Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits
Abstract
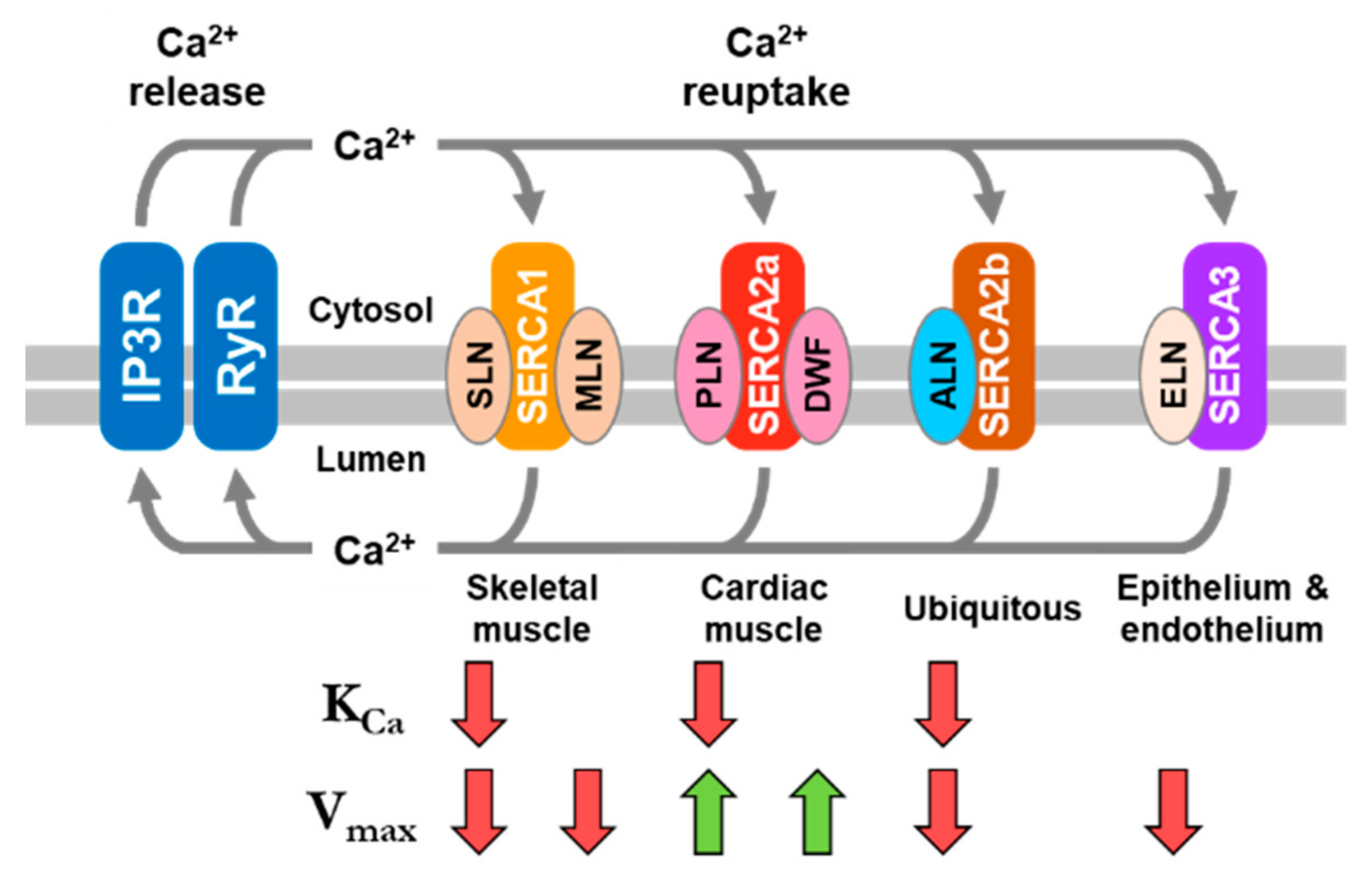
1. Background on SERCA Regulation
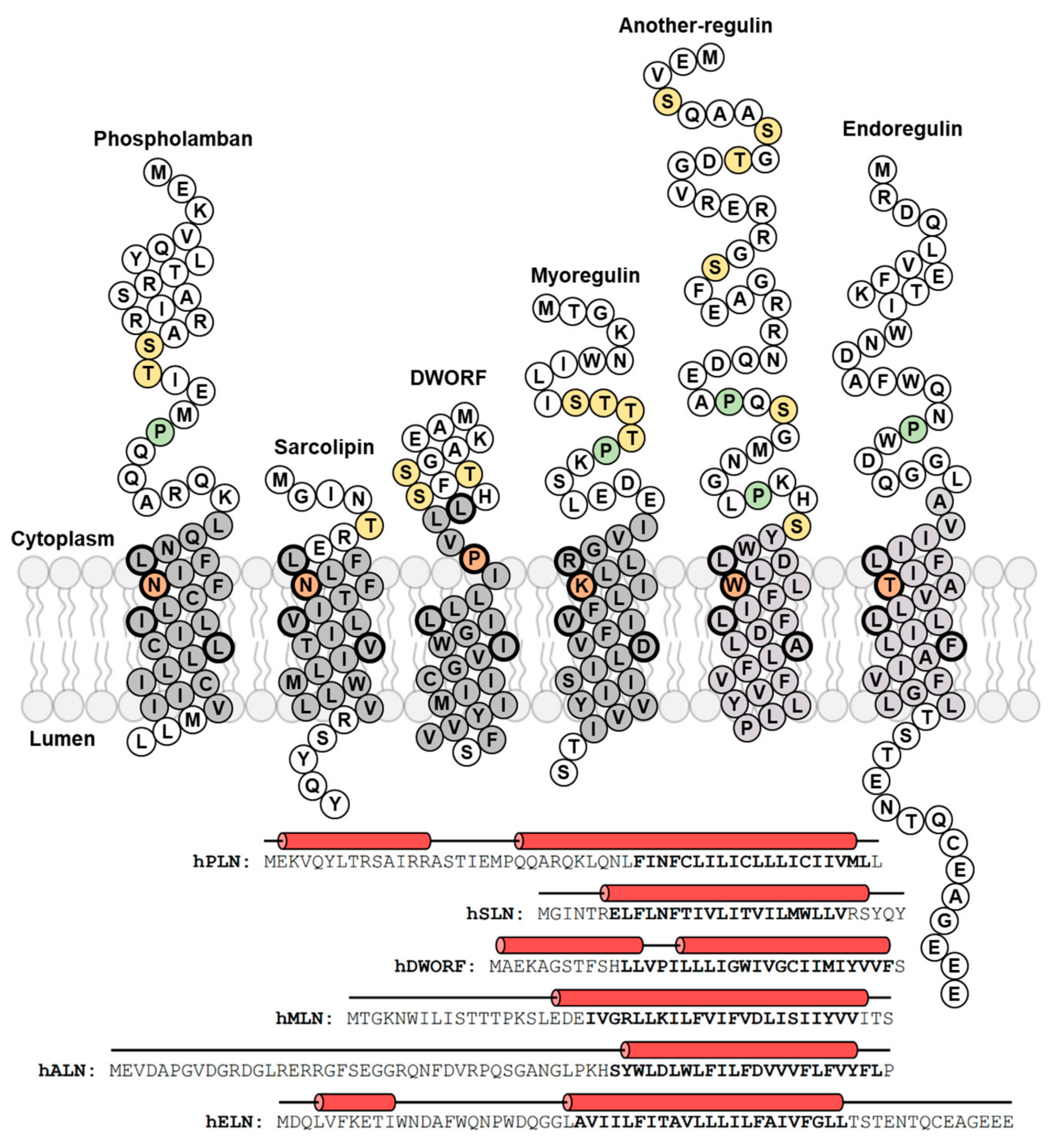
2. Phospholamban (PLN)—The Founding Member of the Regulin Family
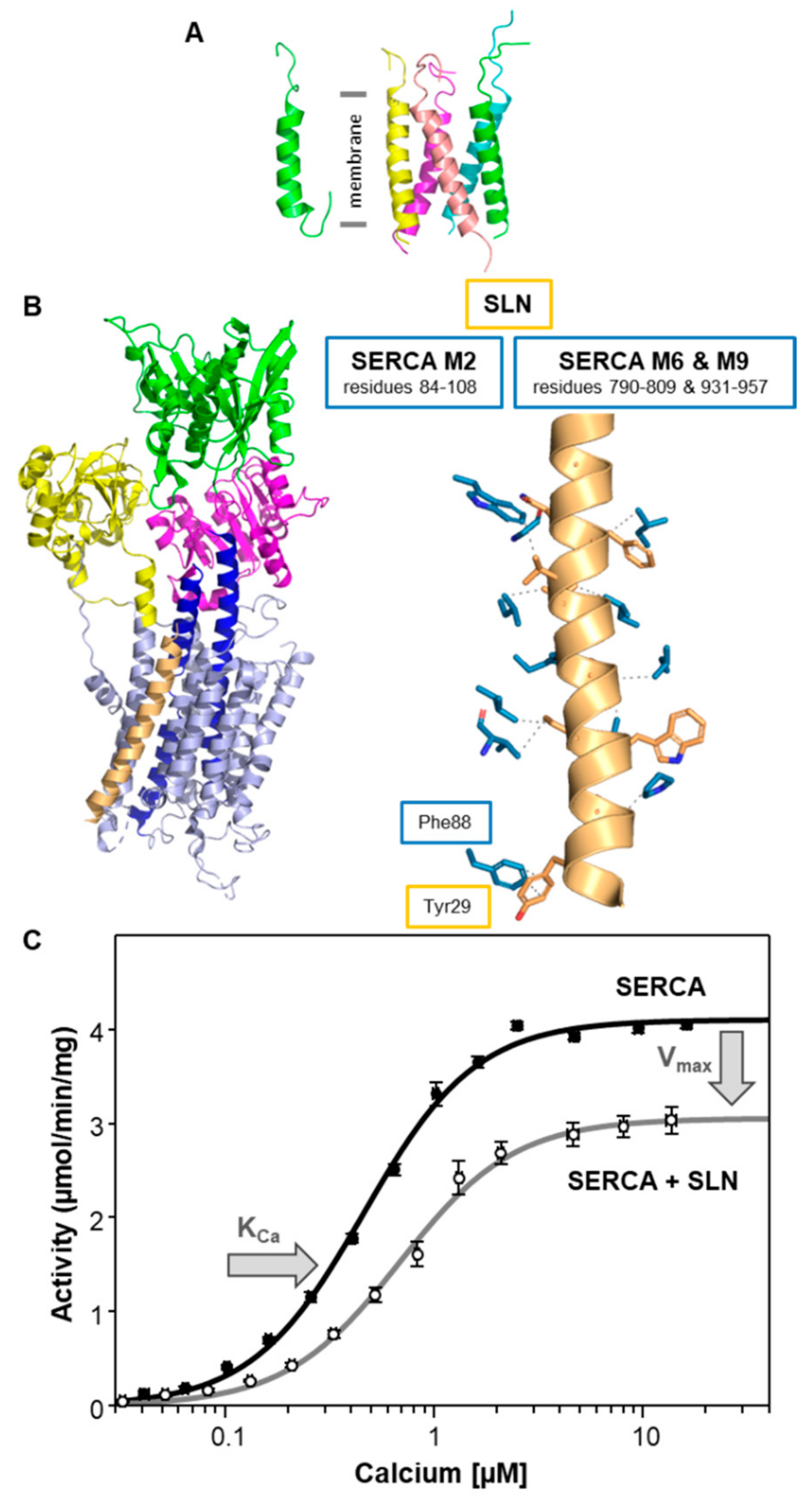
3. Sarcolipin (SLN)—A Proteolipid Becomes a Regulin
4. Dwarf Open Reading Frame (DWORF)—A Little Regulin with a Big Heart
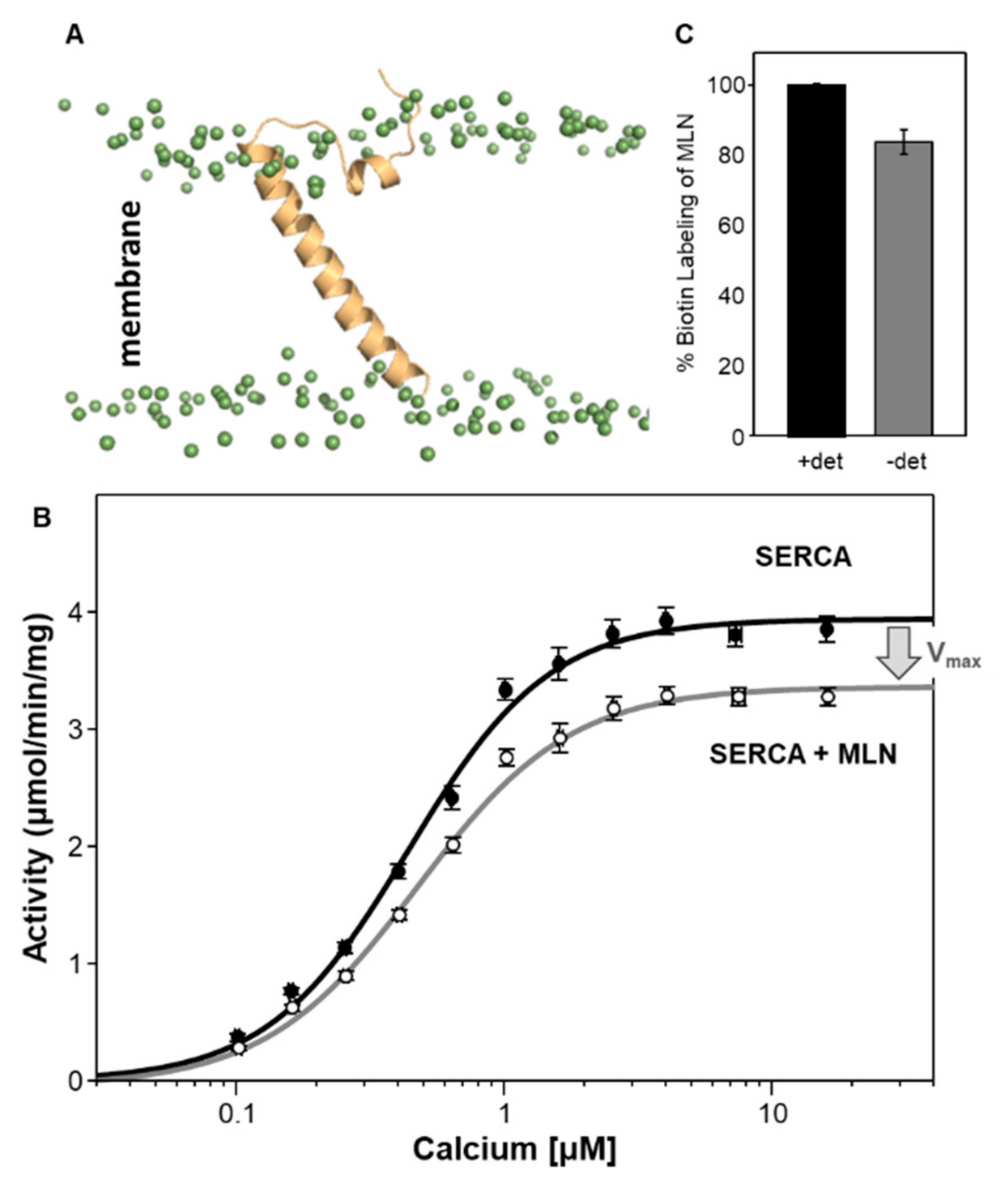
5. Myoregulin (MLN)—The Regulin for Athletes
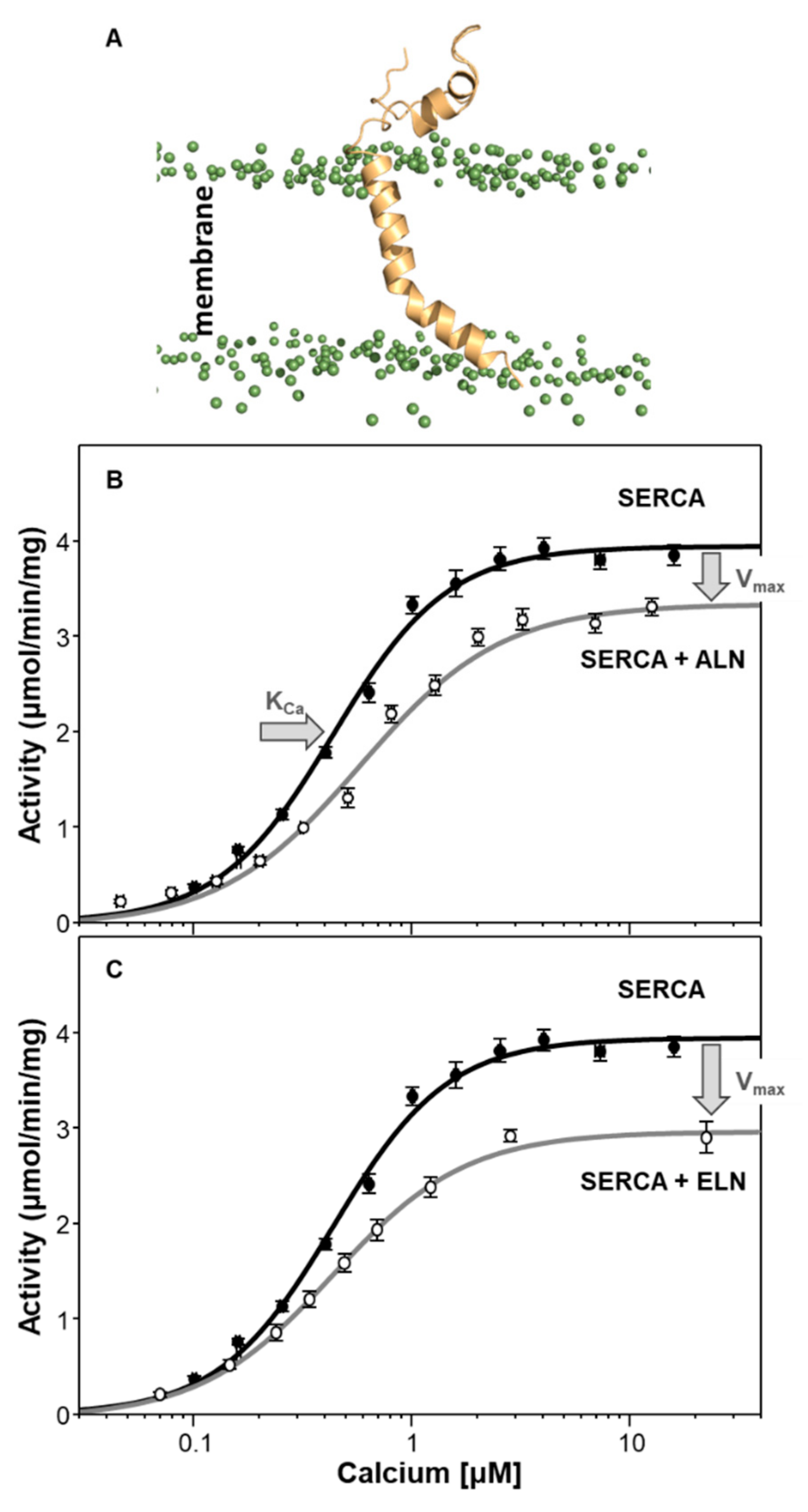
6. Endoregulin (ELN) and Another-Regulin (ALN)—Non-Muscle Cells Finally Get Their Turn
7. There Is Nothing Regular about the Regulins
8. Materials and Methods
8.1. Co-Reconstitution of Regulin Peptides with SERCA
8.2. Activity Assays
8.3. Orientation Assay
8.4. Molecular Modeling of SERCA-Regulin Complexes
8.5. Molecular Dynamics Simulations SERCA-Regulin Complexes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Carafoli, E.; Krebs, J. Why Calcium? How Calcium Became the Best Communicator. J. Biol. Chem. 2016, 291, 20849–20857. [Google Scholar] [CrossRef]
- Brini, M.; Carafoli, E.; Calì, T. The plasma membrane calcium pumps: Focus on the role in (neuro)pathology. Biochem. Biophys. Res. Commun. 2017, 483, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Lytton, J.; Westlin, M.; Burk, S.E.; Shull, G.E.; MacLennan, D.H. Functional comparisons between isoforms of the sarcoplasmic or endoplasmic reticulum family of calcium pumps. J. Biol. Chem. 1992, 267, 14483–14489. [Google Scholar] [CrossRef]
- Vandecaetsbeek, I.; Trekels, M.; De Maeyer, M.; Ceulemans, H.; Lescrinier, E.; Raeymaekers, L.; Wuytack, F.; Vangheluwe, P. Structural basis for the high Ca2+ affinity of the ubiquitous SERCA2b Ca2+ pump. Proc. Natl. Acad. Sci. USA 2009, 106, 18533–18538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Inoue, M.; Tsutsumi, A.; Watanabe, S.; Nishizawa, T.; Nagata, K.; Kikkawa, M.; Inaba, K. Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail. Sci. Adv. 2020, 6, eabb0147. [Google Scholar] [CrossRef]
- Dode, L.; Andersen, J.P.; Leslie, N.; Dhitavat, J.; Vilsen, B.; Hovnanian, A. Dissection of the Functional Differences between Sarco(endo)plasmic Reticulum Ca2+-ATPase (SERCA) 1 and 2 Isoforms and Characterization of Darier Disease (SERCA2) Mutants by Steady-state and Transient Kinetic Analyses. J. Biol. Chem. 2003, 278, 47877–47889. [Google Scholar] [CrossRef]
- MacLennan, D.H.; Kranias, E.G. Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 2003, 4, 566–577. [Google Scholar] [CrossRef]
