, and transfusion medicine. Blood 1992, 80, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Endemann, G.; Stanton, L.W.; Madden, K.S.; Bryant, C.M.; White, R.T.; Protter, A.A. CD36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 1993, 268, 11811–11816. [Google Scholar] [CrossRef]
- Stuart, L.M.; Deng, J.; Silver, J.M.; Takahashi, K.; Tseng, A.A.; Hennessy, E.J.; Ezekowitz, R.A.B.; Moore, K.J. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 2005, 170, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Baranova, I.N.; Kurlander, R.; Bocharov, A.V.; Vishnyakova, T.G.; Chen, Z.; Remaley, A.T.; Csako, G.; Patterson, A.P.; Eggerman, T.L. Role of human CD36 in bacterial recognition, phagocytosis, and pathogen-induced JNK-mediated signaling. J. Immunol. 2008, 181, 7147–7156. [Google Scholar] [CrossRef] [PubMed]
- Savill, J.; Hogg, N.; Haslett, C. Macrophage vitronectin receptor, CD36, and thrombospondin cooperate in recognition of neutrophils undergoing programmed cell death. Chest 1991, 99, 6S–7S. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.L.; Pearce, S.F.; Francisco, L.M.; Sauter, B.; Roy, P.; Silverstein, R.L.; Bhardwaj, N. Immature dendritic cells phagocytose apoptotic cells via alphavbeta5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J. Exp. Med. 1998, 188, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M.; Arai, H.; Inoue, K. Expression cloning of a novel scavenger receptor from human endothelial cells. J. Biol. Chem. 1997, 272, 31217–31220. [Google Scholar] [CrossRef]
- Patten, D.A. SCARF1: A multifaceted, yet largely understudied, scavenger receptor. Inflamm. Res. 2018, 67, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Facciponte, J.G.; Wang, X.-Y.; Subjeck, J.R. Hsp110 and Grp170, members of the Hsp70 superfamily, bind to scavenger receptor-A and scavenger receptor expressed by endothelial cells-I. Eur. J. Immunol. 2007, 37, 2268–2279. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Ortiz, Z.G.; Pendergraft, W.F.; Prasad, A.; Byrne, M.H.; Iram, T.; Blanchette, C.J.; Luster, A.D.; Hacohen, N.; El Khoury, J.; Means, T.K. The scavenger receptor SCARF1 mediates the clearance of apoptotic cells and prevents autoimmunity. Nat. Immunol. 2013, 14, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.D.; Park, S.-Y.; Bae, J.; Yun, Y.; Bae, Y.-C.; Park, R.-W.; Kim, I.-S. MEGF10 functions as a receptor for the uptake of amyloid-β. FEBS Lett. 2010, 584, 3936–3942. [Google Scholar] [CrossRef] [PubMed]
- Iram, T.; Ramirez-Ortiz, Z.; Byrne, M.H.; Coleman, U.A.; Kingery, N.D.; Means, T.K.; Frenkel, D.; El Khoury, J. Megf10 Is a Receptor for C1Q That Mediates Clearance of Apoptotic Cells by Astrocytes. J. Neurosci. 2016, 36, 5185–5192. [Google Scholar] [CrossRef]
- Park, S.-Y.; Jung, M.-Y.; Lee, S.-J.; Kang, K.-B.; Gratchev, A.; Riabov, V.; Kzhyshkowska, J.; Kim, I.-S. Stabilin-1 mediates phosphatidylserine-dependent clearance of cell corpses in alternatively activated macrophages. J. Cell Sci. 2009, 122, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Jung, M.-Y.; Kim, H.-J.; Lee, S.-J.; Kim, S.-Y.; Lee, B.-H.; Kwon, T.-H.; Park, R.-W.; Kim, I.-S. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008, 15, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M. FEEL-1, a novel scavenger receptor with in vitro bacteria-binding and angiogenesis-modulating activities. J. Biol. Chem. 2002, 277, 34264–34270. [Google Scholar] [CrossRef]
- Hirose, Y.; Saijou, E.; Sugano, Y.; Takeshita, F.; Nishimura, S.; Nonaka, H.; Chen, Y.-R.; Sekine, K.; Kido, T.; Nakamura, T.; et al. Inhibition of Stabilin-2 elevates circulating hyaluronic acid levels and prevents tumor metastasis. Proc. Natl. Acad. Sci. USA 2012, 109, 4263–4268. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.-Y.; Kim, S.-Y.; Bae, D.-J.; Pyo, J.-H.; Hong, M.; Kim, I.-S. Cross talk between engulfment receptors stabilin-2 and integrin αvβ5 orchestrates engulfment of phosphatidylserine-exposed erythrocytes. Mol. Cell. Biol. 2012, 32, 2698–2708. [Google Scholar] [CrossRef] [PubMed]
