Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy
Abstract
1. Introduction
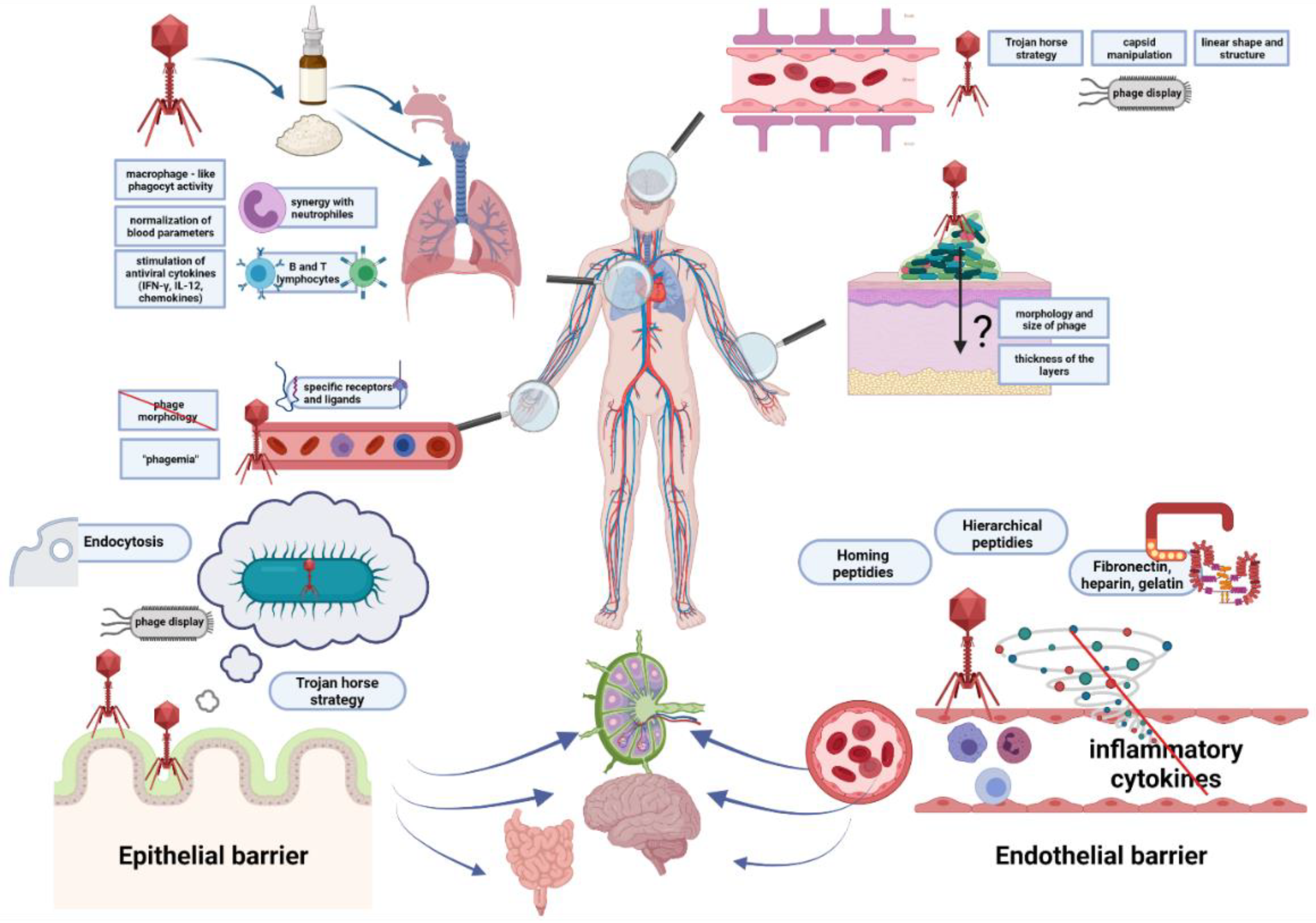
2. Ways for Penetration of Animal and Human Tissues by Bacteriophages
2.1. The Epithelial Barrier
2.2. The Circulatory System
2.3. The Endothelial Barrier
2.4. The Blood–Brain Barrier
2.5. The Skin
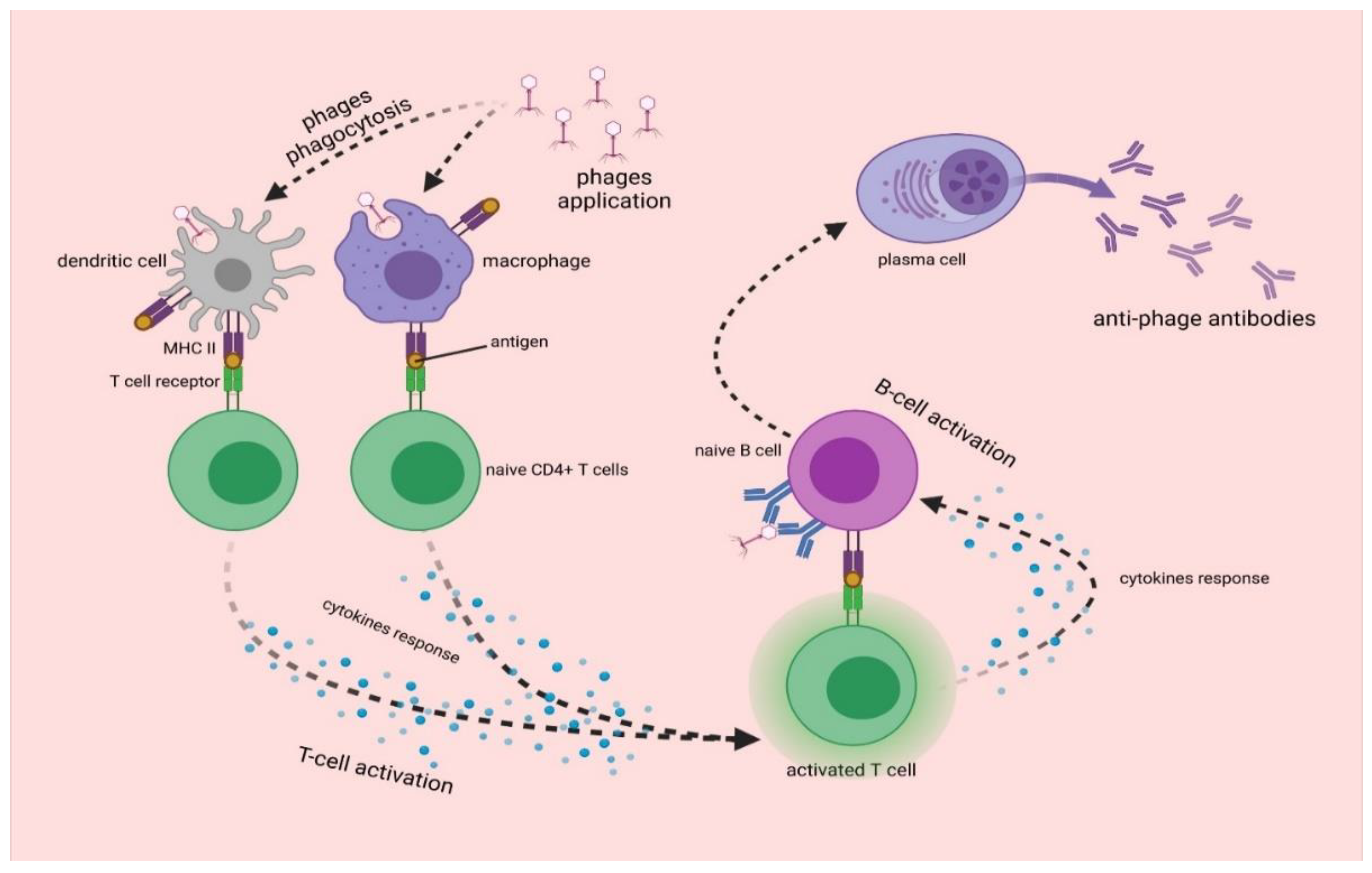
3. Interactions of Bacteriophages with Mammalian Immune System
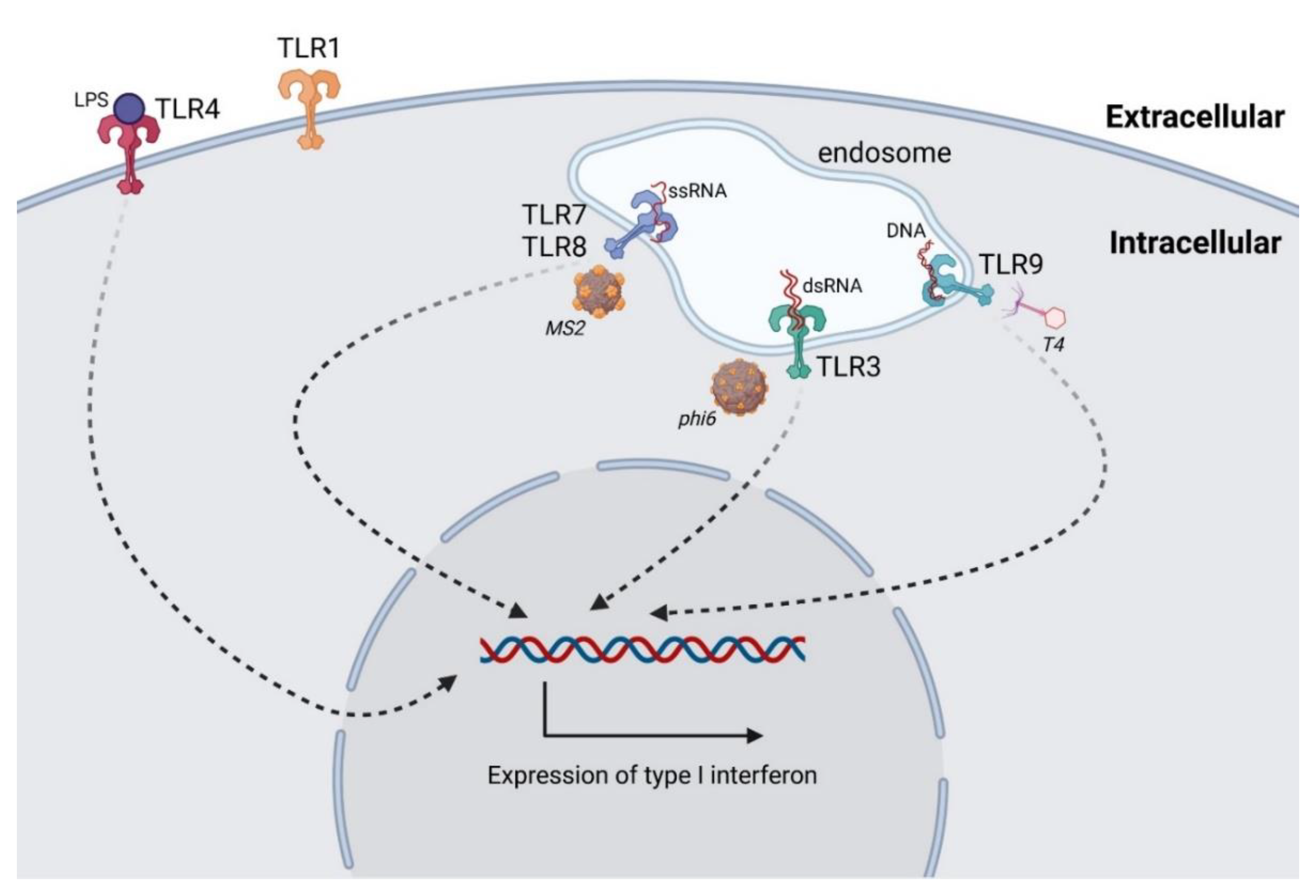
3.1. Antiphage Innate Immune Response
3.1.1. Dendritic Cells
3.1.2. Monocytes and Macrophages
3.1.3. Granulocytes
3.1.4. Foreign Particles vs. Innate Immune Response
3.1.5. Clearance of Bacteriophages
3.1.6. Phage-Induced Cytokine Response
3.1.7. Phages as Factors Increasing Bacterial Phagocytosis
3.2. Antiphage Adaptive Immune Response
4. Interactions of Bacteriophages with the Respiratory System
4.1. Phage Therapy against Bacterial Infections of Lungs
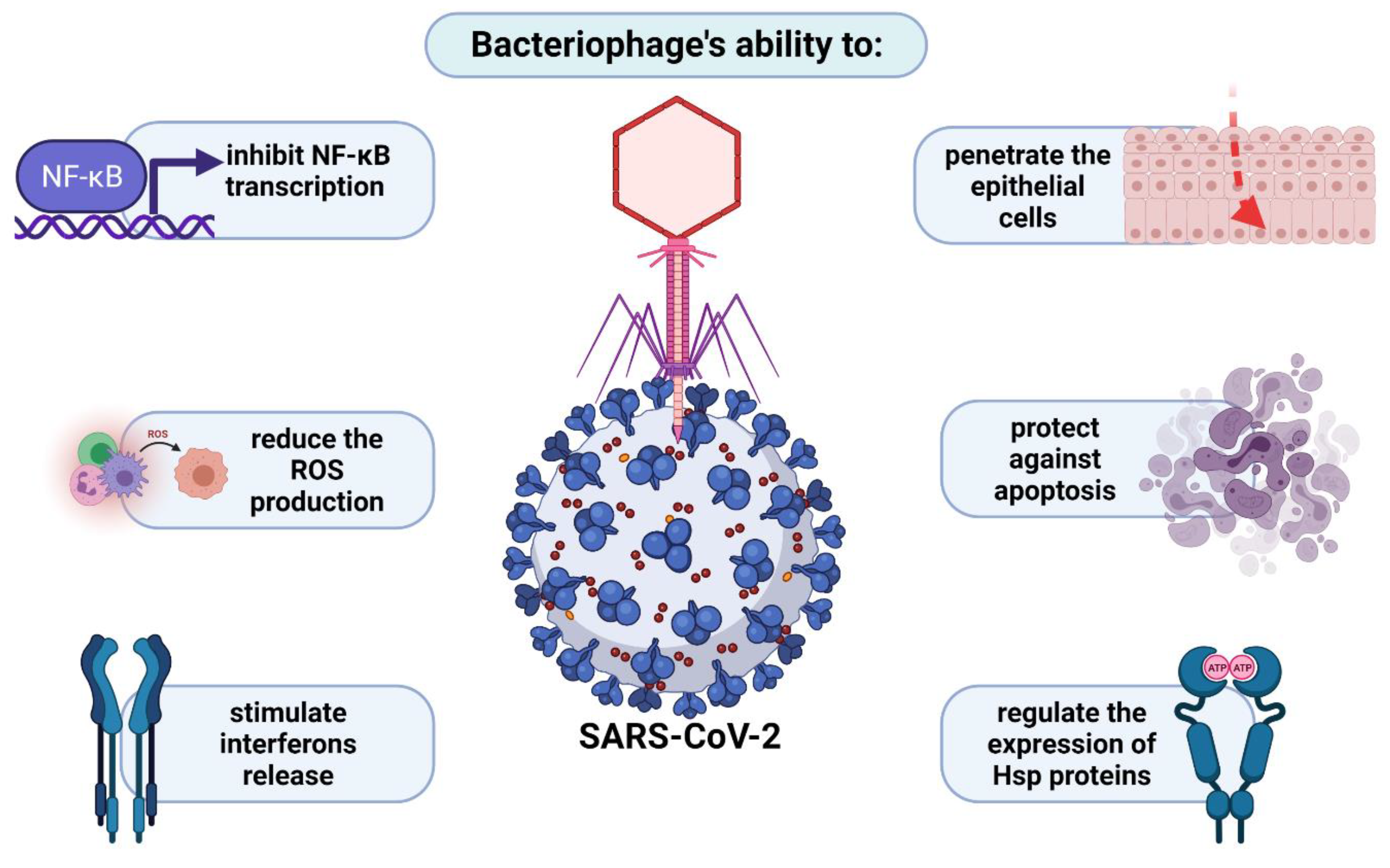
4.2. Penetration of Respiratory System by Phages in the Light of Anti-COVID-19 Therapy
5. Interactions of Bacteriophages with Mammalian Central Nervous System
5.1. Use of Phages in the Treatment of Brain Diseases
5.2. Phages as Central Nervous System Pathogens
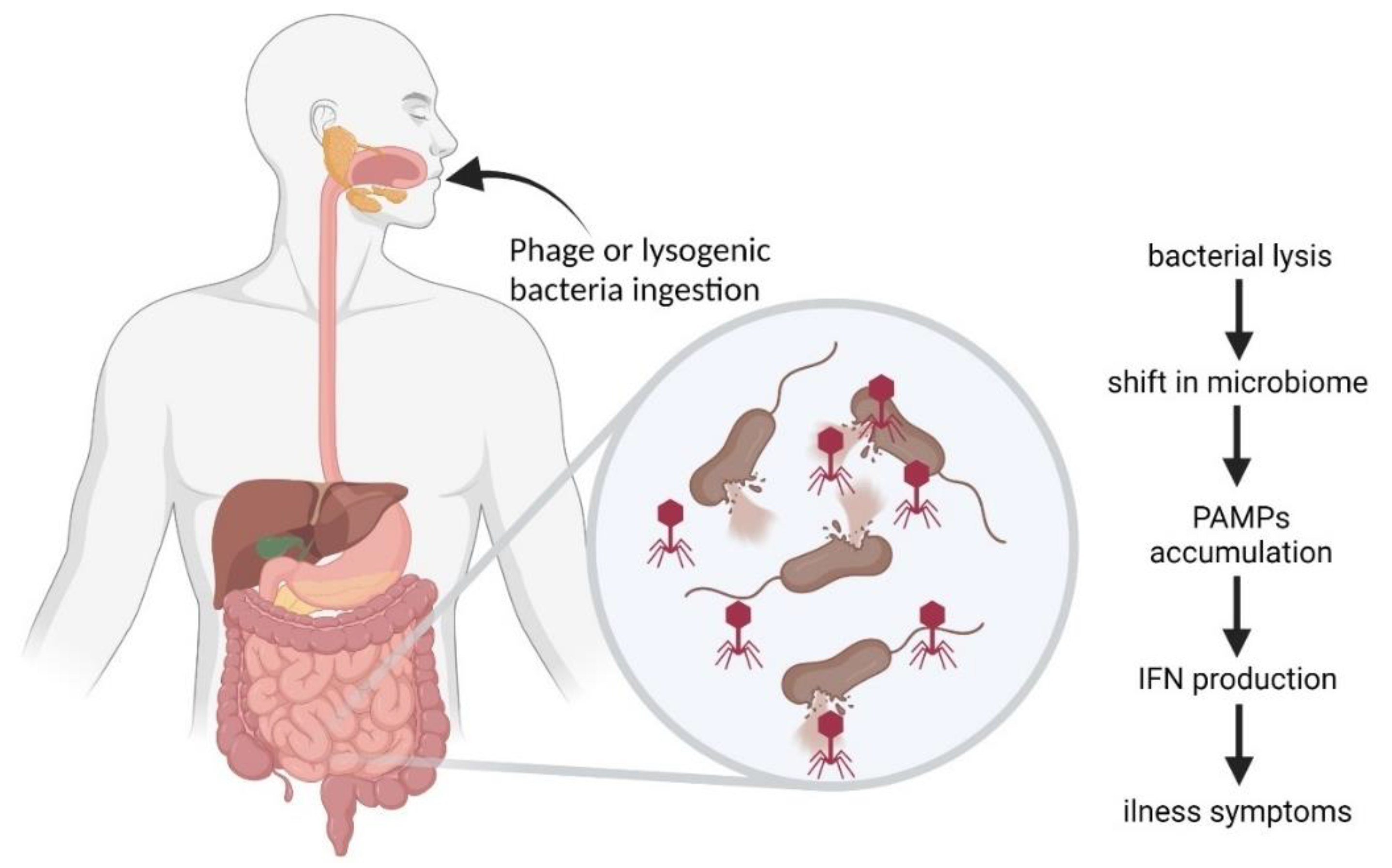
6. Bacteriophages in the Gastrointestinal Tract
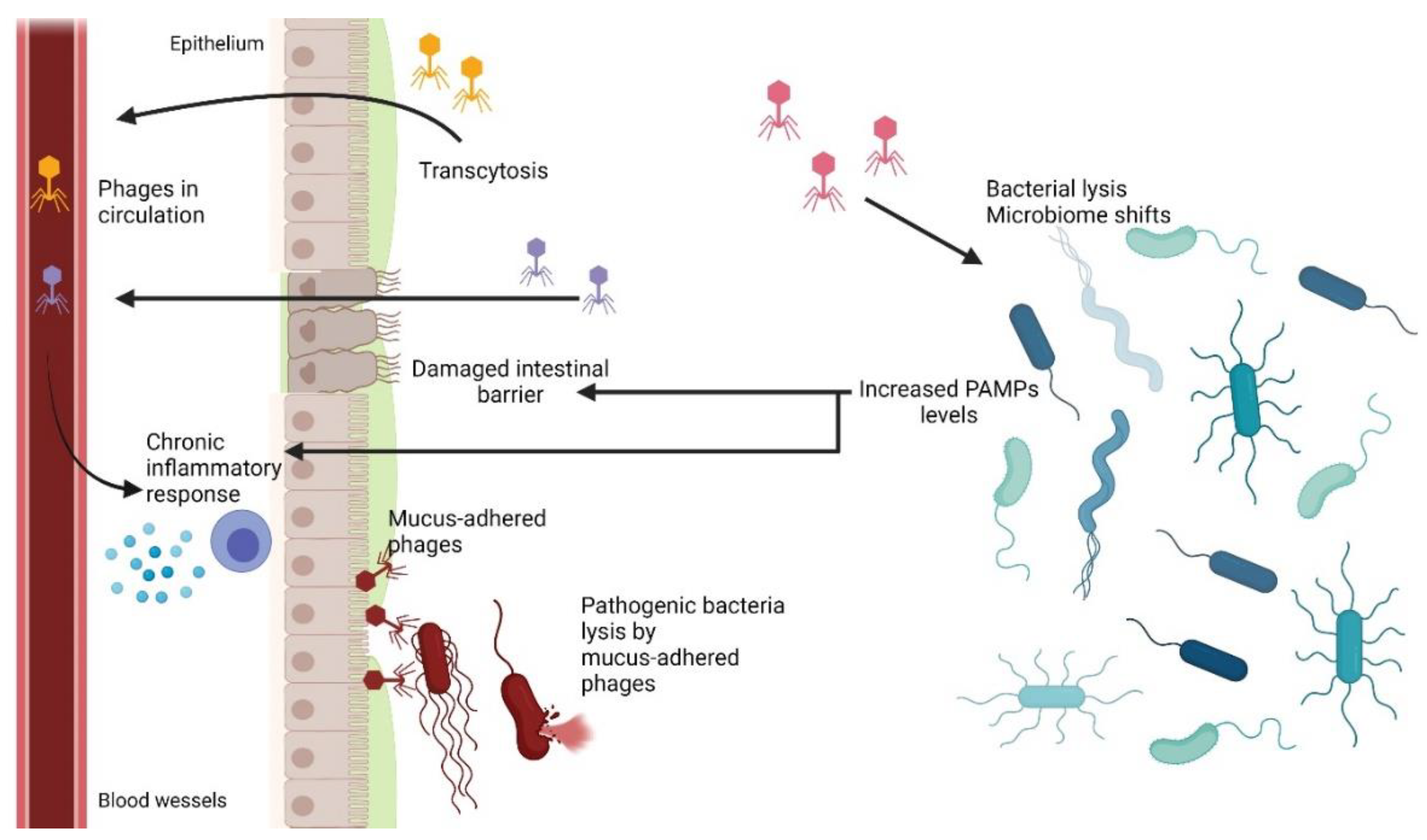
6.1. Phage Adherence to Mucus Layer
6.2. Phage Translocation from Gut to Bloodstream and Other Organs
6.3. Inflammatory Bowel Disease and Other Diseases Influenced by Phages
6.4. Bacteriophages as Gastrointestinal Tract Pathogens
6.5. Phage-Related Bacterial Toxins
6.6. Complexity of Phage-Mediated Effects in Gastrointestinal Tract
7. Bacteriophages in Urinary Tract
8. Bacteriophages in Female Reproductive System and Pregnancy
9. Interactions of Bacteriophages with Cancer
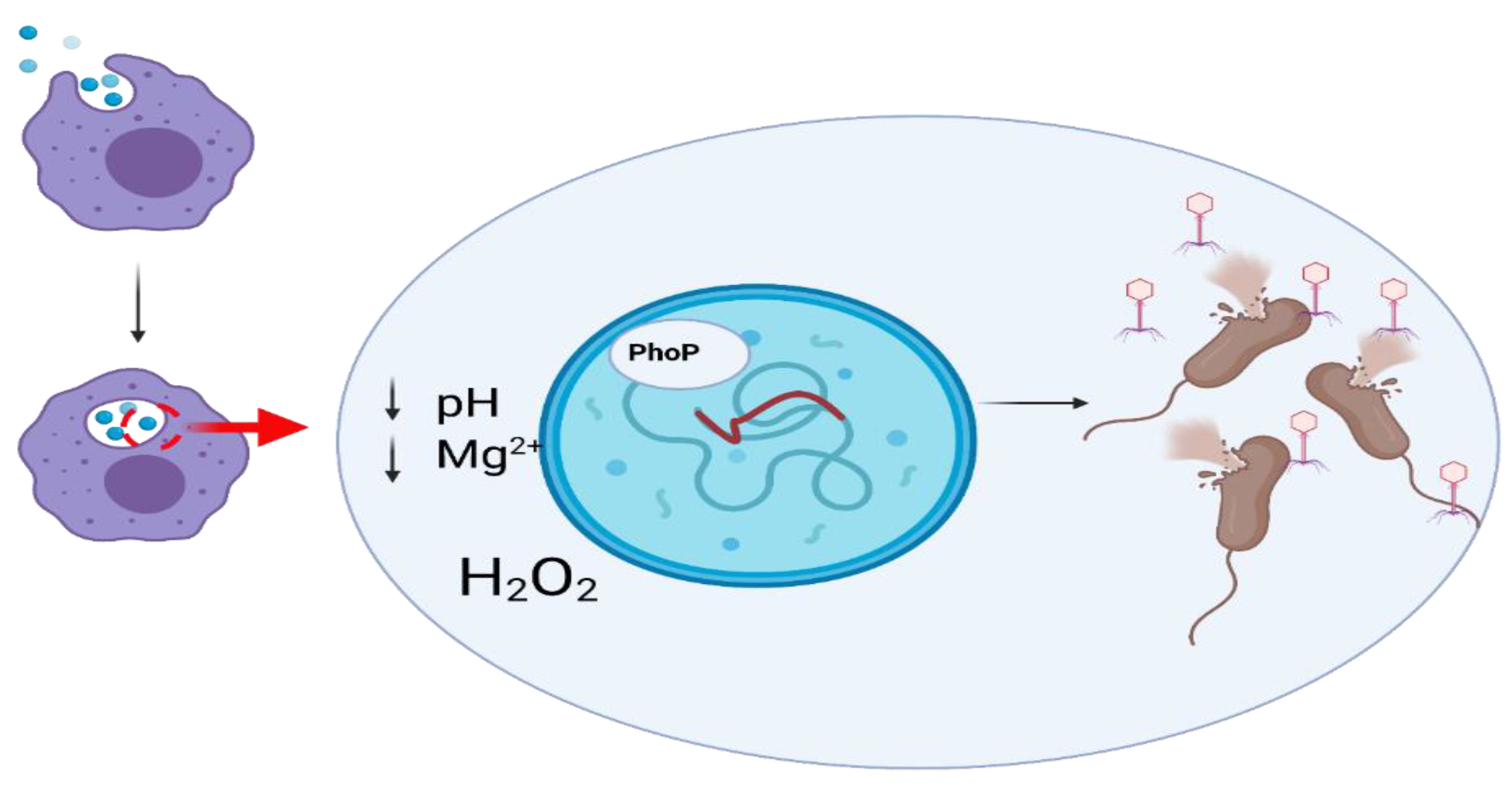
10. Interactions between Prophages and Eukaryotic Cells
11. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Pérez, J.; Contreras-Moreno, F.J.; Marcos-Torres, F.J.; Moraleda-Muñoz, A.; Muñoz-Dorado, J. The Antibiotic Crisis: How Bacterial Predators Can Help. Comput. Struct. Biotechnol. J. 2020, 18, 2547–2555. [Google Scholar] [CrossRef]
- Davies, J. Origins and Evolution of Antibiotic Resistance. Microbiología 1996, 12, 9–16. [Google Scholar] [CrossRef]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-Adhesion Therapy of Bacterial Diseases: Prospects and Problems. FEMS Immunol. Med Microbiol. 2003, 38, 181–191. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage Therapy: Current Status and Perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Jończyk-Matysiak, E.; Borysowski, J.; Letkiewicz, S.; Weber-Dąbrowska, B. The Fall and Rise of Phage Therapy in Modern Medicine. Expert Opin. Biol. Ther. 2019, 19, 1115–1117. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Fernández-Orth, D.; Brown-Jaque, M.; Miró, E.; Espinal, P.; Rodríguez-Rubio, L.; Muniesa, M.; Navarro, F. Unravelling the Consequences of the Bacteriophages in Human Samples. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Dąbrowska, K.; Vaneechoutte, M.; Barr, J.J.; Bollyky, P.L. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Miernikiewicz, P.; Dabrowska, K.; Piotrowicz, A.; Owczarek, B.; Wojas-Turek, J.; Kicielińska, J.; Rossowska, J.; Pajtasz-Piasecka, E.; Hodyra, K.; Macegoniuk, K.; et al. T4 Phage and Its Head Surface Proteins Do Not Stimulate Inflammatory Mediator Production. PLoS ONE 2013, 8, e71036. [Google Scholar] [CrossRef]
- Porayath, C.; Salim, A.; Palillam Veedu, A.; Babu, P.; Nair, B.; Madhavan, A.; Pal, S. Characterization of the Bacteriophages Binding to Human Matrix Molecules. Int. J. Biol. Macromol. 2018, 110, 608–615. [Google Scholar] [CrossRef]
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism to Cross Epithelial Cell Layers. mBio 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Lehti, T.A.; Pajunen, M.I.; Skog, M.S.; Finne, J. Internalization of a Polysialic Acid-Binding Escherichia Coli Bacteriophage into Eukaryotic Neuroblastoma Cells. Nat. Commun. 2017, 8, 1915. [Google Scholar] [CrossRef]
- Redrejo-Rodriǵuez, M.; Munõz-Espín, D.; Holguera, I.; Menciá, M.; Salas, M. Functional Eukaryotic Nuclear Localization Signals Are Widespread in Terminal Proteins of Bacteriophages. Proc. Natl. Acad. Sci. USA 2012, 109, 18482–18487. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive Motion of Bacteriophage in Mucosal Surfaces Increases the Frequency of Bacterial Encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675–13680. [Google Scholar] [CrossRef]
- Bille, E.; Meyer, J.; Jamet, A.; Euphrasie, D.; Barnier, J.P.; Brissac, T.; Larsen, A.; Pelissier, P.; Nassif, X. A Virulence-Associated Filamentous Bacteriophage of Neisseria Meningitidis Increases Host-Cell Colonisation. PLoS Pathog. 2017, 13, 1–24. [Google Scholar] [CrossRef]
- Tuma, P.L.; Hubbard, A.L. Transcytosis: Crossing Cellular Barriers. Physiol. Rev. 2003, 83, 871–932. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wu, M.; Liu, X.; Liu, Z.; Zhou, Q.; Niu, Z.; Huang, Y. Probing the Endocytic Pathways of the Filamentous Bacteriophage in Live Cells Using Ratiometric PH Fluorescent Indicator. Adv. Healthc. Mater. 2015, 4, 413–419. [Google Scholar] [CrossRef]
- Kim, A.; Shin, T.H.; Shin, S.M.; Pham, C.D.; Choi, D.K.; Kwon, M.H.; Kim, Y.S. Cellular Internalization Mechanism and Intracellular Trafficking of Filamentous M13 Phages Displaying a Cell-Penetrating Transbody and TAT Peptide. PLoS ONE 2012, 7, e51813. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Opolski, A.; Wietrzyk, J.; Switala-Jelen, K.; Godlewska, J.; Boratynski, J.; Syper, D.; Weber-Dabrowska, B.; Górski, A. Anticancer Activity of Bacteriophage T4 and Its Mutant HAP1 in Mouse Experimental Tumour Models. Anticancer. Res. 2004, 24, 3991–3995. [Google Scholar]
- Rosenwald, A.G.; Murray, B.; Toth, T.; Madupu, R.; Kyrillos, A.; Arora, G. Evidence for Horizontal Gene Transfer between Chlamydophila Pneumoniae and Chlamydia Phage. Bacteriophage 2014, 4, e965076. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Bordenstein, S.R. Eukaryotic Association Module in Phage WO Genomes from Wolbachia. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Bochkareva, S.S.; Karaulov, A.V.; Aleshkin, A.V.; Novikova, L.I.; Kiseleva, I.A.; Rubal’skii, E.O.; Mekhtiev, E.R.; Styshnev, A.O.; Zul’karneev, E.R.; Anurova, M.N.; et al. Analysis of the Pharmacokinetics of Suppository Forms of Bacteriophages. Bull. Exp. Biol. Med. 2020, 168, 748–752. [Google Scholar] [CrossRef]
- Capparelli, R.; Ventimiglia, I.; Roperto, S.; Fenizia, D.; Iannelli, D. Selection of an Escherichia Coli O157:H7 Bacteriophage for Persistence in the Circulatory System of Mice Infected Experimentally. Clin. Microbiol. Infect. 2006, 12, 248–253. [Google Scholar] [CrossRef]
- Yasuhiko, K.; Toshihiro, N. Infiltration of Bacteriophages from Intestinal Tract to Circulatory System in Goldfish. Fish Pathol. 2012, 47, 1–6. [Google Scholar] [CrossRef][Green Version]
- Chu, F.C.; Johnson, J.B.; Orr, H.C.; Probst, P.G.; Petricciani, J.C. Bacterial Virus Contamination of Fetal Bovine Sera. In Vitro 1973, 9, 31–34. [Google Scholar] [CrossRef]