- Odermatt, A.; Becker, S.; Khanna, V.K.; Kurzydlowski, K.; Leisner, E.; Pette, D.; MacLennan, D.H. Sarcolipin Regulates the Activity of SERCA1, the Fast-twitch Skeletal Muscle Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 1998, 273, 12360–12369. [Google Scholar] [CrossRef]
- Reddy, L.G.; Jones, L.R.; Cala, S.E.; O’Brian, J.J.; Tatulian, S.A.; Stokes, D.L. Functional Reconstitution of Recombinant Phospholamban with Rabbit Skeletal Ca2+-ATPase. J. Biol. Chem. 1995, 270, 9390–9397. [Google Scholar] [CrossRef]
- Bal, N.C.; Maurya, S.K.; Sopariwala, D.H.; Sahoo, S.K.; Gupta, S.C.; Shaikh, S.A.; Pant, M.; Rowland, L.A.; Bombardier, E.; Goonasekera, S.A.; et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat. Med. 2012, 18, 1575–1579. [Google Scholar] [CrossRef]
- Tada, M.; Kirchberger, M.A.; Katz, A.M. Regulation of calcium transport in cardiac sarcoplasmic reticulum by cyclic AMP-dependent protein kinase. Recent Adv. Stud. Card. Struct. Metab. 1976, 9, 225–239. [Google Scholar]
- Simmerman, H.K.; Collins, J.H.; Theibert, J.L.; Wegener, A.D.; Jones, L.R. Sequence analysis of phospholamban. Identification of phosphorylation sites and two major structural domains. J. Biol. Chem. 1986, 261, 13333–13341. [Google Scholar] [CrossRef]
- Catalucci, D.; Latronico, M.V.; Ceci, M.; Rusconi, F.; Young, H.S.; Gallo, P.; Santonastasi, M.; Bellacosa, A.; Brown, J.H.; Condorelli, G. Akt Increases Sarcoplasmic Reticulum Ca2+ Cycling by Direct Phosphorylation of Phospholamban at Thr17. J. Biol. Chem. 2009, 284, 28180–28187. [Google Scholar] [CrossRef]
- Bhupathy, P.; Babu, G.J.; Ito, M.; Periasamy, M. Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. J. Mol. Cell. Cardiol. 2009, 47, 723–729. [Google Scholar] [CrossRef]
- Gramolini, A.O.; Trivieri, M.G.; Oudit, G.Y.; Kislinger, T.; Li, W.; Patel, M.M.; Emili, A.; Kranias, E.G.; Backx, P.H.; MacLennan, D.H. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proc. Natl. Acad. Sci. USA 2006, 103, 2446–2451. [Google Scholar] [CrossRef] [PubMed]
- Makarewich, C.A.; Olson, E.N. Mining for Micropeptides. Trends Cell Biol. 2017, 27, 685–696. [Google Scholar] [CrossRef]
- Tharakan, R.; Sawa, A. Minireview: Novel Micropeptide Discovery by Proteomics and Deep Sequencing Methods. Front. Genet. 2021, 12, 651485. [Google Scholar] [CrossRef]
- Makarewich, C.A. The hidden world of membrane microproteins. Exp. Cell Res. 2020, 388, 111853. [Google Scholar] [CrossRef] [PubMed]
- Kirchberber, M.A.; Tada, M.; Katz, A.M. Phospholamban: A regulatory protein of the cardiac sarcoplasmic reticulum. Recent Adv. Stud. Card. Struct. Metab. 1975, 5, 103–115. [Google Scholar]
- Magny, E.G.; Pueyo, J.I.; Pearl, F.M.; Cespedes, M.A.; Niven, J.E.; Bishop, S.A.; Couso, J.P. Conserved Regulation of Cardiac Calcium Uptake by Peptides Encoded in Small Open Reading Frames. Science 2013, 341, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef]