- Lukácsi, S.; Mácsik-Valent, B.; Nagy-Baló, Z.; Kovács, K.G.; Kliment, K.; Bajtay, Z.; Erdei, A. Utilization of complement receptors in immune cell—Microbe interaction. FEBS Lett. 2020, 1–19. [Google Scholar] [CrossRef] [PubMed]
- van Lookeren Campagne, M.; Wiesmann, C.; Brown, E.J. Macrophage complement receptors and pathogen clearance. Cell. Microbiol. 2007, 9, 2095–2102. [Google Scholar] [CrossRef]
- Cornacoff, J.B.; Hebert, L.A.; Smead, W.L.; VanAman, M.E.; Birmingham, D.J.; Waxman, F.J. Primate erythrocyte-immune complex-clearing mechanism. J. Clin. Invest. 1983, 71, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Ogembo, J.G.; Kannan, L.; Ghiran, I.; Nicholson-Weller, A.; Finberg, R.W.; Tsokos, G.C.; Fingeroth, J.D. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep. 2013, 3, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Thieblemont, N.; Haeffner-Cavaillon, N.; Ledur, A.; L’Age-Stehr, J.; Ziegler-Heitbrock, H.W.L.; Kazatchkine, M.D. CR1 (CD35) and CR3 (CD11b/CD18) mediate infection of human monocytes and monocytic cell lines with complement-opsonized HIV independently of CD4. Clin. Exp. Immunol. 1993, 92, 106–113. [Google Scholar] [CrossRef]
- Fällman, M.; Andersson, R.; Andersson, T. Signaling properties of CR3 (CD11b/CD18) and CR1 (CD35) in relation to phagocytosis of complement-opsonized particles. J. Immunol. 1993, 151, 330–338. [Google Scholar] [PubMed]
- He, J.Q.; Katschke, K.J.; Gribling, P.; Suto, E.; Lee, W.P.; Diehl, L.; Eastham-Anderson, J.; Ponakala, A.; Komuves, L.; Egen, J.G.; et al. CRIg mediates early Kupffer cell responses to adenovirus. J. Leukoc. Biol. 2013, 93, 301–306. [Google Scholar] [CrossRef]
- Helmy, K.Y.; Katschke, K.J.; Gorgani, N.N.; Kljavin, N.M.; Elliott, J.M.; Diehl, L.; Scales, S.J.; Ghilardi, N.; van Lookeren Campagne, M. CRIg: A macrophage complement receptor required for phagocytosis of circulating pathogens. Cell 2006, 124, 915–927. [Google Scholar] [CrossRef]
- Munawara, U.; Small, A.G.; Quach, A.; Gorgani, N.N.; Abbott, C.A.; Ferrante, A. Cytokines regulate complement receptor immunoglobulin expression and phagocytosis of Candida albicans in human macrophages: A control point in anti-microbial immunity. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Erdei, A.; Lukácsi, S.; Mácsik-Valent, B.; Nagy-Baló, Z.; Kurucz, I.; Bajtay, Z. Non-identical twins: Different faces of CR3 and CR4 in myeloid and lymphoid cells of mice and men. Semin. Cell Dev. Biol. 2019, 85, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Garcia-Aguilar, J.; Bickford, J.K.; Corbi, A.L.; Springer, T.A. The I domain is a major recognition site on the leukocyte integrin Mac-1 (CD11b/CD18) for four distinct adhesion ligands. J. Cell Biol. 1993, 120, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, X.M.; Heflin, K.E.; Lavigne, L.M.; Yu, K.; Kim, M.; Salomon, A.R.; Reichner, J.S. Lectin site ligation of CR3 induces conformational changes and signaling. J. Biol. Chem. 2012, 287, 3337–3348. [Google Scholar] [CrossRef]
- Ross, G.D.; Cain, J.A.; Myones, B.L.; Newman, S.L.; Lachmann, P.J. Specificity of membrane complement receptor type three (CR3) for beta-glucans. Complement 1987, 4, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.D.; Levin, S.M.; Jong, M.T.; Chad, Z.; Kabbash, L.G. CR3 (CD11b/CD18) expresses one binding site for Arg-Gly-Asp-containing peptides and a second site for bacterial lipopolysaccharide. J. Exp. Med. 1989, 169, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Ingalls, R.R.; Golenbock, D.T. CD11c/CD18, a transmembrane signaling receptor for lipopolysaccharide. J. Exp. Med. 1995, 181, 1473–1479. [Google Scholar] [CrossRef]
- Jawhara, S.; Pluskota, E.; Cao, W.; Plow, E.F.; Soloviev, D.A. Distinct Effects of Integrins αXβ2 and αMβ2 on Leukocyte Subpopulations during Inflammation and Antimicrobial Responses. Infect. Immun. 2017, 85, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, C.S.; Ellner, J.J.; Russell, D.G.; Rich, E.A. Complement receptor-mediated uptake and tumor necrosis factor-alpha-mediated growth inhibition of Mycobacterium tuberculosis by human alveolar macrophages. J. Immunol. 1994, 152, 743–753. [Google Scholar] [PubMed]
