The Gut-Brain Axis in Inflammatory Bowel Disease—Current and Future Perspectives
Abstract
1. Introduction
2. Genetic Evidence for an Association between Gut and Brain in the Context of Inflammation
3. Evidence from Gut–Brain Communication in Preclinical Mouse Models
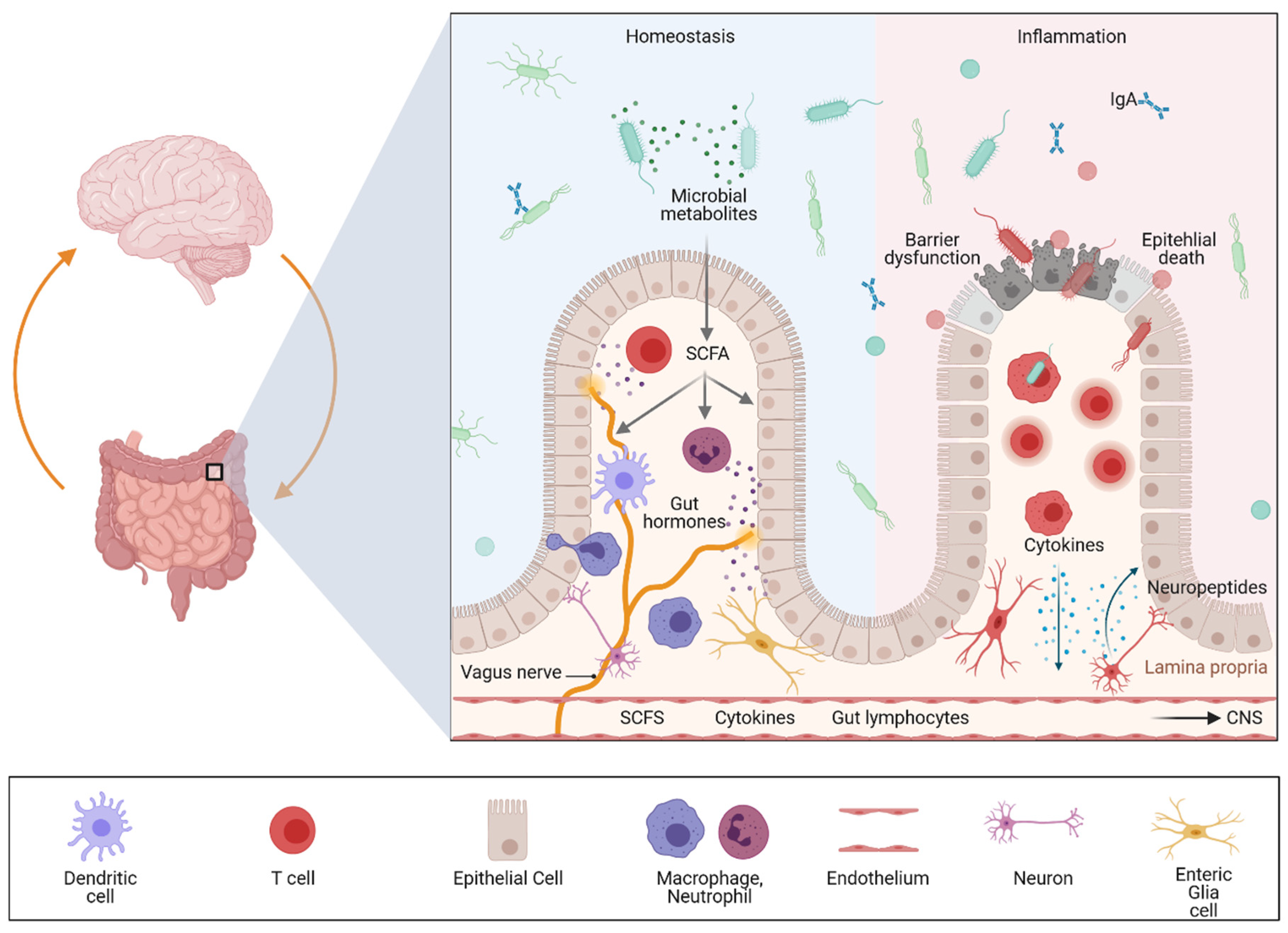
4. The Different Levels of Gut–Brain Communication
4.1. Neuronal Communication
4.2. Microbial Communication
4.3. Immunological Cross Talk
5. Ex-Vivo Organ Models
5.1. Brain Organoids
5.2. Enteric Nervous System (ENS)
5.3. Intestinal Organoids
6. Modeling the Gut–Brain Axis or Barriers in Human Cell-Based Systems
Author Contributions
Funding
Conflicts of Interest
References
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef]
- Rogler, G.; Andus, T. Cytokines in inflammatory bowel disease. World J. Surg. 1998, 22, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Giraldez, M.D.; Carneros, D.; Garbers, C.; Rose-John, S.; Bustos, M. New insights into IL-6 family cytokines in metabolism, hepatology and gastroenterology. Nat. Rev. Gastroenterol. Hepatol. 2021, 5, 1–17. [Google Scholar] [CrossRef]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.B.; Xavier, R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020, 578, 527–539. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.P.; Kugathasan, S.; Cho, J.H. Genetics of Inflammatory Bowel Diseases. Gastroenterology 2015, 149, 1163–1176.e2. [Google Scholar] [CrossRef] [PubMed]
- Gunther, C.; Martini, E.; Wittkopf, N.; Amann, K.; Weigmann, B.; Neumann, H.; Waldner, J.M.; Hedrick, S.M.; Tenzer, S.; Neurath, M.F.; et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature 2011, 477, 335–339. [Google Scholar] [CrossRef]
- Nenci, A.; Becker, C.; Wullaert, A.; Gareus, R.; van Loo, G.; Danese, S.; Huth, M.; Nikolaev, A.; Neufert, C.; Madison, B.; et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 2007, 446, 557–561. [Google Scholar] [CrossRef]
- Pastorelli, L.; De Salvo, C.; Mercado, J.R.; Vecchi, M.; Pizarro, T.T. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: Lessons learned from animal models and human genetics. Front. Immunol. 2013, 4, 280. [Google Scholar] [CrossRef]
- Miyoshi, J.; Sofia, M.A.; Pierre, J.F. The evidence for fungus in Crohn’s disease pathogenesis. Clin. J. Gastroenterol. 2018, 11, 449–456. [Google Scholar] [CrossRef]
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef]
- Neurath, M.F. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 76–77. [Google Scholar] [CrossRef]
- Actis, G.C.; Ribaldone, D.G.; Pellicano, R. The Human Gut: Inflammatory Remote Manifestations Regulated by the Microbiome. J. Gastric Disord. Ther. 2019, 4, 24–27. [Google Scholar]
- Jarret, A.; Jackson, R.; Duizer, C.; Healy, M.E.; Zhao, J.; Rone, J.M.; Bielecki, P.; Sefik, E.; Roulis, M.; Rice, T.; et al. Enteric Nervous System-Derived IL-18 Orchestrates Mucosal Barrier Immunity. Cell 2020, 180, 50–63.e12. [Google Scholar] [CrossRef]