- Anderson, D.M.; Makarewich, C.A.; Anderson, K.M.; Shelton, J.M.; Bezprozvannaya, S.; Bassel-Duby, R.; Olson, E.N. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 2016, 9, ra119. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.E.; Bovo, E.; Aguayo-Ortiz, R.; Cho, E.E.; Pribadi, M.P.; Dalton, M.P.; Rathod, N.; Lemieux, M.J.; Espinoza-Fonseca, L.M.; Robia, S.L.; et al. Dwarf open reading frame (DWORF) is a direct activator of the sarcoplasmic reticulum calcium pump SERCA. eLife 2021, 10, e65545. [Google Scholar] [CrossRef]
- Schmitt, J.P.; Kamisago, M.; Asahi, M.; Li, G.H.; Ahmad, F.; Mende, U.; Kranias, E.G.; MacLennan, D.H.; Seidman, J.G.; Seidman, C.E. Dilated Cardiomyopathy and Heart Failure Caused by a Mutation in Phospholamban. Science 2003, 299, 1410–1413. [Google Scholar] [CrossRef]
- Haghighi, K.; Kolokathis, F.; Pater, L.; Lynch, R.A.; Asahi, M.; Gramolini, A.O.; Fan, G.C.; Tsiapras, D.; Hahn, H.S.; Adamopoulos, S.; et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J. Clin. Investig. 2003, 111, 869–876. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, M.M.; MacLeod, H.M.; Soliven, B.; McNally, E.M. Phospholamban R14 Deletion Results in Late-Onset, Mild, Hereditary Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2006, 48, 1396–1398. [Google Scholar] [CrossRef]
- Haghighi, K.; Kolokathis, F.; Gramolini, A.O.; Waggoner, J.R.; Pater, L.; Lynch, R.A.; Fan, G.-C.; Tsiapras, D.; Parekh, R.R.; Dorn, G.W., 2nd; et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc. Natl. Acad. Sci. USA 2006, 103, 1388–1393. [Google Scholar] [CrossRef]
- Van Der Zwaag, P.A.; Van Rijsingen, I.A.W.; De Ruiter, R.; Nannenberg, E.A.; Groeneweg, J.A.; Post, J.G.; Hauer, R.N.W.; Van Gelder, I.C.; van den Berg, M.; Van Der Harst, P.; et al. Recurrent and founder mutations in the Netherlands—Phospholamban p.Arg14del mutation causes arrhythmogenic cardiomyopathy. Neth. Hear. J. 2013, 21, 286–293. [Google Scholar] [CrossRef]
- Young, H.S.; Ceholski, D.K.; Trieber, C.A. Deception in simplicity: Hereditary phospholamban mutations in dilated cardiomyopathy. Biochem. Cell Biol. 2015, 93, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Verardi, R.; Shi, L.; Traaseth, N.J.; Walsh, N.; Veglia, G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. USA 2011, 108, 9101–9106. [Google Scholar] [CrossRef]
- Akin, B.L.; Hurley, T.D.; Chen, Z.; Jones, L.R. The Structural Basis for Phospholamban Inhibition of the Calcium Pump in Sarcoplasmic Reticulum. J. Biol. Chem. 2013, 288, 30181–30191. [Google Scholar] [CrossRef]