- Sándor, N.; Kristóf, K.; Paréj, K.; Pap, D.; Erdei, A.; Bajtay, Z. CR3 is the dominant phagocytotic complement receptor on human dendritic cells. Immunobiology 2013, 218, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Lukácsi, S.; Nagy-Baló, Z.; Erdei, A.; Sándor, N.; Bajtay, Z. The role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in complement-mediated phagocytosis and podosome formation by human phagocytes. Immunol. Lett. 2017, 189, 64–72. [Google Scholar] [CrossRef]
- Wright, S.D.; Ramos, R.A.; Tobias, P.S.; Ulevitch, R.J.; Mathison, J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990, 249, 1431–1433. [Google Scholar] [CrossRef]
- van Vliet, S.J.; van Liempt, E.; Saeland, E.; Aarnoudse, C.A.; Appelmelk, B.; Irimura, T.; Geijtenbeek, T.B.H.; Blixt, O.; Alvarez, R.; van Die, I.; et al. Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 2005, 17, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Sankala, M.; Brännström, A.; Schulthess, T.; Bergmann, U.; Morgunova, E.; Engel, J.; Tryggvason, K.; Pikkarainen, T. Characterization of recombinant soluble macrophage scavenger receptor MARCO. J. Biol. Chem. 2002, 277, 33378–33385. [Google Scholar] [CrossRef] [PubMed]
- Elomaa, O.; Kangas, M.; Sahlberg, C.; Tuukkanen, J.; Sormunen, R.; Liakka, A.; Thesleff, I.; Kraal, G.; Tryggvason, K. Cloning of a novel bacteria-binding receptor structurally related to scavenger receptors and expressed in a subset of macrophages. Cell 1995, 80, 603–609. [Google Scholar] [CrossRef]
- Harris, E.N.; Weigel, J.A.; Weigel, P.H. The human hyaluronan receptor for endocytosis (HARE/Stabilin-2) is a systemic clearance receptor for heparin. J. Biol. Chem. 2008, 283, 17341–17350. [Google Scholar] [CrossRef]
- Sim, R.B.; Malhotra, V.; Day, A.J.; Erdei, A. Structure and specificity of complement receptors. Immunol. Lett. 1987, 14, 183–190. [Google Scholar] [CrossRef]
- Ghiran, I.; Barbashov, S.F.; Klickstein, L.B.; Tas, S.W.; Jensenius, J.C.; Nicholson-Weller, A. Complement receptor 1/CD35 is a receptor for mannan-binding lectin. J. Exp. Med. 2000, 192, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Losse, J.; Zipfel, P.F.; Józsi, M. Factor H and factor H-related protein 1 bind to human neutrophils via complement receptor 3, mediate attachment to Candida albicans, and enhance neutrophil antimicrobial activity. J. Immunol. 2010, 184, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, V.; Hogg, N.; Sim, R.B. Ligand binding by the p150,95 antigen of U937 monocytic cells: Properties in common with complement receptor type 3 (CR3). Eur. J. Immunol. 1986, 16, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Józsi, M.; Schneider, A.E.; Kárpáti, É.; Sándor, N. Complement factor H family proteins in their non-canonical role as modulators of cellular functions. Semin. Cell Dev. Biol. 2018, 85, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.M.G.; Thomson, J.N. Morphology of programmed cell death in the ventral nerve cord of Caenorhabditis elegans larvae. Development 1982, 67, 89–100. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Pukkila-Worley, R. Surveillance Immunity: An Emerging Paradigm of Innate Defense Activation in Caenorhabditis elegans. PLoS Pathog. 2016, 12, e1005795. [Google Scholar] [CrossRef]
- Huang, G.; Shi, L.Z.; Chi, H. Regulation of JNK and p38 MAPK in the immune system: Signal integration, propagation and termination. Cytokine 2009, 48, 161–169. [Google Scholar] [CrossRef]
- Keshet, Y.; Seger, R. The MAP kinase signaling cascades: A system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 2010, 661, 3–38. [Google Scholar] [CrossRef]
- Kim, D.H.; Feinbaum, R.; Alloing, G.; Emerson, F.E.; Garsin, D.A.; Inoue, H.; Tanaka-Hino, M.; Hisamoto, N.; Matsumoto, K.; Tan, M.-W.; et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 2002, 297, 623–626. [Google Scholar] [CrossRef]