- Merril, C.R.; Friedman, T.B.; Attallah, A.F.M.; Geier, M.R.; Geier, M.R.; Krell, K.; Yarkin, R. Isolation of Bacteriophages from Commercial Sera. In Vitro 1972, 8, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.C.; Sibinovic, K.H.; Probst, P.G.; Hochstein, H.D.; Littlejohn, D.C. Bacteriological Activity in Unfiltered Calf Sera Collected for Tissue Culture Use. In Vitro 1975, 11, 230–233. [Google Scholar] [CrossRef]
- Molander, C.W.; Kniazeff, A.J.; Boone, A.; Paley, A.; Imagawa, D.T. Isolation and Characterization of Viruses from Fetal Calf Serum. In Vitro 1971, 7, 168–173. [Google Scholar] [CrossRef]
- Mankiewicz, E.; Béland, J. The role of Mycobacteriophages and Cortisone in Experimental Tuberculosis and Sarcoidosis. Am. Rev. Respir. Dis. 1963, 89, 707–720. [Google Scholar]
- Parent, K.; Wilson, I.D. Mycobacteriophage in Crohn’ s Disease. Gut 1971, 12, 1019–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoffmann, M. Animal Experiments on Mucosal Passage and Absorption Viraemia of T3 Phages after Oral, Trachéal and Rectal Administration. Zent. Fur Bakteriol. Parasitenkd. Infekt. Und Hyg. 1965, 198, 371–390. [Google Scholar]
- Reynaud, A.; Cloastre, L.; Bernard, J.; Laveran, H.; Ackermann, H.W.; Licois, D.; Joly, B. Characteristics and Diffusion in the Rabbit of a Phage for Escherichia Coli 0103. Attempts to Use This Phage for Therapy. Vet. Microbiol. 1992, 30, 203–212. [Google Scholar] [CrossRef]
- Keller, R.; Engley, F.B. Fate of Bacteriophage Particles Introduced into Mice by Various Routes. Proc. Soc. Exp. Biol. Med. 1958, 98, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Duerr, D.M.; White, S.J.; Schluesener, H.J. Identification of Peptide Sequences That Induce the Transport of Phage across the Gastrointestinal Mucosal Barrier. J. Virol. Methods 2004, 116, 177–180. [Google Scholar] [CrossRef]
- Møller-Olsen, C.; Ross, T.; Leppard, K.N.; Foisor, V.; Smith, C.; Grammatopoulos, D.K.; Sagona, A.P. Bacteriophage K1F Targets Escherichia Coli K1 in Cerebral Endothelial Cells and Influences the Barrier Function. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rajotte, D.; Arap, W.; Hagedorn, M.; Koivunen, E.; Pasqualini, R.; Ruoslahti, E. Molecular Heterogeneity of the Vascular Endothelium Revealed by in Vivo Phage Display. J. Clin. Investig. 1998, 102, 430–437. [Google Scholar] [CrossRef]
- Wu, L.P.; Ahmadvand, D.; Su, J.; Hall, A.; Tan, X.; Farhangrazi, Z.S.; Moghimi, S.M. Crossing the Blood-Brain-Barrier with Nanoligand Drug Carriers Self-Assembled from a Phage Display Peptide. Nat. Commun. 2019, 10, 4635. [Google Scholar] [CrossRef] [PubMed]
- Egawa, G.; Nakamizo, S.; Natsuaki, Y.; Doi, H.; Miyachi, Y.; Kabashima, K. Intravital Analysis of Vascular Permeability in Mice Using Two-Photon Microscopy. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef]
- Górski, A.; Dąbrowska, K.; Hodyra-Stefaniak, K.; Borysowski, J.; Międzybrodzki, R.; Weber-Dąbrowska, B. Phages Targeting Infected Tissues: Novel Approach to Phage Therapy. Future Microbiol. 2015, 10, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Claesson-Welsh, L. Vascular Permeability—The Essentials. Upsala J. Med. Sci. 2015, 120, 135–143. [Google Scholar] [CrossRef]
- Geng, Y.; Dalhaimer, P.; Cai, S.; Tsai, R.; Tewari, M.; Minko, T.; Discher, D.E. Shape Effects of Filaments versus Spherical Particles in Flow and Drug Delivery. Nat. Nanotechnol. 2007, 2, 249–255. [Google Scholar] [CrossRef]
- Berkowitz, S.A.; Day, L.A. Mass, Length, Composition and Structure of the Filamentous Bacterial Virus Fd. J. Mol. Biol. 1976, 102, 531–547. [Google Scholar] [CrossRef]
- Anand, K.; Gengan, R.M.; Phulukdaree, A.; Chuturgoon, A. Agroforestry Waste Moringa Oleifera Petals Mediated Green Synthesis of Gold Nanoparticles and Their Anti-Cancer and Catalytic Activity. J. Ind. Eng. Chem. 2015, 21, 1105–1111. [Google Scholar] [CrossRef]
- Frenkel, D.; Solomon, B. Filamentous Phage as Vector-Mediated Antibody Delivery to the Brain. Proc. Natl. Acad. Sci. USA 2002, 99, 5675–5679. [Google Scholar] [CrossRef] [PubMed]
- Dubos, R.J.; Straus, J.H.; Pierce, C. The Multiplicaton of Bacteriophage In Vivo and Its Protective Effect Against an Experimental Infection with Shigella Dysenteriae. J. Exp. Med. 1943, 78, 161–168. [Google Scholar] [CrossRef]
- Ksendzovsky, A.; Walbridge, S.; Saunders, R.C.; Asthagiri, A.R.; Heiss, J.D.; Lonser, R.R. Convection-Enhanced Delivery of M13 Bacteriophage to the Brain. J. Neurosurg. 2012, 117, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Staquicini, F.I.; Ozawa, M.G.; Moya, C.A.; Driessen, W.H.P.; Barbu, E.M.; Nishimori, H.; Soghomonyan, S.; Flores, L.G.; Liang, X.; Paolillo, V.; et al. Systemic Combinatorial Peptide Selection Yields a Non-Canonical Iron-Mimicry Mechanism for Targeting Tumors in a Mouse Model of Human Glioblastoma. J. Clin. Investig. 2011, 121, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Urich, E.; Schmucki, R.; Ruderisch, N.; Kitas, E.; Certa, U.; Jacobsen, H.; Schweitzer, C.; Bergadano, A.; Ebeling, M.; Loetscher, H.; et al. Cargo Delivery into the Brain by in Vivo Identified Transport Peptides. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ki, V.; Rotstein, C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can. J. Infect. Dis. Med. Microbiol. 2008, 19, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Harjai, K.; Chhibber, S. Bacteriophage versus Antimicrobial Agents for the Treatment of Murine Burn Wound Infection Caused by Klebsiella Pneumoniae B5055. J. Med. Microbiol. 2011, 60, 205–210. [Google Scholar] [CrossRef]
- Pitol, A.K.; Bischel, H.N.; Boehm, A.B.; Kohn, T.; Juliana, T.R. Transfer of Enteric Viruses Adenovirus and Coxsackievirus and Bacteriophage MS2 from Liquid to Human Skin. Appl. Environ. Microbiol. 2018, 84, e01809-18. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Grice, E.A. Microbial Ecology of the Skin in the Era of Metagenomics and Molecular Microbiology. Cold Spring Harb. Perspect. Med. 2013, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J. A Bacteriophages Journey through the Human Body. Immunol. Rev. 2017, 279, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Clement, F.; Merabishvili, M.; Lavigne, R.; Vaneechoutte, M. Pro- and Anti-Inflammatory Responses of Peripheral Blood Mononuclear Cells Induced by Staphylococcus Aureus and Pseudomonas Aeruginosa Phages. Sci. Rep. 2017, 7, 8004. [Google Scholar] [CrossRef] [PubMed]
- Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Owczarek, B.; Międzybrodzki, R.; Łusiak-Szelachowska, M.; Łodej, N.; Górski, A. Phage-Phagocyte Interactions and Their Implications for Phage Application as Therapeutics. Viruses 2017, 9, 150. [Google Scholar] [CrossRef]
- Richards, D.M.; Endres, R.G. How Cells Engulf: A Review of Theoretical Approaches to Phagocytosis. Rep. Prog. Phys. 2017, 80, 126601. [Google Scholar] [CrossRef]
- Barfoot, R.; Denham, S.; Gyure, L.A.; Hall, J.G.; Hobbs, S.M.; Jackson, L.E.; Robertson, D. Some Properties of Dendritic Macrophages from Peripheral Lymph. Immunology 1989, 68, 233–239. [Google Scholar]
- An, T.-W.; Kim, S.-J.; Lee, Y.-D.; Park, J.-H.; Chang, H.-I. The Immune-Enhancing Effect of the Cronobacter Sakazakii ES2 Phage Results in the Activation of Nuclear Factor-ΚB and Dendritic Cell Maturation via the Activation of IL-12p40 in the Mouse Bone Marrow. Immunol. Lett. 2014, 157, 1–8. [Google Scholar] [CrossRef]
- Freyberger, H.R.; He, Y.; Roth, A.L.; Nikolich, M.P.; Filippov, A.A. Effects of Staphylococcus Aureus Bacteriophage K on Expression of Cytokines and Activation Markers by Human Dendritic Cells In Vitro. Viruses 2018, 10, 617. [Google Scholar] [CrossRef]