- Engel, M.A.; Leffler, A.; Niedermirtl, F.; Babes, A.; Zimmermann, K.; Filipovic, M.R.; Izydorczyk, I.; Eberhardt, M.; Kichko, T.I.; Mueller-Tribbensee, S.M.; et al. TRPA1 and substance P mediate colitis in mice. Gastroenterology 2011, 141, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Drokhlyansky, E.; Smillie, C.S.; Van Wittenberghe, N.; Ericsson, M.; Griffin, G.K.; Eraslan, G.; Dionne, D.; Cuoco, M.S.; Goder-Reiser, M.N.; Sharova, T.; et al. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 2020, 182, 1606–1622.e23. [Google Scholar] [CrossRef]
- Hess, A.; Roesch, J.; Saake, M.; Sergeeva, M.; Hirschmann, S.; Neumann, H.; Dorfler, A.; Neurath, M.F.; Atreya, R. Functional Brain Imaging Reveals Rapid Blockade of Abdominal Pain Response Upon Anti-TNF Therapy in Crohn’s Disease. Gastroenterology 2015, 149, 864–866. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Warren, S.; Sommers, S.C. Pathogenesis of ulcerative colitis. Am. J. Pathol. 1949, 25, 657–679. [Google Scholar] [PubMed]
- Truelove, S.C.; Witts, L.J. Cortisone in ulcerative colitis; final report on a therapeutic trial. Br. Med. J. 1955, 2, 1041–1048. [Google Scholar] [CrossRef]
- Truelove, S.C.; Witts, L.J. Cortisone in ulcerative colitis; preliminary report on a therapeutic trial. Br. Med. J. 1954, 2, 375–378. [Google Scholar] [CrossRef]
- Ng, Q.X.; Soh, A.Y.S.; Loke, W.; Lim, D.Y.; Yeo, W.S. The role of inflammation in irritable bowel syndrome (IBS). J. Inflamm. Res. 2018, 11, 345–349. [Google Scholar] [CrossRef]
- Stabler, S.P. Clinical practice. Vitamin B12 deficiency. N. Engl. J. Med. 2013, 368, 149–160. [Google Scholar] [CrossRef]
- Cosma-Grigorov, A.; Meixner, H.; Mrochen, A.; Wirtz, S.; Winkler, J.; Marxreiter, F. Changes in Gastrointestinal Microbiome Composition in PD: A Pivotal Role of Covariates. Front. Neurol. 2020, 11, 1041. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Borucki, D.M.; Tjon, E.C.; Takenaka, M.C.; Chao, C.C.; Ardura-Fabregat, A.; de Lima, K.A.; Gutierrez-Vazquez, C.; Hewson, P.; Staszewski, O.; et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018, 557, 724–728. [Google Scholar] [CrossRef]
- Lin, J.C.; Lin, C.S.; Hsu, C.W.; Lin, C.L.; Kao, C.H. Association Between Parkinson’s Disease and Inflammatory Bowel Disease: A Nationwide Taiwanese Retrospective Cohort Study. Inflamm. Bowel Dis. 2016, 22, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Peter, I.; Dubinsky, M.; Bressman, S.; Park, A.; Lu, C.; Chen, N.; Wang, A. Anti-Tumor Necrosis Factor Therapy and Incidence of Parkinson Disease Among Patients With Inflammatory Bowel Disease. JAMA Neurol. 2018, 75, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Weimers, P.; Halfvarson, J.; Sachs, M.C.; Saunders-Pullman, R.; Ludvigsson, J.F.; Peter, I.; Burisch, J.; Olen, O. Inflammatory Bowel Disease and Parkinson’s Disease: A Nationwide Swedish Cohort Study. Inflamm. Bowel Dis. 2019, 25, 111–123. [Google Scholar] [CrossRef]
- Villumsen, M.; Aznar, S.; Pakkenberg, B.; Jess, T.; Brudek, T. Inflammatory bowel disease increases the risk of Parkinson’s disease: A Danish nationwide cohort study 1977-2014. Gut 2019, 68, 18–24. [Google Scholar] [CrossRef]