- Kim, D.H.; Liberati, N.T.; Mizuno, T.; Inoue, H.; Hisamoto, N.; Matsumoto, K.; Ausubel, F.M. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc. Natl. Acad. Sci. USA 2004, 101, 10990–10994. [Google Scholar] [CrossRef] [PubMed]
- Aballay, A.; Drenkard, E.; Hilbun, L.R.; Ausubel, F.M. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 2003, 13, 47–52. [Google Scholar] [CrossRef]
- Sifri, C.D.; Begun, J.; Ausubel, F.M.; Calderwood, S.B. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 2003, 71, 2208–2217. [Google Scholar] [CrossRef]
- Bolz, D.D.; Tenor, J.L.; Aballay, A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J. Biol. Chem. 2010, 285, 10832–10840. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Ausubel, F.M.; Mylonakis, E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011, 7, e1002074. [Google Scholar] [CrossRef]
- Muhammed, M.; Fuchs, B.B.; Wu, M.P.; Breger, J.; Coleman, J.J.; Mylonakis, E. The role of mycelium production and a MAPK-mediated immune response in the C. elegans-Fusarium model system. Med. Mycol. 2012, 50, 488–496. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huffman, D.L.; Abrami, L.; Sasik, R.; Corbeil, J.; van der Goot, F.G.; Aroian, R.V. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl. Acad. Sci. USA 2004, 101, 10995–11000. [Google Scholar] [CrossRef]
- Hodgkin, J.; Kuwabara, P.E.; Corneliussen, B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 2000, 10, 1615–1618. [Google Scholar] [CrossRef]
- Kurz, C.L.; Ewbank, J.J. Caenorhabditis elegans: An emerging genetic model for the study of innate immunity. Nat. Rev. Genet. 2003, 4, 380–390. [Google Scholar] [CrossRef]
- Nicholas, H.R.; Hodgkin, J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 2004, 41, 479–493. [Google Scholar] [CrossRef]
- Couillault, C.; Pujol, N.; Reboul, J.; Sabatier, L.; Guichou, J.-F.; Kohara, Y.; Ewbank, J.J. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004, 5, 488–494. [Google Scholar] [CrossRef]
- Irazoqui, J.E.; Urbach, J.M.; Ausubel, F.M. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010, 10, 47–58. [Google Scholar] [CrossRef]
- Brandt, J.P.; Ringstad, N. Toll-like Receptor Signaling Promotes Development and Function of Sensory Neurons Required for a C. elegans Pathogen-Avoidance Behavior. Curr. Biol. 2015, 25, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Pujol, N.; Link, E.M.; Liu, L.X.; Kurz, C.L.; Alloing, G.; Tan, M.W.; Ray, K.P.; Solari, R.; Johnson, C.D.; Ewbank, J.J. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 2001, 11, 809–821. [Google Scholar] [CrossRef]
- Liberati, N.T.; Fitzgerald, K.A.; Kim, D.H.; Feinbaum, R.; Golenbock, D.T.; Ausubel, F.M. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 2004, 101, 6593–6598. [Google Scholar] [CrossRef] [PubMed]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.-Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef]
- Pujol, N.; Cypowyj, S.; Ziegler, K.; Millet, A.; Astrain, A.; Goncharov, A.; Jin, Y.; Chisholm, A.D.; Ewbank, J.J. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 2008, 18, 481–489. [Google Scholar] [CrossRef]
- Bischof, L.J.; Kao, C.-Y.; Los, F.C.O.; Gonzalez, M.R.; Shen, Z.; Briggs, S.P.; van der Goot, F.G.; Aroian, R.V. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Zugasti, O.; Bose, N.; Squiban, B.; Belougne, J.; Kurz, C.L.; Schroeder, F.C.; Pujol, N.; Ewbank, J.J. Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nat. Immunol. 2014, 15, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, K.; Kurz, C.L.; Cypowyj, S.; Couillault, C.; Pophillat, M.; Pujol, N.; Ewbank, J.J. Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe 2009, 5, 341–352. [Google Scholar] [CrossRef]