- Bidwell, P.; Blackwell, D.J.; Hou, Z.; Zima, A.V.; Robia, S.L. Phospholamban Binds with Differential Affinity to Calcium Pump Conformers. J. Biol. Chem. 2011, 286, 35044–35050. [Google Scholar] [CrossRef] [PubMed]
- Glaves, J.P.; Primeau, J.O.; Espinoza-Fonseca, L.M.; Lemieux, M.J.; Young, H.S. The Phospholamban Pentamer Alters Function of the Sarcoplasmic Reticulum Calcium Pump SERCA. Biophys. J. 2019, 116, 633–647. [Google Scholar] [CrossRef]
- Alford, R.F.; Smolin, N.; Young, H.S.; Gray, J.J.; Robia, S.L. Protein docking and steered molecular dynamics suggest alternative phospholamban-binding sites on the SERCA calcium transporter. J. Biol. Chem. 2020, 295, 11262–11274. [Google Scholar] [CrossRef]
- Gorski, P.A.; Trieber, C.A.; Ashrafi, G.; Young, H.S. Regulation of the Sarcoplasmic Reticulum Calcium Pump by Divergent Phospholamban Isoforms in Zebrafish. J. Biol. Chem. 2015, 290, 6777–6788. [Google Scholar] [CrossRef]
- Verboomen, H.; Wuytack, F.; De Smedt, H.; Himpens, B.; Casteels, R. Functional difference between SERCA2a and SERCA2b Ca2+ pumps and their modulation by phospholamban. Biochem. J. 1992, 286 Pt 2, 591–595. [Google Scholar] [CrossRef]
- Reddy, L.G.; Cornea, R.L.; Winters, D.L.; McKenna, E.; Thomas, D.D. Defining the Molecular Components of Calcium Transport Regulation in a Reconstituted Membrane System. Biochemistry 2003, 42, 4585–4592. [Google Scholar] [CrossRef]
- Ceholski, D.K.; Trieber, C.; Young, H.S. Hydrophobic Imbalance in the Cytoplasmic Domain of Phospholamban Is a Determinant for Lethal Dilated Cardiomyopathy. J. Biol. Chem. 2012, 287, 16521–16529. [Google Scholar] [CrossRef] [PubMed]
- Trieber, C.A.; Afara, M.; Young, H.S. Effects of Phospholamban Transmembrane Mutants on the Calcium Affinity, Maximal Activity, and Cooperativity of the Sarcoplasmic Reticulum Calcium Pump. Biochemistry 2009, 48, 9287–9296. [Google Scholar] [CrossRef] [PubMed]
- Trieber, C.A.; Douglas, J.L.; Afara, M.; Young, H.S. The Effects of Mutation on the Regulatory Properties of Phospholamban in Co-Reconstituted Membranes. Biochemistry 2005, 44, 3289–3297. [Google Scholar] [CrossRef] [PubMed]
- Glaves, J.P.; Trieber, C.A.; Ceholski, D.K.; Stokes, D.L.; Young, H.S. Phosphorylation and Mutation of Phospholamban Alter Physical Interactions with the Sarcoplasmic Reticulum Calcium Pump. J. Mol. Biol. 2011, 405, 707–723. [Google Scholar] [CrossRef]
- Stokes, D.L.; Pomfret, A.J.; Rice, W.J.; Glaves, J.P.; Young, H.S. Interactions between Ca2+-ATPase and the Pentameric Form of Phospholamban in Two-Dimensional Co-Crystals. Biophys. J. 2006, 90, 4213–4223. [Google Scholar] [CrossRef][Green Version]