- Zhu, F.; Li, C.; Gong, J.; Zhu, W.; Gu, L.; Li, N. The risk of Parkinson’s disease in inflammatory bowel disease: A systematic review and meta-analysis. Dig. Liver Dis. 2019, 51, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Krainc, D. alpha-synuclein toxicity in neurodegeneration: Mechanism and therapeutic strategies. Nat. Med. 2017, 23, 1–13. [Google Scholar] [CrossRef]
- Olanow, C.W.; Tatton, W.G. Etiology and pathogenesis of Parkinson’s disease. Annu. Rev. Neurosci. 1999, 22, 123–144. [Google Scholar] [CrossRef]
- Kosmidou, M.; Katsanos, A.H.; Katsanos, K.H.; Kyritsis, A.P.; Tsivgoulis, G.; Christodoulou, D.; Giannopoulos, S. Multiple sclerosis and inflammatory bowel diseases: A systematic review and meta-analysis. J. Neurol. 2017, 264, 254–259. [Google Scholar] [CrossRef]
- Avasarala, J.; Guduru, Z.; McLouth, C.J.; Wilburn, A.; Talbert, J.; Sutton, P.; Sokola, B.S. Use of anti-TNF-alpha therapy in Crohn’s disease is associated with increased incidence of multiple sclerosis. Mult. Scler. Relat. Disord. 2021, 51, 102942. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Gelfand, J.M.; Lewis, J.D. Increased risk for demyelinating diseases in patients with inflammatory bowel disease. Gastroenterology 2005, 129, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Alkhawajah, M.M.; Caminero, A.B.; Freeman, H.J.; Oger, J.J. Multiple sclerosis and inflammatory bowel diseases: What we know and what we would need to know! Mult. Scler. 2013, 19, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kumar, N.; Loftus, E.V.; Jr Kane, S.V. Neurologic complications in patients with inflammatory bowel disease: Increasing relevance in the era of biologics. Inflamm. Bowel Dis. 2013, 19, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Pokorny, C.S.; Beran, R.G.; Pokorny, M.J. Association between ulcerative colitis and multiple sclerosis. Intern. Med. J. 2007, 37, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Jovani, M.; Danese, S. Vedolizumab for the treatment of IBD: A selective therapeutic approach targeting pathogenic a4b7 cells. Curr. Drug Targets 2013, 14, 1433–1443. [Google Scholar] [CrossRef]
- Lamb, C.A.; O’Byrne, S.; Keir, M.E.; Butcher, E.C. Gut-Selective Integrin-Targeted Therapies for Inflammatory Bowel Disease. J. Crohns. Colitis. 2018, 12 (Suppl. 2), S653–S668. [Google Scholar] [CrossRef]
- Perez-Jeldres, T.; Alvarez-Lobos, M.; Rivera-Nieves, J. Targeting Sphingosine-1-Phosphate Signaling in Immune-Mediated Diseases: Beyond Multiple Sclerosis. Drugs 2021, 81, 985–1002. [Google Scholar] [CrossRef]
- Argollo, M.; Furfaro, F.; Gilardi, D.; Roda, G.; Allocca, M.; Peyrin-Biroulet, L.; Danese, S. Modulation of sphingosine-1-phosphate in ulcerative colitis. Expert Opin. Biol. Ther. 2020, 20, 413–420. [Google Scholar] [CrossRef]
- Karuppuchamy, T.; Tyler, C.J.; Lundborg, L.R.; Perez-Jeldres, T.; Kimball, A.K.; Clambey, E.T.; Jedlicka, P.; Rivera-Nieves, J. Sphingosine-1-Phosphate Lyase Inhibition Alters the S1P Gradient and Ameliorates Crohn’s-Like Ileitis by Suppressing Thymocyte Maturation. Inflamm. Bowel Dis. 2020, 26, 216–228. [Google Scholar] [CrossRef]