- Bocian, K.; Borysowski, J.; Zarzycki, M.; Pacek, M.; Weber-Dąbrowska, B.; Machcińska, M.; Korczak-Kowalska, G.; Górski, A. The Effects of T4 and A3/R Bacteriophages on Differentiation of Human Myeloid Dendritic Cells. Front. Microbiol. 2016, 7, 1267. [Google Scholar] [CrossRef]
- Aronow, R.; Danon, D.; Shahar, A.; Aronson, M. Electron Microscopy of In Vitro Endocytosis Of T2 Phage by Cells From Rabbit Peritoneal Exudate. J. Exp. Med. 1964, 120, 943–954. [Google Scholar] [CrossRef]
- Kaur, T.; Nafissi, N.; Wasfi, O.; Sheldon, K.; Wettig, S.; Slavcev, R. Immunocompatibility of Bacteriophages as Nanomedicines. J. Nanotechnol. 2012, 2012, 1–13. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yıldızlı, G.; Coral, G.; Ayaz, F. Immunostimulatory Activities of Coliphages on In Vitro Activated Mammalian Macrophages. Inflammation 2020, 43, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; Voyich, J.M.; Burlak, C.; DeLeo, F.R. Neutrophils in the Innate Immune Response. Arch. Immunol. Et Ther. Exp. 2005, 53, 505–517. [Google Scholar]
- Roach, D.R.; Chollet-Martin, S.; Noël, B.; Granger, V.; Debarbieux, L.; Chaisemartin, L. Human Neutrophil Response to Pseudomonas Bacteriophages. bioRxiv 2019, 786905. [Google Scholar] [CrossRef]
- Borysowski, J.; Wierzbicki, P.; Kłosowska, D.; Korczak-Kowalska, G.; Weber-Dąbrowska, B.; Górski, A. The Effects of T4 and A3/R Phage Preparations on Whole-Blood Monocyte and Neutrophil Respiratory Burst. Viral Immunol. 2010, 23, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Borysowski, J.; Międzybrodzki, R.; Wierzbicki, P.; Kłosowska, D.; Korczak-Kowalska, G.; Weber-Dąbrowska, B.; Górski, A. A3R Phage and Staphylococcus Aureus Lysate Do Not Induce Neutrophil Degranulation. Viruses 2017, 9, 36. [Google Scholar] [CrossRef]
- Ramirez, G.A.; Yacoub, M.-R.; Ripa, M.; Mannina, D.; Cariddi, A.; Saporiti, N.; Ciceri, F.; Castagna, A.; Colombo, G.; Dagna, L. Eosinophils from Physiology to Disease: A Comprehensive Review. BioMed Res. Int. 2018, 2018, e9095275. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Karasuyama, H. Emerging Roles of Basophils in Allergic Inflammation. Allergol. Int. 2017, 66, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, E.; Yang, L.; Song, J.; Wu, B. Therapeutic Application of Bacteriophage PHB02 and Its Putative Depolymerase Against Pasteurella Multocida Capsular Type A in Mice. Front. Microbiol. 2018, 9, 1678. [Google Scholar] [CrossRef]
- Krut, O.; Bekeredjian-Ding, I. Contribution of the Immune Response to Phage Therapy. J. Immunol. 2018, 200, 3037–3044. [Google Scholar] [CrossRef] [PubMed]
- Carroll-Portillo, A.; Lin, H.C. Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms 2019, 7, 625. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and Immunomodulation. Future Microbiol. 2017, 12, 905–914. [Google Scholar] [CrossRef]
- Elkins, K.L.; Rhinehart-Jones, T.R.; Stibitz, S.; Conover, J.S.; Klinman, D.M. Bacterial DNA containing CpG motifs stimulates lymphocyte-dependent protection of mice against lethal infection with intracellular bacteria. J. Immunol. 1999, 162, 2291–2298. [Google Scholar]
- Krieg, A.M. The Role of CpG Motifs in Innate Immunity. Curr. Opin. Immunol. 2000, 12, 35–43. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299.e8. [Google Scholar] [CrossRef]
- Lee, M.; Hosseindoust, A.; Oh, S.; Ko, H.; Cho, E.; Sa, S.; Kim, Y.; Choi, J.; Kim, J. Impact of an Anti-Salmonella. Typhimurium Bacteriophage on Intestinal Microbiota and Immunity Status of Laying Hens. J. Anim. Physiol. Anim. Nutr. 2020, 8. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, Z.; Zou, T.; Chen, J.; Li, G.; Zheng, L.; Li, S.; You, J. Bacteriophage as an Alternative to Antibiotics Promotes Growth Performance by Regulating Intestinal Inflammation, Intestinal Barrier Function and Gut Microbiota in Weaned Piglets. Front. Vet. Sci. 2021, 8, 623899. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.; Wong, S.; Jean, J.S.; Slavcev, R. Bacteriophage Interactions with Mammalian Tissue: Therapeutic Applications. Adv. Drug Deliv. Rev. 2019, 145, 4–17. [Google Scholar] [CrossRef]
- Dąbrowska, K. Phage Therapy: What Factors Shape Phage Pharmacokinetics and Bioavailability? Systematic and Critical Review. Med. Res. Rev. 2019, 39, 2000–2025. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.Y.T.; Chang, R.Y.K.; Li, M.; Wang, Y.; Lin, Y.; Morales, S.; McLachlan, A.J.; Kutter, E.; Li, J.; Chan, H.K. Pharmacokinetics and Time-Kill Study of Inhaled Antipseudomonal Bacteriophage Therapy in Mice. Antimicrob. Agents Chemother. 2020, 65, e01470-20. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas Aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.R.; Kim, S.; Rahman, M.; Kim, J. Antibacterial Efficacy of Lytic Pseudomonas Bacteriophage in Normal and Neutropenic Mice Models. J. Microbiol. 2011, 49, 994–999. [Google Scholar] [CrossRef]
- Hájek, P. The Elimination of Bacteriophages PhiX 174 and T2 from the Circulating Blood of Newborn Precolostral Pigs. Folia Microbiol. (Praha) 1970, 15, 125–128. [Google Scholar] [CrossRef]
- Otero, J.; García-Rodríguez, A.; Cano-Sarabia, M.; Maspoch, D.; Marcos, R.; Cortés, P.; Llagostera, M. Biodistribution of Liposome-Encapsulated Bacteriophages and Their Transcytosis During Oral Phage Therapy. Front. Microbiol. 2019, 10, 689. [Google Scholar] [CrossRef]
- Rouse, M.D.; Stanbro, J.; Roman, J.A.; Lipinski, M.A.; Jacobs, A.; Biswas, B.; Regeimbal, J.; Henry, M.; Stockelman, M.G.; Simons, M.P. Impact of Frequent Administration of Bacteriophage on Therapeutic Efficacy in an A. Baumannii Mouse Wound Infection Model. Front. Microbiol. 2020, 11, 414. [Google Scholar] [CrossRef] [PubMed]
- Naghizadeh, M.; Karimi Torshizi, M.A.; Rahimi, S.; Engberg, R.M.; Sørensen Dalgaard, T. Effect of Serum Anti-Phage Activity on Colibacillosis Control by Repeated Phage Therapy in Broilers. Vet. Microbiol. 2019, 234, 61–71. [Google Scholar] [CrossRef]
- Prazak, J.; Valente, L.G.; Iten, M.; Federer, L.; Grandgirard, D.; Soto, S.; Resch, G.; Leib, S.L.; Jakob, S.M.; Haenggi, M.; et al. Benefits of Aerosolized Phages for the Treatment of Pneumonia Due to Methicillin-Resistant Staphylococcus Aureus: An Experimental Study in Rats. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Chadha, P.; Katare, O.P.; Chhibber, S. Liposome Loaded Phage Cocktail: Enhanced Therapeutic Potential in Resolving Klebsiella Pneumoniae Mediated Burn Wound Infections. Burns 2017, 43, 1532–1543. [Google Scholar] [CrossRef]
- Kaźmierczak, Z.; Majewska, J.; Milczarek, M.; Owczarek, B.; Dąbrowska, K. Circulation of Fluorescently Labelled Phage in a Murine Model. Viruses 2021, 13, 297. [Google Scholar] [CrossRef]
- Łobocka, M.; Dąbrowska, K.; Górski, A. Engineered Bacteriophage Therapeutics: Rationale, Challenges and Future. BioDrugs 2021, 35, 255–280. [Google Scholar] [CrossRef]