- Dierking, K.; Polanowska, J.; Omi, S.; Engelmann, I.; Gut, M.; Lembo, F.; Ewbank, J.J.; Pujol, N. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 2011, 9, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Pujol, N.; Zugasti, O.; Wong, D.; Couillault, C.; Kurz, C.L.; Schulenburg, H.; Ewbank, J.J. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008, 4, e1000105. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.A.; García-Silva, S.; Mazariegos, M.; González de la Aleja, A.; Nicolás-Ávila, J.A.; Walter, W.; Adrover, J.M.; Crainiciuc, G.; Kuchroo, V.K.; Rothlin, C.V.; et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 2017, 214, 1281–1296. [Google Scholar] [CrossRef]
- Roberts, A.W.; Lee, B.L.; Deguine, J.; John, S.; Shlomchik, M.J.; Barton, G.M. Tissue-Resident Macrophages Are Locally Programmed for Silent Clearance of Apoptotic Cells. Immunity 2017, 47, 913–927.e6. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Voelker, D.R.; Campbell, P.A.; Cohen, J.J.; Bratton, D.L.; Henson, P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992, 148, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.P.; Vestergaard, A.L.; Mikkelsen, S.A.; Mogensen, L.S.; Chalat, M.; Molday, R.S. P4-ATPases as Phospholipid Flippases-Structure, Function, and Enigmas. Front. Physiol. 2016, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- Segawa, K.; Kurata, S.; Yanagihashi, Y.; Brummelkamp, T.R.; Matsuda, F.; Nagata, S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 2014, 344, 1164–1168. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Denning, D.P.; Imanishi, E.; Horvitz, H.R.; Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013, 341, 403–406. [Google Scholar] [CrossRef]
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.; Henson, P.M. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405, 85–90. [Google Scholar] [CrossRef]
- Williamson, P.; Schlegel, R.A. Hide and seek: The secret identity of the phosphatidylserine receptor. J. Biol. 2004, 3, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Böse, J.; Gruber, A.D.; Helming, L.; Schiebe, S.; Wegener, I.; Hafner, M.; Beales, M.; Köntgen, F.; Lengeling, A. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004, 3, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Umetsu, S.E.; Lee, W.-L.; McIntire, J.J.; Downey, L.; Sanjanwala, B.; Akbari, O.; Berry, G.J.; Nagumo, H.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat. Immunol. 2005, 6, 447–454. [Google Scholar] [CrossRef]
- Ichimura, T.; Asseldonk, E.J.P.V.; Humphreys, B.D.; Gunaratnam, L.; Duffield, J.S.; Bonventre, J.V. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J. Clin. Invest. 2008, 118, 1657–1668. [Google Scholar] [CrossRef]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- de Mingo Pulido, Á.; Gardner, A.; Hiebler, S.; Soliman, H.; Rugo, H.S.; Krummel, M.F.; Coussens, L.M.; Ruffell, B. TIM-3 Regulates CD103+ Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018, 33, 60–74.e6. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Karisola, P.; Peña-Cruz, V.; Dorfman, D.M.; Jinushi, M.; Umetsu, S.E.; Butte, M.J.; Nagumo, H.; Chernova, I.; Zhu, B.; et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007, 27, 927–940. [Google Scholar] [CrossRef] [PubMed]
- Miyanishi, M.; Tada, K.; Koike, M.; Uchiyama, Y.; Kitamura, T.; Nagata, S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G. How macrophages deal with death. Nat. Rev. Immunol. 2019, 19, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Lew, E.D.; Oh, J.; Burrola, P.G.; Lax, I.; Zagórska, A.; Través, P.G.; Schlessinger, J.; Lemke, G. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. Elife 2014, 3, 1–23. [Google Scholar] [CrossRef]
- Seitz, H.M.; Camenisch, T.D.; Lemke, G.; Earp, H.S.; Matsushima, G.K. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J. Immunol. 2007, 178, 5635–5642. [Google Scholar] [CrossRef] [PubMed]
- Rothlin, C.V.; Ghosh, S.; Zuniga, E.I.; Oldstone, M.B.A.; Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 2007, 131, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, J.H.M.; van der Poll, T.; van ’t Veer, C. TAM receptors, Gas6, and protein S: Roles in inflammation and hemostasis. Blood 2014, 123, 2460–2469. [Google Scholar] [CrossRef]