- Katsanos, A.H.; Katsanos, K.H. Inflammatory bowel disease and demyelination: More than just a coincidence? Expert Rev. Clin. Immunol. 2014, 10, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Natale, G.; Limanaqi, F.; Busceti, C.L.; Mastroiacovo, F.; Nicoletti, F.; Puglisi-Allegra, S.; Fornai, F. Glymphatic System as a Gateway to Connect Neurodegeneration From Periphery to CNS. Front. Neurosci. 2021, 15, 639140. [Google Scholar] [CrossRef] [PubMed]
- Natale, G.; Biagioni, F.; Busceti, C.L.; Gambardella, S.; Limanaqi, F.; Fornai, F. TREM Receptors Connecting Bowel Inflammation to Neurodegenerative Disorders. Cells 2019, 8, 1124. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E.; Vila, M.; Klein, C.; Rascol, O. LRRK2 in Parkinson disease: Challenges of clinical trials. Nat. Rev. Neurol. 2020, 16, 97–107. [Google Scholar] [CrossRef]
- Umeno, J.; Asano, K.; Matsushita, T.; Matsumoto, T.; Kiyohara, Y.; Iida, M.; Nakamura, Y.; Kamatani, N.; Kubo, M. Meta-analysis of published studies identified eight additional common susceptibility loci for Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 2011, 17, 2407–2415. [Google Scholar] [CrossRef]
- Michail, S.; Bultron, G.; Depaolo, R.W. Genetic variants associated with Crohn’s disease. Appl. Clin. Genet. 2013, 6, 25–32. [Google Scholar] [CrossRef]
- Witoelar, A.; Jansen, I.E.; Wang, Y.; Desikan, R.S.; Gibbs, J.R.; Blauwendraat, C.; Thompson, W.K.; Hernandez, D.G.; Djurovic, S.; Schork, A.J.; et al. Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases. JAMA Neurol. 2017, 74, 780–792. [Google Scholar] [CrossRef]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Franke, A.; McGovern, D.P.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; Roberts, R.; et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010, 42, 1118–1125. [Google Scholar] [CrossRef]
- Yang, E.; Shen, J. The roles and functions of Paneth cells in Crohn’s disease: A critical review. Cell Prolif. 2021, 54, e12958. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, Y.; Yan, R.; Zeng, B.; Wang, H.; Zhang, X.; Li, W.; Wei, H.; Liu, Z. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat. Immunol. 2015, 16, 918–926. [Google Scholar] [CrossRef]
- Liu, T.C.; Naito, T.; Liu, Z.; VanDussen, K.L.; Haritunians, T.; Li, D.; Endo, K.; Kawai, Y.; Nagasaki, M.; Kinouchi, Y.; et al. LRRK2 but not ATG16L1 is associated with Paneth cell defect in Japanese Crohn’s disease patients. JCI Insight 2017, 2, e91917. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Patel, K.K.; Komatsu, M.; Virgin HWt Stappenbeck, T.S. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 2009, 5, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Patel, K.K.; Maloney, N.S.; Liu, T.; Ng, A.C.Y.; Storer, C.E.; Hear, R.D.; Xavier, R.; Stappenbeck, T.S.; Virgin, H.W. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010, 141, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Loh, J.; Lenners, J.K.; Kiski, C.; Kc, W.; Carrero, J.A.; Hunt, S.; Stone, C.D.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Adolph, T.E.; Tomczak, M.F.; Niederreiter, L.; Ko, H.; Böck, J.; Martinez-Naves, E.; Glickman, J.N.; Tschurtschenthaler, M.; Hartwig, J.; Hosomi, S.; et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013, 503, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef]