- Merril, C.R.; Biswas, B.; Carlton, R.; Jensen, N.C.; Creed, G.J.; Zullo, S.; Adhya, S. Long-Circulating Bacteriophage as Antibacterial Agents. Proc. Natl. Acad. Sci. USA 1996, 93, 3188–3192. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, C.L.; Merril, C.R.; Adhya, S. An Amino Acid Substitution in a Capsid Protein Enhances Phage Survival in Mouse Circulatory System More than a 1000-Fold. Virus Res. 2005, 114, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Hodyra-Stefaniak, K.; Lahutta, K.; Majewska, J.; Kaźmierczak, Z.; Lecion, D.; Harhala, M.; Kęska, W.; Owczarek, B.; Jończyk-Matysiak, E.; Kłopot, A.; et al. Bacteriophages Engineered to Display Foreign Peptides May Become Short-Circulating Phages. Microb. Biotechnol. 2019, 12, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Buttaro, B.A.; Fouts, D.E.; Sanjari, S.; Evans, B.S.; Stevens, R.H. Bacteriophage ΦEf11 ORF28 Endolysin, a Multifunctional Lytic Enzyme with Properties Distinct from All Other Identified Enterococcus Faecalis Phage Endolysins. Appl. Environ. Microbiol. 2019, 85, e00555-19. [Google Scholar] [CrossRef] [PubMed]
- Miernikiewicz, P.; Kłopot, A.; Soluch, R.; Szkuta, P.; Kęska, W.; Hodyra-Stefaniak, K.; Konopka, A.; Nowak, M.; Lecion, D.; Kaźmierczak, Z.; et al. T4 Phage Tail Adhesin Gp12 Counteracts LPS-Induced Inflammation In Vivo. Front. Microbiol. 2016, 7, 1112. [Google Scholar] [CrossRef]
- Xue, Y.; Zhai, S.; Wang, Z.; Ji, Y.; Wang, G.; Wang, T.; Wang, X.; Xi, H.; Cai, R.; Zhao, R.; et al. The Yersinia Phage X1 Administered Orally Efficiently Protects a Murine Chronic Enteritis Model Against Yersinia Enterocolitica Infection. Front. Microbiol. 2020, 11, 351. [Google Scholar] [CrossRef]
- Pjanova, D.; Mandrika, L.; Petrovska, R.; Vaivode, K.; Donina, S. Comparison of the Effects of Bacteriophage-Derived DsRNA and Poly(I:C) on Ex Vivo Cultivated Peripheral Blood Mononuclear Cells. Immunol. Lett. 2019, 212, 114–119. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Haileselassie, Y.; Navis, M.; Cooper, C.; Sverremark-Ekström, E.; Nilsson, A.S. Morphologically Distinct Escherichia Coli Bacteriophages Differ in Their Efficacy and Ability to Stimulate Cytokine Release In Vitro. Front. Microbiol. 2016, 7, 437. [Google Scholar] [CrossRef]
- Górski, A.; Borysowski, J.; Miȩdzybrodzki, R. Bacteriophage Interactions With Epithelial Cells: Therapeutic Implications. Front. Microbiol. 2021, 11, 631161. [Google Scholar] [CrossRef]
- Jariah, R.O.A.; Hakim, M.S. Interaction of Phages, Bacteria, and the Human Immune System: Evolutionary Changes in Phage Therapy. Rev. Med. Virol. 2019, 29, e2055. [Google Scholar] [CrossRef]
- Przerwa, A.; Zimecki, M.; Switała-Jeleń, K.; Dabrowska, K.; Krawczyk, E.; Łuczak, M.; Weber-Dabrowska, B.; Syper, D.; Miedzybrodzki, R.; Górski, A. Effects of Bacteriophages on Free Radical Production and Phagocytic Functions. Med. Microbiol. Immunol. 2006, 195, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kabwe, M.; Meehan-Andrews, T.; Ku, H.; Petrovski, S.; Batinovic, S.; Chan, H.T.; Tucci, J. Lytic Bacteriophage EFA1 Modulates HCT116 Colon Cancer Cell Growth and Upregulates ROS Production in an Enterococcus Faecalis Co-Culture System. Front. Microbiol. 2021, 12, 650849. [Google Scholar] [CrossRef]
- Kennedy, M.A. A Brief Review of the Basics of Immunology: The Innate and Adaptive Response. Vet. Clin. N. Am. Small Anim. Pract. 2010, 40, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Gembara, K.; Dąbrowska, K. Phage-Specific Antibodies. Curr. Opin. Biotechnol. 2021, 68, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity Studies of Proteins Forming the T4 Phage Head Surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef] [PubMed]
- Hodyra-Stefaniak, K.; Kaźmierczak, Z.; Majewska, J.; Sillankorva, S.; Miernikiewicz, P.; Międzybrodzki, R.; Górski, A.; Azeredo, J.; Lavigne, R.; Lecion, D.; et al. Natural and Induced Antibodies against Phages in Humans: Induction Kinetics and Immunogenicity for Structural Proteins of PB1-Related Phages. PHAGE 2020, 1, 91–99. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Borysowski, J.; Dąbrowska, K.; Wierzbicki, P.; Ohams, M.; Korczak-Kowalska, G.; Olszowska-Zaremba, N.; Łusiak-Szelachowska, M.; Kłak, M.; et al. Phage as a Modulator of Immune Responses: Practical Implications for Phage Therapy. Adv. Virus Res. 2012, 83, 41–71. [Google Scholar] [CrossRef]
- Majewska, J.; Kaźmierczak, Z.; Lahutta, K.; Lecion, D.; Szymczak, A.; Miernikiewicz, P.; Drapała, J.; Harhala, M.; Marek-Bukowiec, K.; Jędruchniewicz, N.; et al. Induction of Phage-Specific Antibodies by Two Therapeutic Staphylococcal Bacteriophages Administered per Os. Front. Immunol. 2019, 10, 2607. [Google Scholar] [CrossRef]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef]
- Oli, A.K.; Shivshetty, N.; Ahmed, L.; Chavadi, M.; Kambar, R.N.; Chandrakanth, R.K. Efficacy of Bacteriophage Therapy against Vancomycin-Resistant Enterococcus Feacalis in Induced and Non-Induced Diabetic Mice. bioRxiv 2021, 427594. [Google Scholar] [CrossRef]
- Alomari, M.M.M.; Dec, M.; Nowaczek, A.; Puchalski, A.; Wernicki, A.; Kowalski, C.; Urban-Chmiel, R. Therapeutic and Prophylactic Effect of the Experimental Bacteriophage Treatment to Control Diarrhea Caused by E. Coli in Newborn Calves. ACS Infect. Dis. 2021, 7, 2093–2101. [Google Scholar] [CrossRef]
- Archana, A.; Patel, P.S.; Kumar, R.; Nath, G. Neutralizing Antibody Response against Subcutaneously Injected Bacteriophages in Rabbit Model. Virusdisease 2021, 32, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak, Z.; Majewska, J.; Miernikiewicz, P.; Międzybrodzki, R.; Nowak, S.; Harhala, M.; Lecion, D.; Kęska, W.; Owczarek, B.; Ciekot, J.; et al. Immune Response to Therapeutic Staphylococcal Bacteriophages in Mammals: Kinetics of Induction, Immunogenic Structural Proteins, Natural and Induced Antibodies. Front. Immunol. 2021, 12, 639570. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage Treatment of Human Infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef]
- Sadoff, H.L.; Almlof, J.W. Testing of Filters for Phage Removal. Ind. Eng. Chem. 1956, 48, 2199–2203. [Google Scholar] [CrossRef]
- Debarbieux, L.; Leduc, D.; Maura, D.; Morello, E.; Criscuolo, A.; Grossi, O.; Balloy, V.; Touqui, L. Bacteriophages Can Treat and Prevent Pseudomonas Aeruginosa Lung Infections. J. Infect. Dis. 2010, 201, 1096–1104. [Google Scholar] [CrossRef]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 2017, 22, 38–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Dufour, N.; Delattre, R.; Chevallereau, A.; Ricard, J.D.; Debarbieux, L. Phage Therapy of Pneumonia Is Not Associated with an Overstimulation of the Inflammatory Response Compared to Antibiotic Treatment in Mice. Antimicrob. Agents Chemother. 2019, 63, 1–12. [Google Scholar] [CrossRef]
- Górski, A.; Miedzybrodzki, R.; Żaczek, M.; Borysowski, J. Phages in the Fight against COVID-19? Future Microbiol. 2020, 15. [Google Scholar] [CrossRef]
- Wu, Y.; Li, C.; Xia, S.; Tian, X.; Kong, Y.; Wang, Z.; Gu, C.; Zhang, R.; Tu, C.; Xie, Y.; et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe 2020, 27, 891–898.e5. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.; Frenkel, D. Generation and Brain Delivery of Anti-Aggregating Antibodies against Beta-Amyloid Plaques Using Phage Display Technology. J. Neural Transm. Suppl. 2002, 321–325. [Google Scholar] [CrossRef]
- Dimant, H.; Sharon, N.; Solomon, B. Modulation Effect of Filamentous Phage on α-Synuclein Aggregation. Biochem. Biophys. Res. Commun. 2009, 383, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Dimant, H.; Solomon, B. Filamentous Phages Reduce α-Synuclein Oligomerization in the Membrane Fraction of SH-SY5Y Cells. Neurodegener. Dis. 2010, 7, 203–205. [Google Scholar] [CrossRef]