- Hanayama, R.; Tanaka, M.; Miwa, K.; Shinohara, A.; Iwamatsu, A.; Nagata, S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002, 417, 182–187. [Google Scholar] [CrossRef]
- Stern, M.; Savill, J.; Haslett, C. Human monocyte-derived macrophage phagocytosis of senescent eosinophils undergoing apoptosis. Mediation by alpha v beta 3/CD36/thrombospondin recognition mechanism and lack of phlogistic response. Am. J. Pathol. 1996, 149, 911–921. [Google Scholar]
- Savill, J.; Hogg, N.; Ren, Y.; Haslett, C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Invest. 1992, 90, 1513–1522. [Google Scholar] [CrossRef]
- Albert, M.L.; Kim, J.I.; Birge, R.B. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2000, 2, 899–905. [Google Scholar] [CrossRef]
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 2001, 107, 27–41. [Google Scholar] [CrossRef]
- Shah, P.P.; Fong, M.Y.; Kakar, S.S. PTTG induces EMT through integrin αVβ3-focal adhesion kinase signaling in lung cancer cells. Oncogene 2012, 31, 3124–3135. [Google Scholar] [CrossRef]
- Liang, Y.Y.; Arnold, T.; Michlmayr, A.; Rainprecht, D.; Perticevic, B.; Spittler, A.; Oehler, R. Serum-dependent processing of late apoptotic cells for enhanced efferocytosis. Cell Death Dis. 2014, 5, e1264. [Google Scholar] [CrossRef]
- Nauta, A.J.; Castellano, G.; Xu, W.; Woltman, A.M.; Borrias, M.C.; Daha, M.R.; van Kooten, C.; Roos, A. Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J. Immunol. 2004, 173, 3044–3050. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.A.; Laust, A.K.; Nelson, E.L.; Tenner, A.J. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J. Immunol. 2009, 183, 6175–6185. [Google Scholar] [CrossRef]
- Takizawa, F.; Tsuji, S.; Nagasawa, S. Enhancement of macrophage phagocytosis upon iC3b deposition on apoptotic cells. FEBS Lett. 1996, 397, 269–272. [Google Scholar] [CrossRef]
- Matsui, H.; Tsuji, S.; Nishimura, H.; Nagasawa, S. Activation of the alternative pathway of complement by apoptotic Jurkat cells. FEBS Lett. 1994, 351, 419–422. [Google Scholar] [CrossRef]
- Mevorach, D.; Mascarenhas, J.O.; Gershov, D.; Elkon, K.B. Complement-dependent clearance of apoptotic cells by human macrophages. J. Exp. Med. 1998, 188, 2313–2320. [Google Scholar] [CrossRef]
- Benoit, M.E.; Clarke, E.V.; Morgado, P.; Fraser, D.A.; Tenner, A.J. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J. Immunol. 2012, 188, 5682–5693. [Google Scholar] [CrossRef]
- Fraser, D.A.; Arora, M.; Bohlson, S.S.; Lozano, E.; Tenner, A.J. Generation of inhibitory NFkappaB complexes and phosphorylated cAMP response element-binding protein correlates with the anti-inflammatory activity of complement protein C1q in human monocytes. J. Biol. Chem. 2007, 282, 7360–7367. [Google Scholar] [CrossRef] [PubMed]
- Bratton, D.L.; Fadok, V.A.; Richter, D.A.; Kailey, J.M.; Guthrie, L.A.; Henson, P.M. Appearance of phosphatidylserine on apoptotic cells requires calcium-mediated nonspecific flip-flop and is enhanced by loss of the aminophospholipid translocase. J. Biol. Chem. 1997, 272, 26159–26165. [Google Scholar] [CrossRef]
- Lu, N.; Zhou, Z. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int. Rev. Cell Mol. Biol. 2012, 293, 269–309. [Google Scholar] [CrossRef] [PubMed]
- Mangahas, P.M.; Zhou, Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin. Cell Dev. Biol. 2005, 16, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, J.M.; Cabello, J.; Klingele, D.; Wong, K.; Feichtinger, R.; Schnabel, H.; Schnabel, R.; Hengartner, M.O. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 2005, 434, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; He, B.; Lu, N.; Conradt, B.; Grant, B.D.; Zhou, Z. Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans. Development 2013, 140, 3230–3243. [Google Scholar] [CrossRef]
- Zhou, Z.; Hartwieg, E.; Horvitz, H.R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 2001, 104, 43–56. [Google Scholar] [CrossRef]