- Kimura, Y.; Kurzydlowski, K.; Tada, M.; MacLennan, D.H. Phospholamban Inhibitory Function Is Activated by Depolymerization. J. Biol. Chem. 1997, 272, 15061–15064. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, C.; Asahi, M.; Sugita, Y.; Khanna, R.; Tsuda, T.; MacLennan, D.H. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl. Acad. Sci. USA 2003, 100, 467–472. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, D.H.; Yip, C.C.; Iles, G.H.; Seeman, P. Isolation of Sarcoplasmic Reticulum Proteins. Cold Spring Harb. Symp. Quant. Biol. 1972, 37, 469–477. [Google Scholar] [CrossRef]
- Wawrzynow, A.; Theibert, J.L.; Murphy, C.; Jona, I.; Martonosi, A.; Collins, J.H. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch. Biochem. Biophys. 1992, 298, 620–623. [Google Scholar] [CrossRef]
- Odermatt, A.; Taschner, P.E.; Scherer, S.W.; Beatty, B.; Khanna, V.K.; Cornblath, D.R.; Chaudhry, V.; Yee, W.-C.; Schrank, B.; Karpati, G.; et al. Characterization of the Gene Encoding Human Sarcolipin (SLN), a Proteolipid Associated with SERCA1: Absence of Structural Mutations in Five Patients with Brody Disease. Genomics 1997, 45, 541–553. [Google Scholar] [CrossRef]
- Xie, L.-H.; Shanmugam, M.; Park, J.Y.; Zhao, Z.; Wen, H.; Tian, B.; Periasamy, M.; Babu, G.J. Ablation of sarcolipin results in atrial remodeling. Am. J. Physiol. Physiol. 2012, 302, C1762–C1771. [Google Scholar] [CrossRef]
- Shanmugam, M.; Molina, C.E.; Gao, S.; Severac-Bastide, R.; Fischmeister, R.; Babu, G.J. Decreased sarcolipin protein expression and enhanced sarco(endo)plasmic reticulum Ca2+ uptake in human atrial fibrillation. Biochem. Biophys. Res. Commun. 2011, 410, 97–101. [Google Scholar] [CrossRef]
- Nyberg, M.T.; Stoevring, B.; Behr, E.R.; Ravn, L.S.; McKenna, W.J.; Christiansen, M. The variation of the sarcolipin gene (SLN) in atrial fibrillation, long QT syndrome and sudden arrhythmic death syndrome. Clin. Chim. Acta 2007, 375, 87–91. [Google Scholar] [CrossRef]
- Gorski, P.A.; Glaves, J.P.; Vangheluwe, P.; Young, H.S. Sarco(endo)plasmic Reticulum Calcium ATPase (SERCA) Inhibition by Sarcolipin Is Encoded in Its Luminal Tail. J. Biol. Chem. 2013, 288, 8456–8467. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, C.; Iwasawa, S.; Ogawa, H.; Hirata, A.; Tsueda, J.; Inesi, G. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 2013, 495, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Winther, A.-M.L.; Bublitz, M.; Karlsen, J.L.; Møller, J.V.; Hansen, J.B.; Nissen, P.; Buch-Pedersen, M.J. The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nat. Cell Biol. 2013, 495, 265–269. [Google Scholar] [CrossRef]