- Krishnan, R.; Tsubery, H.; Proschitsky, M.Y.; Asp, E.; Lulu, M.; Gilead, S.; Gartner, M.; Waltho, J.P.; Davis, P.J.; Hounslow, A.M.; et al. A Bacteriophage Capsid Protein Provides a General Amyloid Interaction Motif (GAIM) That Binds and Remodels Misfolded Protein Assemblies. J. Mol. Biol. 2014, 426, 2500–2519. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.R.A.; Kaufmann, G.F.; Mee, J.M.; Meijler, M.M.; Koob, G.F.; Janda, K.D. Treating Cocaine Addiction with Viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 10416–10421. [Google Scholar] [CrossRef]
- Eriksson, F.; Tsagozis, P.; Lundberg, K.; Parsa, R.; Mangsbo, S.M.; Persson, M.A.A.; Harris, R.A.; Pisa, P. Tumor-Specific Bacteriophages Induce Tumor Destruction through Activation of Tumor-Associated Macrophages. J. Immunol. 2009, 182, 3105–3111. [Google Scholar] [CrossRef]
- Eriksson, F.; Culp, W.D.; Massey, R.; Egevad, L.; Garland, D.; Persson, M.A.A.; Pisa, P. Tumor Specific Phage Particles Promote Tumor Regression in a Mouse Melanoma Model. Cancer Immunol. Immunother. 2007, 56, 677–687. [Google Scholar] [CrossRef]
- Dor-On, E.; Solomon, B. Targeting Glioblastoma via Intranasal Administration of Ff Bacteriophages. Front. Microbiol. 2015, 6, 530. [Google Scholar] [CrossRef]
- Rieder, R.; Wisniewski, P.J.; Alderman, B.L.; Campbell, S.C. Microbes and Mental Health: A Review. Brain Behav. Immun. 2017, 66, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Brown, S.M.; Hao, Y.; Tetz, V. Parkinson’s Disease and Bacteriophages as Its Overlooked Contributors. Sci. Rep. 2018, 8, 10812. [Google Scholar] [CrossRef] [PubMed]
- Yolken, R.H.; Severance, E.G.; Sabunciyan, S.; Gressitt, K.L.; Chen, O.; Stallings, C.; Origoni, A.; Katsafanas, E.; Schweinfurth, L.A.B.; Savage, C.L.G.; et al. Metagenomic Sequencing Indicates That the Oropharyngeal Phageome of Individuals With Schizophrenia Differs From That of Controls. Schizophr. Bull. 2015, 41, 1153–1161. [Google Scholar] [CrossRef]
- Tetz, G.; Tetz, V. Bacteriophages as New Human Viral Pathogens. Microorganisms 2018, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Mizee, M.; van Doorn, R.; Vries, A.; de Vries, H.E. Blood-Brain Barrier Disruption in Multiple Sclerosis. Blood-Brain Barrier Health Dis. 2015, 2, 22. [Google Scholar]
- Sutton, T.D.S.; Hill, C. Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. (Lausanne) 2019, 10, 784. [Google Scholar] [CrossRef]
- Guerin, E.; Hill, C. Shining Light on Human Gut Bacteriophages. Front. Cell. Infect. Microbiol. 2020, 10, 481. [Google Scholar] [CrossRef] [PubMed]
- Virgin, H.W. The Virome in Mammalian Physiology and Disease. Cell 2014, 157, 142–150. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage Adhering to Mucus Provide a Non-Host-Derived Immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Switała-Jelen, K.; Opolski, A.; Weber-Dabrowska, B.; Gorski, A. Bacteriophage Penetration in Vertebrates. J. Appl. Microbiol. 2005, 98, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Myelnikov, D. An Alternative Cure: The Adoption and Survival of Bacteriophage Therapy in the USSR, 1922–1955. J. Hist. Med. Allied Sci. 2018, 73, 385–411. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Wojciechowska, R.; Górski, A. Bacteriophages in the Gastrointestinal Tract and Their Implications. Gut Pathog. 2017, 9, 44. [Google Scholar] [CrossRef]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of Bacteriophages: Morphological and Biological Properties of a Large Group of Phages Isolated from Urban Sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Wang, D.; Holtz, L.R. The Bacterial Microbiome and Virome Milestones of Infant Development. Trends Microbiol. 2016, 24, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the Faecal Microbiota of Monozygotic Twins and Their Mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef]
- 1Yutin, N.; Makarova, K.S.; Gussow, A.B.; Krupovic, M.; Segall, A.; Edwards, R.A.; Koonin, E.V. Discovery of an Expansive Bacteriophage Family That Includes the Most Abundant Viruses from the Human Gut. Nat. Microbiol. 2018, 3, 38–46. [Google Scholar] [CrossRef]
- Marbouty, M.; Thierry, A.; Millot, G.A.; Koszul, R. MetaHiC Phage-Bacteria Infection Network Reveals Active Cycling Phages of the Healthy Human Gut. eLife 2021, 10, e60608. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527–541.e5. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef]
- Hsu, B.B.; Plant, I.N.; Lyon, L.; Anastassacos, F.M.; Way, J.C.; Silver, P.A. In Situ Reprogramming of Gut Bacteria by Oral Delivery. Nat. Commun. 2020, 11, 5030. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and Their Potential to Modulate the Microbiome and Immunity. Cell. Mol. Immunol. 2021, 18, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, S.; Nakazono, N.; Sugino, Y. The So-Called Chromosomal Verotoxin Genes Are Actually Carried by Defective Prophages. DNA Res. 1999, 6, 141–143. [Google Scholar] [CrossRef][Green Version]
- Zhu, Q.; Li, L.; Guo, Z.; Yang, R. Identification of Shiga-like Toxin Escherichia Coli Isolated from Children with Diarrhea by Polymerase Chain Reaction. Chin. Med. J. Engl. 2002, 115, 815–818. [Google Scholar]
- Moreno-Gallego, J.L.; Chou, S.-P.; Rienzi, S.C.D.; Goodrich, J.K.; Spector, T.D.; Bell, J.T.; Youngblut, N.D.; Hewson, I.; Reyes, A.; Ley, R.E. Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins. Cell Host Microbe 2019, 25, 261–272.e5. [Google Scholar] [CrossRef]
- Vaga, S.; Lee, S.; Ji, B.; Andreasson, A.; Talley, N.J.; Agreus, L.; Bidkhori, G.; Kovatcheva-Datchary, P.; Park, J.; Lee, D.; et al. Compositional and Functional Differences of the Mucosal Microbiota along the Intestine of Healthy Individuals. Sci. Rep. 2020, 10, 14977. [Google Scholar] [CrossRef]
- Sicard, J.-F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Kim, M.; Ashida, H.; Ogawa, M.; Yoshikawa, Y.; Mimuro, H.; Sasakawa, C. Bacterial Interactions with the Host Epithelium. Cell Host Microbe 2010, 8, 20–35. [Google Scholar] [CrossRef]
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-Like Domains on Bacteriophages: A Tale of Promiscuity and Deceit. J. Mol. Biol. 2006, 359, 496–507. [Google Scholar] [CrossRef]
- Fraser, J.S.; Maxwell, K.L.; Davidson, A.R. Immunoglobulin-like Domains on Bacteriophage: Weapons of Modest Damage? Curr. Opin. Microbiol. 2007, 10, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Wazna, E.; Dabrowska, B.-W.; Dabrowska, K.; Switała-Jeleń, K.; Miedzybrodzki, R. Bacteriophage Translocation. FEMS Immunol. Med. Microbiol. 2006, 46, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Żaczek, M.; Górski, A.; Skaradzińska, A.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B. Phage Penetration of Eukaryotic Cells: Practical Implications. Future Virol. 2020, 14, 11. [Google Scholar] [CrossRef]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Bäckhed, F. The Gut Microbiota—Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Focà, A.; Liberto, M.C.; Quirino, A.; Marascio, N.; Zicca, E.; Pavia, G. Gut Inflammation and Immunity: What Is the Role of the Human Gut Virome? Mediat. Inflamm. 2015, 2015, 326032. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Maksimovic, J.; Farries, G.; Sim, W.H.; Bishop, R.F.; Cameron, D.J.; Catto-Smith, A.G.; Kirkwood, C.D. Bacteriophages in Gut Samples from Pediatric Crohn’s Disease Patients: Metagenomic Analysis Using 454 Pyrosequencing. Inflamm. Bowel Dis. 2013, 19, 1598–1608. [Google Scholar] [CrossRef]