- Hampe, J.; Cuthbert, A.; Croucher, P.J.; Mirza, M.M.; Mascheretti, S.; Fisher, S.; Frenzel, H.; King, K.; Hasselmeyer, A.; MacPherson, A.J.; et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 2001, 357, 1925–1928. [Google Scholar] [CrossRef]
- Hernandez, D.G.; Reed, X.; Singleton, A.B. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016, 139 (Suppl. 1), 59–74. [Google Scholar] [CrossRef]
- Hamza, T.H.; Zabetian, C.P.; Tenesa, A.; Laederach, A.; Montimurro, J.; Yearout, D.; Kay, D.M.; Doheny, K.F.; Paschall, J.; Pugh, E.; et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010, 42, 781–785. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; Freeman, C.; Hunt, S.E.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar] [PubMed]
- Dempsey, E.; Abautret-Daly, A.; Docherty, N.G.; Medina, C.; Harkin, A. Persistent central inflammation and region specific cellular activation accompany depression- and anxiety-like behaviours during the resolution phase of experimental colitis. Brain Behav. Immun. 2019, 80, 616–632. [Google Scholar] [CrossRef] [PubMed]
- Villaran, R.F.; Espinosa-Oliva, A.M.; Sarmiento, M.; De Pablos, R.M.; Arguelles, S.; Delgado-Cortes, M.J.; Sobrino, V.; Van Rooijen, N.; Venero, J.L.; Herrera, A.J.; et al. Ulcerative colitis exacerbates lipopolysaccharide-induced damage to the nigral dopaminergic system: Potential risk factor in Parkinson‘s disease. J. Neurochem. 2010, 114, 1687–1700. [Google Scholar] [CrossRef]
- Kishimoto, Y.; Zhu, W.; Hosoda, W.; Sen, J.M.; Mattson, M.P. Chronic Mild Gut Inflammation Accelerates Brain Neuropathology and Motor Dysfunction in alpha-Synuclein Mutant Mice. Neuromolecular. Med. 2019, 21, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Resnikoff, H.; Metzger, J.M.; Lopez, M.; Bondarenko, V.; Mejia, A.; Simmons, H.A.; Emborg, M.E. Colonic inflammation affects myenteric alpha-synuclein in nonhuman primates. J. Inflamm. Res. 2019, 12, 113–126. [Google Scholar] [CrossRef]
- Han, Y.; Zhao, T.; Cheng, X.; Zhao, M.; Gong, S.H.; Zhao, Y.Q.; Wu, H.T.; Fan, M.; Zhu, L.L. Cortical Inflammation is Increased in a DSS-Induced Colitis Mouse Model. Neurosci. Bull. 2018, 34, 1058–1066. [Google Scholar] [CrossRef]
- Nouri, M.; Bredberg, A.; Westrom, B.; Lavasani, S. Intestinal barrier dysfunction develops at the onset of experimental autoimmune encephalomyelitis, and can be induced by adoptive transfer of auto-reactive T cells. PLoS ONE 2014, 9, e106335. [Google Scholar] [CrossRef]
- Kunze, W.A.; Bornstein, J.C.; Furness, J.B. Identification of sensory nerve cells in a peripheral organ (the intestine) of a mammal. Neuroscience 1995, 66, 1–4. [Google Scholar] [CrossRef]