- Venegas, V.; Zhou, Z. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol. Biol. Cell 2007, 18, 3180–3192. [Google Scholar] [CrossRef]
- Yu, X.; Odera, S.; Chuang, C.-H.; Lu, N.; Zhou, Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 2006, 10, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Farkas, Z.; Petric, M.; Liu, X.; Herit, F.; Rajnavölgyi, É.; Szondy, Z.; Budai, Z.; Orbán, T.I.; Sándor, S.; Mehta, A.; et al. The nucleoside diphosphate kinase NDK-1/NME1 promotes phagocytosis in concert with DYN-1/Dynamin. FASEB J. 2019, 33, 11606–11614. [Google Scholar] [CrossRef]
- Yu, X.; Lu, N.; Zhou, Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008, 6, e61. [Google Scholar] [CrossRef]
- Reddien, P.W.; Horvitz, H.R. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2004, 20, 193–221. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.-Y.; Wu, Y.-C. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr. Biol. 2010, 20, 477–486. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Y.-C.; Fadok, V.A.; Lee, M.-C.; Gengyo-Ando, K.; Cheng, L.-C.; Ledwich, D.; Hsu, P.-K.; Chen, J.-Y.; Chou, B.-K.; et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 2003, 302, 1563–1566. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-H.; Hsu, T.-Y.; Jiang, H.-S.; Wu, Y.-C. Integrin α PAT-2/CDC-42 signaling is required for muscle-mediated clearance of apoptotic cells in Caenorhabditis elegans. PLoS Genet. 2012, 8, e1002663. [Google Scholar] [CrossRef]
- Neukomm, L.J.; Zeng, S.; Frei, A.P.; Huegli, P.A.; Hengartner, M.O. Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ. 2014, 21, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Cabello, J.; Neukomm, L.J.; Günesdogan, U.; Burkart, K.; Charette, S.J.; Lochnit, G.; Hengartner, M.O.; Schnabel, R. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 2010, 8, e1000297. [Google Scholar] [CrossRef] [PubMed]
- Klinkert, K.; Echard, A. Rab35 GTPase: A Central Regulator of Phosphoinositides and F-actin in Endocytic Recycling and Beyond. Traffic 2016, 17, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Haley, R.; Wang, Y.; Zhou, Z. The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells. PLoS Genet. 2018, 14, e1007558. [Google Scholar] [CrossRef]
- Kawane, K.; Ohtani, M.; Miwa, K.; Kizawa, T.; Kanbara, Y.; Yoshioka, Y.; Yoshikawa, H.; Nagata, S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006, 443, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lai, H.-J.; Lin, T.-W.; Chen, C.-S.; Lo, S.J. Loss of DNase II function in the gonad is associated with a higher expression of antimicrobial genes in Caenorhabditis elegans. Biochem. J. 2015, 470, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Mukae, N.; Yokoyama, H.; Yokokura, T.; Sakoyama, Y.; Nagata, S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002, 16, 2662–2671. [Google Scholar] [CrossRef] [PubMed]
- Seong, C.-S.; Varela-Ramirez, A.; Aguilera, R.J. DNase II deficiency impairs innate immune function in Drosophila. Cell. Immunol. 2006, 240, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, J. Detection of Pathogens and Regulation of Immunity by the Caenorhabditis elegans Nervous System. MBio 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, E.M. Neural regulation of innate immunity: A coordinated nonspecific host response to pathogens. Nat. Rev. Immunol. 2006, 6, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Steinman, L. Elaborate interactions between the immune and nervous systems. Nat. Immunol. 2004, 5, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Tracey, K.J. The inflammatory reflex. Nature 2002, 420, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Giunti, S.; Andersen, N.; Rayes, D.; De Rosa, M.J. Drug discovery: Insights from the invertebrate Caenorhabditis elegans. Pharmacol. Res. Perspect. 2021, 9, e00721. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, R. Neuroimmune Interactions: From the Brain to the Immune System and Vice Versa. Physiol. Rev. 2018, 98, 477–504. [Google Scholar] [CrossRef] [PubMed]