- Pérez-Brocal, V.; García-López, R.; Nos, P.; Beltrán, B.; Moret, I.; Moya, A. Metagenomic Analysis of Crohn’s Disease Patients Identifies Changes in the Virome and Microbiome Related to Disease Status and Therapy, and Detects Potential Interactions and Biomarkers. Inflamm. Bowel Dis. 2015, 21, 2515–2532. [Google Scholar] [CrossRef]
- Ma, Y.; You, X.; Mai, G.; Tokuyasu, T.; Liu, C. A Human Gut Phage Catalog Correlates the Gut Phageome with Type 2 Diabetes. Microbiome 2018, 6, 24. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Żaczek, M.; Borysowski, J.; Górski, A. The Presence of Bacteriophages in the Human Body: Good, Bad or Neutral? Microorganisms 2020, 8, 2012. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of Shiga Toxin-Producing Escherichia Coli and Their Contribution to Pathogenicity. Pathogens 2021, 10, 404. [Google Scholar] [CrossRef]
- De Paepe, M.; Leclerc, M.; Tinsley, C.R.; Petit, M.-A. Bacteriophages: An Underestimated Role in Human and Animal Health? Front. Cell. Infect. Microbiol. 2014, 4, 39. [Google Scholar] [CrossRef]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and Antibiotics on Biofilm Characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F. Bacteriophage-Encoded Bacterial Virulence Factors and Phage-Pathogenicity Island Interactions. Adv. Virus Res. 2012, 82, 91–118. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.-C.; Sekulovic, O. Importance of Prophages to Evolution and Virulence of Bacterial Pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Sandvig, K.; Bergan, J.; Dyve, A.-B.; Skotland, T.; Torgersen, M.L. Endocytosis and Retrograde Transport of Shiga Toxin. Toxicon 2010, 56, 1181–1185. [Google Scholar] [CrossRef]
- Boyd, E.F. Efficiency and Specificity of CTXϕ Chromosomal Integration: Dif Makes All the Difference. Proc. Natl. Acad. Sci. USA 2010, 107, 3951–3952. [Google Scholar] [CrossRef]
- Davis, B.M.; Waldor, M.K. Filamentous Phages Linked to Virulence of Vibrio Cholerae. Curr. Opin. Microbiol. 2003, 6, 35–42. [Google Scholar] [CrossRef]
- Dirita, V.J. Molecular Basis of Vibrio cholerae Pathogenesis. In Principles of Bacterial Pathogenesis; Groisman, E.A., Ed.; Academic Press: San Diego, CA, USA, 2001; pp. 457–508. ISBN 978-0-12-304220-0. [Google Scholar]
- Bharati, K.; Ganguly, N.K. Cholera Toxin: A Paradigm of a Multifunctional Protein. Indian J. Med Res. 2011, 133, 179–187. [Google Scholar]
- Govind, R.; Vediyappan, G.; Rolfe, R.D.; Dupuy, B.; Fralick, J.A. Bacteriophage-Mediated Toxin Gene Regulation in Clostridium Difficile. J. Virol. 2009, 83, 12037–12045. [Google Scholar] [CrossRef] [PubMed]
- Larkum, N.W. Bacteriophagy in Urinary Infection Part I. The Incidence of Bacteriophage and of Bacillus coli Susceptible to Dissolution by the Bacteriophage in Urines. Presentation of Cases of Reneal Infection in Which Bacteriophage Was Used Therapeutically. J. Bacteriol. 1926, 12, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Larkum, N.W. Bacteriophagy in Urinary Infection Part II. Bacteriophagy in the Bladder. J. Bacteriol. 1926, 12, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Brown-Jaque, M.; Muniesa, M.; Navarro, F. Bacteriophages in Clinical Samples Can Interfere with Microbiological Diagnostic Tools. Sci. Rep. 2016, 6, 33000. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Rodriguez, T.M.; Ly, M.; Bonilla, N.; Pride, D.T. The Human Urine Virome in Association with Urinary Tract Infections. Front. Microbiolol. 2015, 6, 14. [Google Scholar] [CrossRef]
- Ghose, C.; Ly, M.; Schwanemann, L.K.; Shin, J.H.; Atab, K.; Barr, J.J.; Little, M.; Schooley, R.T.; Chopyk, J.; Pride, D.T. The Virome of Cerebrospinal Fluid: Viruses Where We Once Thought There Were None. Front. Microbiol. 2019, 10, 2061. [Google Scholar] [CrossRef]
- Malki, K.; Sible, E.; Cooper, A.; Garretto, A.; Bruder, K.; Watkins, S.C.; Putonti, C. Seven Bacteriophages Isolated from the Female Urinary Microbiota. Genome Announc. 2016, 4, e01003-16. [Google Scholar] [CrossRef]
- Garretto, A.; Thomas-White, K.; Wolfe, A.J.; Putonti, C. Detecting Viral Genomes in the Female Urinary Microbiome. J. Gen. Virol. 2018, 99, 1141–1146. [Google Scholar] [CrossRef]
- Garretto, A.; Miller-Ensminger, T.; Wolfe, A.J.; Putonti, C. Bacteriophages of the Lower Urinary Tract. Nat. Rev. Urol. 2019, 16, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Miller-Ensminger, T.; Garretto, A.; Brenner, J.; Thomas-White, K.; Zambom, A.; Wolfe, A.J.; Putonti, C. Bacteriophages of the Urinary Microbiome. J. Bacteriol. 2018, 200, e00738-17. [Google Scholar] [CrossRef]
- Miller-Ensminger, T.; Garretto, A.; Stark, N.; Putonti, C. Mimicking Prophage Induction in the Body: Induction in the Lab with PH Gradients. PeerJ 2020, 8, e9718. [Google Scholar] [CrossRef]
- Lai, H.-C.; Chang, S.-N.; Lin, H.-C.; Hsu, Y.-L.; Wei, H.-M.; Kuo, C.-C.; Hwang, K.-P.; Chiang, H.-Y. Association between Urine PH and Common Uropathogens in Children with Urinary Tract Infections. J. Microbiol. Immunol. Infect. 2021, 54, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.W.; Putonti, C. UPΦ Phages, a New Group of Filamentous Phages Found in Several Members of Enterobacteriales. Virus Evol. 2020, 6, veaa030. [Google Scholar] [CrossRef] [PubMed]
- Miedzybrodzki, R.; Switala-Jelen, K.; Fortuna, W.; Weber-Dabrowska, B.; Przerwa, A.; Lusiak-Szelachowska, M.; Dabrowska, K.; Kurzepa, A.; Boratynski, J.; Syper, D.; et al. Bacteriophage Preparation Inhibition of Reactive Oxygen Species Generation by Endotoxin-Stimulated Polymorphonuclear Leukocytes. Virus Res. 2008, 10, 233–242. [Google Scholar] [CrossRef]
- Tóthová, L.; Celec, P.; Bábíčková, J.; Gajdošová, J.; Al-Alami, H.; Kamodyova, N.; Drahovská, H.; Liptáková, A.; Turňa, J.; Hodosy, J. Phage Therapy of Cronobacter-Induced Urinary Tract Infection in Mice. Med. Sci. Monit. 2011, 17, 173–178. [Google Scholar] [CrossRef]
- Goetsch, H.E.; Zhao, L.; Gnegy, M.; Imperiale, M.J.; Love, N.G.; Wigginton, K.R. Fate of the Urinary Tract Virus BK Human Polyomavirus in Source-Separated Urine. Appl. Environ. Microbiol. 2018, 84, 12. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A.; Pradhan, S.K.; Heinonen-Tanski, H. Survival of Enteric Bacteria and Coliphage MS2 in Pure Human Urine. J. Appl. Microbiol. 2009, 107, 1651–1657. [Google Scholar] [CrossRef]
- Tan, D.; Zhang, Y.; Cheng, M.; Le, S.; Gu, J. Characterization of Klebsiella Pneumoniae ST11 Isolates and Their Interactions with Lytic Phages. Viruses 2019, 11, 1080. [Google Scholar] [CrossRef]
- Pereira, S.; Pereira, C.; Santos, L.; Klumpp, J.; Almeida, A. Potential of Phage Cocktails in the Inactivation of Enterobacter Cloacae—An in Vitro Study in a Buffer Solution and in Urine Samples. Virus Res. 2016, 211, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Elena Cardoso, M.; Fernández, L.; Tejería, E.; Esperón, P.; Terán, M. Evaluation of a Labelled Bacteriophage with 99mTc as a Potential Agent for Infection Diagnosis. Curr. Radiopharm. 2016, 9, 137–142. [Google Scholar] [CrossRef]
- Holman, D.; Lungren, M.P.; Hardy, J.; Contag, C.; Blankenberg, F. Preparation of Tc99m-Labeled Pseudomonas Bacteriophage without Adversely Impacting Infectivity or Biodistribution. Bioconjugate Chem. 2017, 28, 2698–2706. [Google Scholar] [CrossRef]
- Letarov, A.V.; Golomidova, A.K.; Tarasyan, K.K. Ecological Basis for Rational Phage Therapy. Acta Nat. 2010, 2, 60–72. [Google Scholar] [CrossRef]