- Spencer, N.J.; Hu, H. Enteric nervous system: Sensory transduction, neural circuits and gastrointestinal motility. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 338–351. [Google Scholar] [CrossRef]
- Uesaka, T.; Young, H.M.; Pachnis, V.; Enomoto, H. Development of the intrinsic and extrinsic innervation of the gut. Dev. Biol. 2016, 417, 158–167. [Google Scholar] [CrossRef]
- Di Giovangiulio, M.; Verheijden, S.; Bosmans, G.; Stakenborg, N.; Boeckxstaens, G.E.; Matteoli, G. The Neuromodulation of the Intestinal Immune System and Its Relevance in Inflammatory Bowel Disease. Front. Immunol. 2015, 6, 590. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Gershon, M.D. The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol Hepatol 2016, 13, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.A.; Becker, C.; Reeh, P.W.; Neurath, M.F. Role of sensory neurons in colitis: Increasing evidence for a neuroimmune link in the gut. Inflamm. Bowel. Dis. 2011, 17, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.A.; Khalil, M.; Mueller-Tribbensee, S.M.; Becker, C.; Neuhuber, W.L.; Neurath, M.F.; Reeh, P.W. The proximodistal aggravation of colitis depends on substance P released from TRPV1-expressing sensory neurons. J. Gastroenterol. 2012, 47, 256–265. [Google Scholar] [CrossRef]
- Berer, K.; Mues, M.; Koutrolos, M.; Rasbi, Z.A.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Therap. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Boertien, J.M.; Pereira, P.A.B.; Aho, V.T.E.; Scheperjans, F. Increasing Comparability and Utility of Gut Microbiome Studies in Parkinson’s Disease: A Systematic Review. J. Parkinsons. Dis. 2019, 9, S297–S312. [Google Scholar] [CrossRef] [PubMed]
- Barichella, M.; Severgnini, M.; Cilia, R.; Cassani, E.; Bolliri, C.; Caronni, S.; Ferri, V.; Cancello, R.; Ceccarani, C.; Faierman, S.; et al. Unraveling gut microbiota in Parkinson’s disease and atypical parkinsonism. Mov. Disord. 2019, 34, 396–405. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Doring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef]
- Lubomski, M.; Tan, A.H.; Lim, S.Y.; Holmes, A.J.; Davis, R.L.; Sue, C.M. Parkinson’s disease and the gastrointestinal microbiome. J. Neurol. 2020, 267, 2507–2523. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Kawasumi, M.; Onawa, S.; Taguchi-Atarashi, N.; Morita, H.; Taylor, T.D.; Hattori, M.; Ohno, H. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Hart, J.; Roalstad, S.; Graves, J.; Spencer, C.M.; Lynch, S.V.; Zamvil, S.S.; Waubant, E.; et al. Gut microbiota in early pediatric multiple sclerosis: A case-control study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.L.; Kunstner, A.; Kennedy, J.J.; Fang, Q.; Asarian, L.; Culp-Hill, R.; D’Alessandro, A.; Teuscher, C.; Busch, H.; Krementsov, D.N. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc. Natl. Acad. Sci. USA 2020, 117, 27516–27527. [Google Scholar] [CrossRef]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Hart, J.; Roalstad, S.; Graves, J.; Spencer, C.M.; Lynch, S.V.; Zamvil, S.S.; Waubant, E.; et al. Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 2016, 16, 182. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef] [PubMed]