- Sahoo, S.K.; Shaikh, S.A.; Sopariwala, D.H.; Bal, N.C.; Periasamy, M. Sarcolipin Protein Interaction with Sarco(endo)plasmic Reticulum Ca2+ATPase (SERCA) Is Distinct from Phospholamban Protein, and Only Sarcolipin Can Promote Uncoupling of the SERCA Pump. J. Biol. Chem. 2013, 288, 6881–6889. [Google Scholar] [CrossRef]
- Autry, J.M.; Rubin, J.E.; Pietrini, S.D.; Winters, D.L.; Robia, S.L.; Thomas, D.D. Oligomeric Interactions of Sarcolipin and the Ca-ATPase. J. Biol. Chem. 2011, 286, 31697–31706. [Google Scholar] [CrossRef] [PubMed]
- Glaves, J.P.; Primeau, J.O.; Gorski, P.A.; Espinoza-Fonseca, L.M.; Lemieux, M.J.; Young, H.S. Interaction of a Sarcolipin Pentamer and Monomer with the Sarcoplasmic Reticulum Calcium Pump, SERCA. Biophys. J. 2020, 118, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, V.A.; Bombardier, E.; Vigna, C.; Devji, T.; Bloemberg, D.; Gamu, D.; Gramolini, A.O.; Quadrilatero, J.; Tupling, A.R. Co-Expression of SERCA Isoforms, Phospholamban and Sarcolipin in Human Skeletal Muscle Fibers. PLoS ONE 2013, 8, e84304. [Google Scholar] [CrossRef]
- Vangheluwe, P.; Schuermans, M.; Zádor, E.; Waelkens, E.; Raeymaekers, L.; Wuytack, F. Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem. J. 2005, 389, 151–159. [Google Scholar] [CrossRef]
- Zheng, J.; Yancey, D.M.; Ahmed, M.I.; Wei, C.-C.; Powell, P.C.; Shanmugam, M.; Gupta, H.; Lloyd, S.G.; McGiffin, D.C.; Schiros, C.G.; et al. Increased Sarcolipin Expression and Adrenergic Drive in Humans With Preserved Left Ventricular Ejection Fraction and Chronic Isolated Mitral Regurgitation. Circ. Hear. Fail. 2014, 7, 194–202. [Google Scholar] [CrossRef]
- Morales Rodriguez, B.; Domínguez-Rodríguez, A.; Benitah, J.-P.; Lefebvre, F.; Marais, T.; Mougenot, N.; Beauverger, P.; Bonne, G.; Briand, V.; Gómez, A.-M.; et al. Activation of sarcolipin expression and altered calcium cycling in LMNA cardiomyopathy. Biochem. Biophys. Rep. 2020, 22, 100767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Qiu, C.; Zhu, H.; Pan, S.; Jia, H.; Kang, H.; Guan, G.; Hui, R.; Zhu, L.; et al. Diabetes mellitus exacerbates post-myocardial infarction heart failure by reducing sarcolipin promoter methylation. ESC Hear. Fail. 2020, 7, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.S.; Shanmugam, M.; Gonzalez, J.P.; Lopez, H.; Gordan, R.; Fraidenraich, D.; Babu, G.J. Increased sarcolipin expression and decreased sarco(endo)plasmic reticulum Ca2+ uptake in skeletal muscles of mouse models of Duchenne muscular dystrophy. J. Muscle Res. Cell Motil. 2013, 34, 349–356. [Google Scholar] [CrossRef]
- Smith, W.S.; Broadbridge, R.; East, J.M.; Lee, A.G. Sarcolipin uncouples hydrolysis of ATP from accumulation of Ca2+ by the Ca2+-ATPase of skeletal-muscle sarcoplasmic reticulum. Biochem. J. 2002, 361, 277–286. [Google Scholar] [CrossRef]