- Sillankorva, S.; Oliveira, D.; Moura, A.; Henriques, M.; Faustino, A.; Nicolau, A.; Azeredo, J. Efficacy of a Broad Host Range Lytic Bacteriophage Against, E. Coli Adhered to Urothelium. Curr. Microbiol. 2011, 62, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Blanco, C.; Chen, I.A. Phage Therapy Administered Noninvasively Could Be Effective in Thin Tubes Subject to Episodic Flow despite Washout: A Simulation Study. Phys. Biolol. 2019, 16, 054001. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.S.; Lehman, S.M.; Tweardy, D.J.; Donlan, R.M.; Trautner, B.W. Bacteriophages Are Synergistic with Bacterial Interference for the Prevention of Pseudomonas Aeruginosa Biofilm Formation on Urinary Catheters. J. Appl. Microbiol. 2012, 113, 1530–1539. [Google Scholar] [CrossRef]
- Rakov, C.; Ben Porat, S.; Alkalay-Oren, S.; Yerushalmy, O.; Abdalrhman, M.; Gronovich, N.; Huang, L.; Pride, D.; Coppenhagen-Glazer, S.; Nir-Paz, R.; et al. Targeting Biofilm of MDR Providencia Stuartii by Phages Using a Catheter Model. Antibiotics 2021, 10, 375. [Google Scholar] [CrossRef]
- Sybesma, W.; Zbinden, R.; Chanishvili, N.; Kutateladze, M.; Chkhotua, A.; Ujmajuridze, A.; Mehnert, U.; Kessler, T.M. Bacteriophages as Potential Treatment for Urinary Tract Infections. Front. Microbiol. 2016, 7, 465. [Google Scholar] [CrossRef] [PubMed]
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P.; et al. Intravesical Bacteriophages for Treating Urinary Tract Infections in Patients Undergoing Transurethral Resection of the Prostate: A Randomised, Placebo-Controlled, Double-Blind Clinical Trial. Lancet Infect. Dis. 2021, 21, 427–436. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Wojciuk, B.; Roszak, M.; Łubowska, N.; Błażejczak, P.; Jursa-Kulesza, J.; Rakoczy, R.; Masiuk, H.; Dołęgowska, B. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter Baumannii Biofilm in a Human Urine Model. Microb. Drug Resist. 2021, 27, 25–35. [Google Scholar] [CrossRef]
- Townsend, E.M.; Moat, J.; Jameson, E. CAUTI’s next Top Model—Model Dependent Klebsiella Biofilm Inhibition by Bacteriophages and Antimicrobials. Biofilm 2020, 2, 100038. [Google Scholar] [CrossRef]
- Moradpour, Z.; Yousefi, N.; Sadeghi, D.; Ghasemian, A. Synergistic Bactericidal Activity of a Naturally Isolated Phage and Ampicillin against Urinary Tract Infecting Escherichia Coli O157. Iran. J. Basic Med. Sci. 2020, 23, 257–263. [Google Scholar] [CrossRef]
- Kuipers, S.; Ruth, M.M.; Mientjes, M.; de Sévaux, R.G.L.; van Ingen, J.A. Dutch Case Report of Successful Treatment of Chronic Relapsing Urinary Tract Infection with Bacteriophages in a Renal Transplant Patient. Antimicrob. Agents Chemother. 2019, 64, e01281-19. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J.; et al. Non-Active Antibiotic and Bacteriophage Synergism to Successfully Treat Recurrent Urinary Tract Infection Caused by Extensively Drug-Resistant Klebsiella Pneumoniae. Emerg. Microbes Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef]
- Zalewska-Piątek, B.; Piątek, R. Phage Therapy as a Novel Strategy in the Treatment of Urinary Tract Infections Caused by E. Coli. Antibiotics 2020, 9, 304. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, L.L.; Chang, B.J.; Payne, M.S. Applications for Bacteriophage Therapy during Pregnancy and the Perinatal Period. Front. Microbiol. 2018, 8, 2660. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, M.E.; Tezuka, E.; Devkota, B.; Izaike, Y.; Osawa, T. Persistence of Uterine Bacterial Infection, and Its Associations with Endometritis and Ovarian Function in Postpartum Dairy Cows. J. Reprod. Dev. 2015, 61, 54–60. [Google Scholar] [CrossRef]
- Mitchell, C.; Prabhu, M. Pelvic Inflammatory Disease: Current Concepts in Pathogenesis, Diagnosis and Treatment. Infect. Dis. Clin. N. Am. 2013, 27, 793–809. [Google Scholar] [CrossRef]
- Dunlop, A.L.; Mulle, J.G.; Ferranti, E.P.; Edwards, S.; Dunn, A.B.; Corwin, E.J. Maternal Microbiome and Pregnancy Outcomes That Impact Infant Health: A Review. Adv. Neonatal Care 2015, 15, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Loebstein, R.; Lalkin, A.; Koren, G. Pharmacokinetic Changes during Pregnancy and Their Clinical Relevance. Clin. Pharmacokinet. 1997, 33, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Lohy Das, J.; Rulisa, S.; de Vries, P.J.; Mens, P.F.; Kaligirwa, N.; Agaba, S.; Tarning, J.; Karlsson, M.O.; Dorlo, T.P.C. Population Pharmacokinetics of Artemether, Dihydroartemisinin, and Lumefantrine in Rwandese Pregnant Women Treated for Uncomplicated Plasmodium Falciparum Malaria. Antimicrob. Agents Chemother. 2018, 62, e00518-18. [Google Scholar] [CrossRef] [PubMed]
- Bookstaver, P.B.; Bland, C.M.; Griffin, B.; Stover, K.R.; Eiland, L.S.; McLaughlin, M. A Review of Antibiotic Use in Pregnancy. Pharmacotherapy 2015, 35, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Almaramhy, H.H.; Al-Zalabani, A.H. The Association of Prenatal and Postnatal Macrolide Exposure with Subsequent Development of Infantile Hypertrophic Pyloric Stenosis: A Systematic Review and Meta-Analysis. Ital. J. Pediatrics 2019, 45, 20. [Google Scholar] [CrossRef]
- Machado, V.S.; Bicalho, M.L.S.; Pereira, R.V.; Caixetta, L.S.; Bittar, J.H.J.; Oikonomou, G.; Gilbert, R.O.; Bicalho, R.C. The Effect of Intrauterine Administration of Mannose or Bacteriophage on Uterine Health and Fertility of Dairy Cows with Special Focus on Escherichia Coli and Arcanobacterium Pyogenes. J. Dairy Sci. 2012, 95, 3100–3109. [Google Scholar] [CrossRef] [PubMed]
- Meira, E.B.S.; Rossi, R.S.; Teixeira, A.G.; Kaçar, C.; Oikonomou, G.; Gregory, L.; Bicalho, R.C. The Effect of Prepartum Intravaginal Bacteriophage Administration on the Incidence of Retained Placenta and Metritis. J. Dairy Sci. 2013, 96, 7658–7665. [Google Scholar] [CrossRef] [PubMed]
- Sume, M.R.; Paxton, J.W.; Keelan, J.A. Drug Transfer and Metabolism by the Human Placenta. Clin. Pharmacokinet. 2004, 43, 487–514. [Google Scholar] [CrossRef]
- Arora, N.; Sadovsky, Y.; Dermody, T.S.; Coyne, C.B. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017, 21, 561–567. [Google Scholar] [CrossRef]
- Bayer, A.; Delorme-Axford, E.; Sleigher, C.; Frey, T.K.; Trobaugh, D.W.; Klimstra, W.B.; Emert-Sedlak, L.A.; Smithgall, T.E.; Kinchington, P.R.; Vadia, S.; et al. Human Trophoblasts Confer Resistance to Viruses Implicated in Perinatal Infection. Am. J. Obstet. Gynecol. 2015, 212, 71.e1–71.e8. [Google Scholar] [CrossRef]
- Blair, J.E.; Reeves, D.L. The Placental Transmission of Bacteriophage. J. Infect. Dis. 1928, 42, 440–443. [Google Scholar] [CrossRef]
- Kulangara, A.C.; Sellers, M.I. Passage of Bacteriophages from Mother to Foetus in the Rat. Proc. Soc. Exp. Biol. Med. 1959, 101, 207–211. [Google Scholar] [CrossRef]
- Uhr, J.W.; Dancis, J.; Finkelstein, M.S. Passage of Bacteriophage ΦX 174 Across the Placenta in Guinea Pigs. Proc. Soc. Exp. Biol. Med. 1963, 113, 391–394. [Google Scholar] [CrossRef]
- Srivastava, A.S.; Chauhan, D.P.; Carrier, E. In Utero Detection of T7 Phage after Systemic Administration to Pregnant Mice. BioTechniques 2004, 37, 81–83. [Google Scholar] [CrossRef]
- Sheridan, M.A.; Yunusov, D.; Balaraman, V.; Alexenko, A.P.; Yabe, S.; Verjovski-Almeida, S.; Schust, D.J.; Franz, A.W.; Sadovsky, Y.; Ezashi, T.; et al. Vulnerability of Primitive Human Placental Trophoblast to Zika Virus. Proc. Natl. Acad. Sci. USA 2017, 114, E1587–E1596. [Google Scholar] [CrossRef]