| Receptors | Recognized Molecules | Distribution |
|---|---|---|
| Pattern recognition receptors | ||
| CD14 | LPS [155] | Mo, Mφ, DC, Neu |
| C-type lectins | ||
| langerin (CD207) | sulfated and mannosylated glycans [104,105], fungal β-glucans [106], viral envelope glycoproteins [107,108] | LC |
| DC-SIGN (CD209) | fucosylated-, oligomannose- and N-glycans [87], HIV-1 [94] | DC |
| Dectin-1 (CD369) | fungal β-glucans [70,71], Leishmania [72], Mycobacterium | Mo, Mφ, DC, B cell, PMN |
| Dectin-2 | mannose in fungal and bacterial cell walls [75,76,77] | Mo, Mφ, DC, Neu, B cell |
| MMR (CD206) | mannose, fucose, GlcNAc [100,101] | Mφ, DC |
| Mincle-MCL | fungal and bacterial glycolipids [81,82] | Mo, Mφ, DC, B cell |
| MGL | terminal GalNAc residues [156] | DC |
| DCIR | fucose and mannose containing glycans, HIV-1 [96,97] | Mo, Mφ, DC, PMN |
| Scavenger receptors | ||
| MSR1 (SR-A1) | modified LDL, β-amyloid, heat-shock proteins [113,114], bacterial wall components [115,116] | Mo, Mφ, DC, MC |
| MARCO (SR-A6) | LPS, acetylated LDL [157,158] | Mφ |
| CD36 (SR-B2) | oxidized LDL, β-amyloids [120], polyanionic bacterial ligands [122], apoptotic cells [123] | Mo, Mφ, platelets |
| SCARF-1 (SR-F1) | bacterial, fungal pathogens, heat-shock proteins, apoptotic cells [126,127,128] | Mφ, DC, endothelial cells |
| MEGF10 (SR-F2) | C1q on apoptotic cells, β-amyloids [129,130] | astrocytes |
| Stabilin-1 (SR-H1) | PtdSer [131], bacterial components [133] | macrophages, endothelial cells |
| Stabilin-2 (SR-H2) | PtdSer [132], hyaluronic acid [134], heparin [159] | sinusoidal endothelial cells. |
| Complement receptors | ||
| CR1 (CD35) | C3b, C4b [160], EBV [139], MBL, C1q [161] | Mo, Mφ, DC, PMN, NK, B cell, T cell subsets, RBC, FDC |
| CRIg | C3b, iC3b [143] | KC and tissue-resident macrophage subsets |
| CR3 (αMβ2, CD11b/CD18) | iC3b [146], FH, FHR-1 [162], β-glucan [148], LPS [149] | Mo, Mφ, DC, PMN, NK, lymphocyte subsets |
| CR4 (αXβ2, CD11c/CD18) | iC3b [163], FH [164], LPS [150] | Mo, Mφ, DC, PMN, NK, lymphocyte subsets |
| Human Protein | Function | C. elegans Protein |
|---|---|---|
| ATP8A2 | P4-type ATPase/flippase | TAT-1 |
| XKR8 | scramblase | CED-8 |
| SCARF1, MEGF10 and 11, LRP1 (CD91), Jedi-1 | phagocytic receptor | CED-1 |
| GULP | adaptor | CED-6 |
| NME1 | nucleoside diphosphate kinase | NDK-1 |
| Dynamin | large GTPase | DYN-1 |
| ABCA1 and ABCA7 | ABC transporter | CED-7 |
| JMJD6 (PSR?) | receptor | PSR-1 |
| FZD1 and 7 (Frizzled class receptor 1 and 7) | receptor | MOM-5 |
| integrin α/β chain | receptor | INA-1/PAT-3 |
| SRC | non-receptor tyrosine kinase | SRC-1 |
| CrkII | adaptor | CED-2 |
| ELMO | adaptor | CED-12 |
| Dock180 | Rac GEF | CED-5 |
| Rac1 | Rho family GTPase | CED-10 |
| MFG-E8 | bridging between PtdSer and integrins | - |
| TIM1, 3, 4 | PtdSer receptor | - |
| Protein S, GAS6 | bridging between PtdSer and TAM receptors (MER, AXL, TYRO3) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukácsi, S.; Farkas, Z.; Saskői, É.; Bajtay, Z.; Takács-Vellai, K. Conserved and Distinct Elements of Phagocytosis in Human and C. elegans. Int. J. Mol. Sci. 2021, 22, 8934. https://doi.org/10.3390/ijms22168934
Lukácsi S, Farkas Z, Saskői É, Bajtay Z, Takács-Vellai K. Conserved and Distinct Elements of Phagocytosis in Human and C. elegans. International Journal of Molecular Sciences. 2021; 22(16):8934. https://doi.org/10.3390/ijms22168934
Chicago/Turabian StyleLukácsi, Szilvia, Zsolt Farkas, Éva Saskői, Zsuzsa Bajtay, and Krisztina Takács-Vellai. 2021. "Conserved and Distinct Elements of Phagocytosis in Human and C. elegans" International Journal of Molecular Sciences 22, no. 16: 8934. https://doi.org/10.3390/ijms22168934
APA StyleLukácsi, S., Farkas, Z., Saskői, É., Bajtay, Z., & Takács-Vellai, K. (2021). Conserved and Distinct Elements of Phagocytosis in Human and C. elegans. International Journal of Molecular Sciences, 22(16), 8934. https://doi.org/10.3390/ijms22168934