- Mall, S.; Broadbridge, R.; Harrison, S.L.; Gore, M.G.; Lee, A.G.; East, J.M. The Presence of Sarcolipin Results in Increased Heat Production by Ca2+-ATPase. J. Biol. Chem. 2006, 281, 36597–36602. [Google Scholar] [CrossRef]
- Autry, J.M.; Thomas, D.D.; Espinoza-Fonseca, L.M. Sarcolipin Promotes Uncoupling of the SERCA Ca2+ Pump by Inducing a Structural Rearrangement in the Energy-Transduction Domain. Biochemistry 2016, 55, 6083–6086. [Google Scholar] [CrossRef]
- Smeazzetto, S.; Armanious, G.P.; Moncelli, M.R.; Bak, J.J.; Lemieux, M.J.; Young, H.S.; Tadini-Buoninsegni, F. Conformational memory in the association of the transmembrane protein phospholamban with the sarcoplasmic reticulum calcium pump SERCA. J. Biol. Chem. 2017, 292, 21330–21339. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Munir, A.Z.; Schiattarella, G.G.; Bezprozvannaya, S.; Raguimova, O.N.; Cho, E.E.; Vidal, A.H.; Robia, S.L.; Bassel-Duby, R.; Olson, E.N. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 2018, 7, e38319. [Google Scholar] [CrossRef] [PubMed]
- Makarewich, C.A.; Bezprozvannaya, S.; Gibson, A.M.; Bassel-Duby, R.; Olson, E.N. Gene Therapy with the DWORF Micropeptide Attenuates Cardiomyopathy in Mice. Circ. Res. 2020, 127, 1340–1342. [Google Scholar] [CrossRef]
- Gopinath, T.; Weber, D.; Wang, S.; Larsen, E.; Veglia, G. Solid-State NMR of Membrane Proteins in Lipid Bilayers: To Spin or Not To Spin? Accounts Chem. Res. 2021, 54, 1430–1439. [Google Scholar] [CrossRef]
- Singh, D.R.; Dalton, M.P.; Cho, E.E.; Pribadi, M.P.; Zak, T.J.; Šeflová, J.; Makarewich, C.A.; Olson, E.N.; Robia, S.L. Newly Discovered Micropeptide Regulators of SERCA Form Oligomers but Bind to the Pump as Monomers. J. Mol. Biol. 2019, 431, 4429–4443. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Yao, Q.; Squier, T.C.; Bigelow, D.J. Comparable Levels of Ca-ATPase Inhibition by Phospholamban in Slow-Twitch Skeletal and Cardiac Sarcoplasmic Reticulum. Biochemistry 2002, 41, 13289–13296. [Google Scholar] [CrossRef]
- Negash, S.; Chen, L.T.; Bigelow, D.J.; Squier, T.C. Phosphorylation of Phospholamban by cAMP-Dependent Protein Kinase Enhances Interactions between Ca-ATPase Polypeptide Chains in Cardiac Sarcoplasmic Reticulum Membranes. Biochemistry 1996, 35, 11247–11259. [Google Scholar] [CrossRef]
- Tupling, A.R.; Bombardier, E.; Gupta, S.C.; Hussain, D.; Vigna, C.; Bloemberg, D.; Quadrilatero, J.; Trivieri, M.G.; Babu, G.J.; Backx, P.H.; et al. Enhanced Ca2+ transport and muscle relaxation in skeletal muscle from sarcolipin-null mice. Am. J. Physiol. Cell Physiol. 2011, 301, C841–C849. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.; Middleton, D.A. Comparison of the Structure and Function of Phospholamban and the Arginine-14 Deficient Mutant Associated with Dilated Cardiomyopathy. PLoS ONE 2014, 9, e106746. [Google Scholar] [CrossRef] [PubMed]