- Robbins, J.R.; Zeldovich, V.B.; Poukchanski, A.; Boothroyd, J.C.; Bakardjiev, A.I. Tissue Barriers of the Human Placenta to Infection with Toxoplasma Gondii. Infect. Immun. 2012, 80, 418–428. [Google Scholar] [CrossRef]
- Kurzepa-Skaradzinska, A.; Skaradzinski, G.; Weber, B.; Zaczek, M.; Maj, T.; Slawek, A.; Switalska, M.; Maciejewska, M.; Wietrzyk, J.; Rymowicz, W.; et al. Influence of Bacteriophage Preparations on Migration of HL-60 Leukemia Cells In Vitro. Anticancer. Res. 2013, 33, 1569–1574. [Google Scholar] [PubMed]
- Dąbrowska, K.; Opolski, A.; Wietrzyk, J.; Switala-Jelen, K.; Boratynski, J.; Nasulewicz, A.; Lipinska, L.; Chybicka, A.; Kujawa, M.; Zabel, M.; et al. Antitumor Activity of Bacteriophages in Murine Experimental Cancer Models Caused Possibly by Inhibition of Β3 Integrin Signaling Pathway. Acta Virol. 2004, 48, 241–248. [Google Scholar] [PubMed]
- Dąbrowska, K.; Skaradziński, G.; Jończyk, P.; Kurzępa, A.; Wietrzyk, J.; Owczarek, B.; Żaczek, M.; Świtała-Jeleń, K.; Boratyński, J.; Poźniak, G.; et al. The Effect of Bacteriophages T4 and HAP1 on in Vitro Melanoma Migration. BMC Microbiol. 2009, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Opolski, A.; Wietrzyk, J.; Nevozhay, D.; Szczaurska, K.; Świtała-Jeleń, K.; Boratyński, J.; Górski, A. Activity of Bacteriophages in Murine Tumor Models Depends on the Route of Phage Administration. Oncol. Res. 2005, 15, 183–187. [Google Scholar] [CrossRef]
- Trikha, M.; Zhou, Z.; Timar, J.; Raso, E.; Kennel, M.; Emmell, E.; Nakada, M.T. Multiple Roles for Platelet GPIIb/IIIa and alphavbeta3 Integrins in Tumor Growth, Angiogenesis, and Metastasis. Cancer Res. 2002, 62, 2824–2833. [Google Scholar] [PubMed]
- Dąbrowska, K.; Skaradziński, G.; Kurzępa, A.; Owczarek, B.; Żaczek, M.; Weber-Dąbrowska, B.; Wietrzyk, J.; Maciejewska, M.; Budynek, P.; Górski, A. The Effects of Staphylococcal Bacteriophage Lysates on Cancer Cells in Vitro. Clin. Exp. Med. 2010, 10, 81–85. [Google Scholar] [CrossRef]
- Felisbino, S.L. Natural Bacteriophages T4 and M13 Down-Regulates Hsp90 Gene Expression in Human Prostate Cancer Cells (PC-3) Representing a Potential Nanoparticle against Cancer. Virol. Res. J. 2017, 1, 21–23. [Google Scholar]
- Murgas, P.; Bustamante, N.; Araya, N.; Cruz-Gómez, S.; Durán, E.; Gaete, D.; Oyarce, C.; López, E.; Herrada, A.A.; Ferreira, N.; et al. A Filamentous Bacteriophage Targeted to Carcinoembryonic Antigen Induces Tumor Regression in Mouse Models of Colorectal Cancer. Cancer Immunol. Immunother. 2018, 67, 183–193. [Google Scholar] [CrossRef]
- Pajtasz-Piasecka, E.; Rossowska, J.; Duś, D.; Weber-Dąbrowska, B.; Zabłocka, A.; Górski, A. Bacteriophages Support Anti-Tumor Response Initiated by DC-Based Vaccine against Murine Transplantable Colon Carcinoma. Immunol. Lett. 2008, 116, 24–32. [Google Scholar] [CrossRef]
- Gambashidze, K.; Khorava, P.; Azaladze, T.; Kalandarishvili, K.; Jaiani, E.; Lasareishvil, B.; Azaladze, A.; Tediashvili, M. Antitumor and Adjuvant Effects of Phagelysates of E. coli in mice with Ehrlich Carcinoma. Exp. Oncol. 2012, 34, 107–111. [Google Scholar]
- Fluckiger, A.; Daillère, R.; Sassi, M.; Sixt, B.S.; Liu, P.; Loos, F.; Richard, C.; Rabu, C.; Alou, M.T.; Goubet, A.-G.; et al. Cross-Reactivity between Tumor MHC Class I–Restricted Antigens and an Enterococcal Bacteriophage. Science 2020, 369, 936–942. [Google Scholar] [CrossRef]
- Wagner, P.L.; Acheson, D.W.K.; Waldor, M.K. Human Neutrophils and Their Products Induce Shiga Toxin Production by Enterohemorrhagic Escherichia Coli. Infect. Immun. 2001, 69, 1934–1937. [Google Scholar] [CrossRef]
- Carrolo, M.; Frias, M.J.; Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. Prophage Spontaneous Activation Promotes DNA Release Enhancing Biofilm Formation in Streptococcus Pneumoniae. PLoS ONE 2010, 5, e15678. [Google Scholar] [CrossRef]
- Müller, M.G.; Ing, J.Y.; Cheng, M.K.-W.; Flitter, B.A.; Moe, G.R. Identification of a Phage-Encoded Ig-Binding Protein from Invasive Neisseria Meningitidis. J. Immunol. 2013, 191, 3287–3296. [Google Scholar] [CrossRef]
- François, P. Temperate Prophages Increase Bacterial Adhesin Expression and Virulence in an Experimental Model of Endocarditis Due to Staphylococcus Aureus from the CC398 Lineage. Front. Microbiol. 2019, 10, 742. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Marshall, M.L.; Fry, A.J.; Kim, U.; Wernegreen, J.J. The Tripartite Associations between Bacteriophage, Wolbachia, and Arthropods. PLoS Pathog. 2006, 2, e43. [Google Scholar] [CrossRef]
- LePage, D.P.; Metcalf, J.A.; Bordenstein, S.R.; On, J.; Perlmutter, J.I.; Shropshire, J.D.; Layton, E.M.; Funkhouser-Jones, L.J.; Beckmann, J.F.; Bordenstein, S.R. Prophage WO Genes Recapitulate and Enhance Wolbachia-Induced Cytoplasmic Incompatibility. Nature 2017, 543, 243–247. [Google Scholar] [CrossRef]
- Korczynska, J.E.; Turkenburg, J.P.; Taylor, E.J. The Structural Characterization of a Prophage-Encoded Extracellular DNase from Streptococcus Pyogenes. Nucleic Acids Res. 2012, 40, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Sela, U.; Euler, C.W.; Correa da Rosa, J.; Fischetti, V.A. Strains of Bacterial Species Induce a Greatly Varied Acute Adaptive Immune Response: The Contribution of the Accessory Genome. PLoS Pathog. 2018, 14, e1006726. [Google Scholar] [CrossRef]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage Excision Activates Listeria Competence Genes That Promote Phagosomal Escape and Virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef]
- Bodner, K.; Melkonian, A.L.; Barth, A.I.M.; Kudo, T.; Tanouchi, Y.; Covert, M.W. Engineered Fluorescent E. Coli Lysogens Allow Live-Cell Imaging of Functional Prophage Induction Triggered inside Macrophages. Cell Syst. 2020, 10, 254–264.e9. [Google Scholar] [CrossRef]
| Bacteriophage | Detected Antibody | In Vivo Model | Source of Isolation | Administration Route | Reference |
|---|---|---|---|---|---|
| GACP | IgG, IgM | mice | blood | intraperitoneally | [111] |
| φ26, φ27, φ29 | IgG, IgA | calf | sera | suppositories | [112] |
| bacteriophage specific to E. coli | undefined | rabbit | blood | subcutaneous injection | [113] |
| AbArmy ϕ1, AbNavy ϕ1, AbNavy ϕ2, AbNavy ϕ3, AbNavy ϕ4 | IgG2a, IgG2b | mice | serum | intraperitoneally | [86] |
| A3R, 676Z | IgM, IgG | mice | plasma | per os | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 2021, 22, 8937. https://doi.org/10.3390/ijms22168937
Podlacha M, Grabowski Ł, Kosznik-Kawśnicka K, Zdrojewska K, Stasiłojć M, Węgrzyn G, Węgrzyn A. Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy. International Journal of Molecular Sciences. 2021; 22(16):8937. https://doi.org/10.3390/ijms22168937
Chicago/Turabian StylePodlacha, Magdalena, Łukasz Grabowski, Katarzyna Kosznik-Kawśnicka, Karolina Zdrojewska, Małgorzata Stasiłojć, Grzegorz Węgrzyn, and Alicja Węgrzyn. 2021. "Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy" International Journal of Molecular Sciences 22, no. 16: 8937. https://doi.org/10.3390/ijms22168937
APA StylePodlacha, M., Grabowski, Ł., Kosznik-Kawśnicka, K., Zdrojewska, K., Stasiłojć, M., Węgrzyn, G., & Węgrzyn, A. (2021). Interactions of Bacteriophages with Animal and Human Organisms—Safety Issues in the Light of Phage Therapy. International Journal of Molecular Sciences, 22(16), 8937. https://doi.org/10.3390/ijms22168937