- Primeau, J.O.; Armanious, G.P.; Fisher, M.E.; Young, H.S. The SarcoEndoplasmic Reticulum Calcium ATPase. Membr. Protein Complexes Struct. Funct. 2018, 87, 229–258. [Google Scholar] [CrossRef]
- Toyoshima, C.; Nakasako, M.; Nomura, H.; Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 2000, 405, 647–655. [Google Scholar] [CrossRef]
- Douglas, J.L.; Trieber, C.A.; Afara, M.; Young, H.S. Rapid, high-yield expression and purification of Ca2+-ATPase regulatory proteins for high-resolution structural studies. Protein Expr. Purif. 2005, 40, 118–125. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger, L. The PyMol Molecular Graphics System, Version 1.7; Schrodinger, LLC: New York, NY, USA, 2015. [Google Scholar]



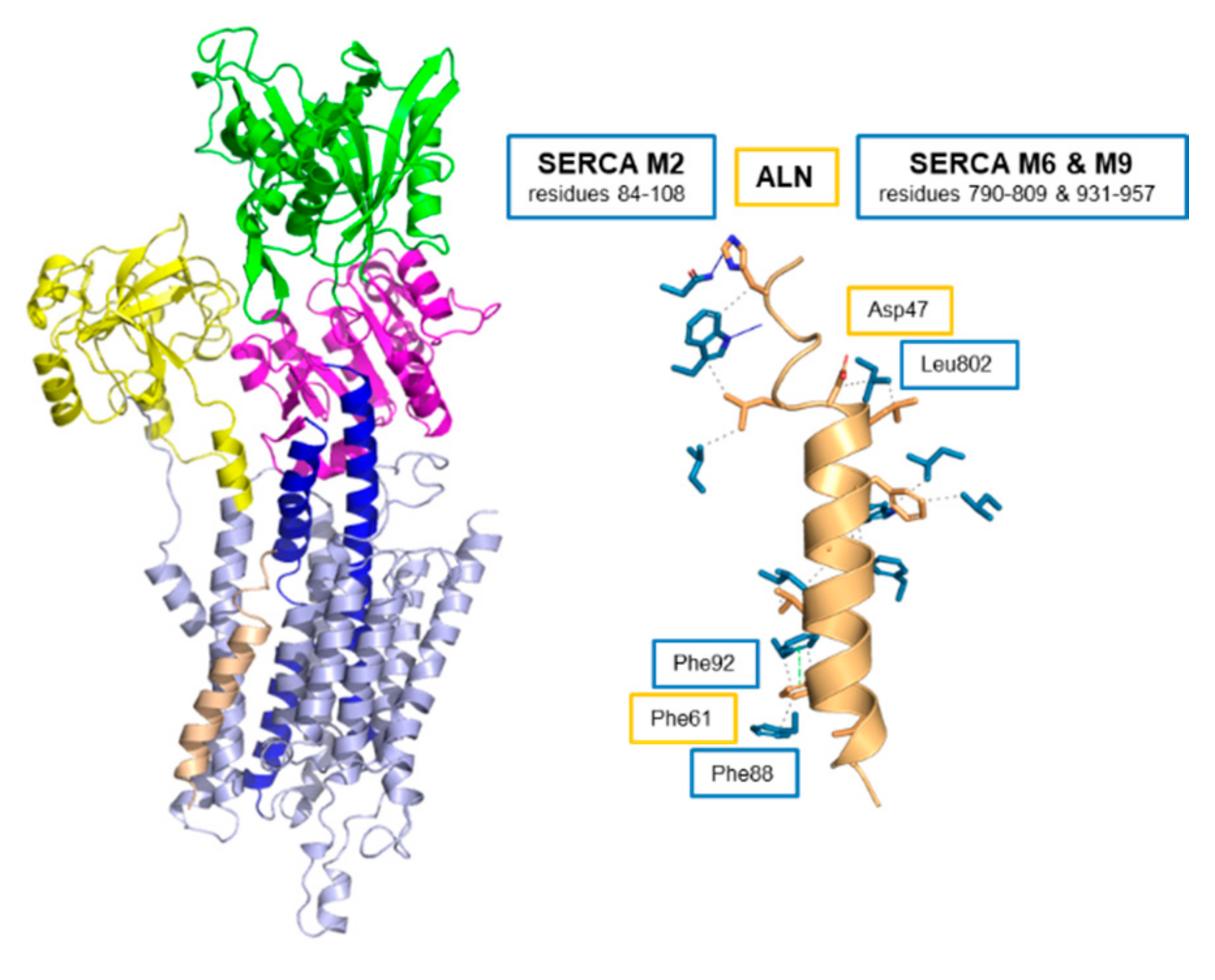
| Regulin | KCa (µM Calcium) | Vmax (µmol/min/mg) |
|---|---|---|
| SERCA alone | 0.44 ± 0.02 | 4.0 ± 0.1 |
| PLN | 0.89 ± 0.03 a | 6.1 ± 0.2 a |
| SLN | 0.74 ± 0.03 a | 3.1 ± 0.1 a |
| DWORF | 0.48 ± 0.03 | 6.9 ± 0.1 a |
| MLN | 0.47 ± 0.02 | 3.4 ± 0.1 a |
| ALN | 0.58 ± 0.05 b | 3.4 ± 0.1 a |
| ELN | 0.44 ± 0.02 | 3.1 ± 0.1 a |
| Regulin | Number of SERCA Contacts | ||
|---|---|---|---|
| M2 | M6 | M9 | |
| PLN | 1 | 5 | 7 |
| SLN | 4 | 6 | 6 |
| DWORF | 9 | 1 | 7 |
| MLN | 6 | 1 | 5 |
| ALN | 10 | 3 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathod, N.; Bak, J.J.; Primeau, J.O.; Fisher, M.E.; Espinoza-Fonseca, L.M.; Lemieux, M.J.; Young, H.S. Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits. Int. J. Mol. Sci. 2021, 22, 8891. https://doi.org/10.3390/ijms22168891
Rathod N, Bak JJ, Primeau JO, Fisher ME, Espinoza-Fonseca LM, Lemieux MJ, Young HS. Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits. International Journal of Molecular Sciences. 2021; 22(16):8891. https://doi.org/10.3390/ijms22168891
Chicago/Turabian StyleRathod, Nishadh, Jessi J. Bak, Joseph O. Primeau, M’Lynn E. Fisher, Lennane Michel Espinoza-Fonseca, Mary Joanne Lemieux, and Howard S. Young. 2021. "Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits" International Journal of Molecular Sciences 22, no. 16: 8891. https://doi.org/10.3390/ijms22168891
APA StyleRathod, N., Bak, J. J., Primeau, J. O., Fisher, M. E., Espinoza-Fonseca, L. M., Lemieux, M. J., & Young, H. S. (2021). Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits. International Journal of Molecular Sciences, 22(16), 8891. https://doi.org/10.3390/ijms22168891