- Oliveras-Salva, M.; Van Rompuy, A.S.; Heeman, B.; Van den Haute, C.; Baekelandt, V. Loss-of-function rodent models for parkin and PINK1. J. Parkinsons. Dis. 2011, 1, 229–251. [Google Scholar] [CrossRef]
- Matheoud, D.; Cannon, T.; Voisin, A.; Penttinen, A.M.; Ramet, L.; Fahmy, A.M.; Ducrot, C.; Laplante, A.; Bourque, M.J.; Zhu, L.; et al. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1(-/-) mice. Nature 2019, 571, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Derkinderen, P.; Shannon, K.M.; Brundin, P. Gut feelings about smoking and coffee in Parkinson’s disease. Mov. Disord. 2014, 29, 976–979. [Google Scholar] [CrossRef]
- Van Den Berge, N.; Ferreira, N.; Gram, H.; Mikkelsen, T.W.; Alstrup, A.K.O.; Casadei, N.; Tsung-Pin, P.; Riess, O.; Nyengaard, J.R.; Tamguney, G.; et al. Evidence for bidirectional and trans-synaptic parasympathetic and sympathetic propagation of alpha-synuclein in rats. Acta Neuropathol. 2019, 138, 535–550. [Google Scholar] [CrossRef]
- Kim, S.; Kwon, S.H.; Kam, T.I.; Panicker, N.; Karuppagounder, S.S.; Lee, S.; Lee, J.H.; Kim, W.R.; Kook, M.; Foss, C.A.; et al. Transneuronal Propagation of Pathologic alpha-Synuclein from the Gut to the Brain Models Parkinson’s Disease. Neuron 2019, 103, 627–641.e7. [Google Scholar] [CrossRef]
- Cosorich, I.; Dalla-Costa, G.; Sorini, C.; Ferrarese, R.; Messina, M.J.; Dolpady, J.; Radice, E.; Mariani, A.; Testoni, P.A.; Canducci, F.; et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci. Adv. 2017, 3, e1700492. [Google Scholar] [CrossRef]
- Sommer, A.; Marxreiter, F.; Krach, F.; Fadler, T.; Grosch, J.; Maroni, M.; Graef, D.; Eberhardt, E.; Riemenschneider, M.J.; Yeo, G.W.; et al. Th17 Lymphocytes Induce Neuronal Cell Death in a Human iPSC-Based Model of Parkinson’s Disease. Cell Stem. Cell 2018, 23, 123–131.e6. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber HLt Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Erny, D.; Hrabe de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4615–4622. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Sugisawa, E.; Takayama, Y.; Takemura, N.; Kondo, T.; Hatakeyama, S.; Kumagai, Y.; Sunagawa, M.; Tominaga, M.; Maruyama, K. RNA Sensing by Gut Piezo1 Is Essential for Systemic Serotonin Synthesis. Cell 2020, 182, 609–624.e21. [Google Scholar] [CrossRef]
- Bellono, N.W.; Bayrer, J.R.; Leitch, D.B.; Castro, J.; Zhang, C.; O’Donnel, T.A.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell 2017, 170, 185–198.e16. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.A.; Nguyen, J.C.; Polglaze, K.E.; Bertrand, P.P. Influence of Tryptophan and Serotonin on Mood and Cognition with a Possible Role of the Gut-Brain Axis. Nutrients 2016, 8, 56. [Google Scholar] [CrossRef]
- Maini Rekdal, V.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, 6445. [Google Scholar] [CrossRef]
- Powley, T.L.; Jaffey, D.M.; McAdams, J.; Baronowsky, E.A.; Black, D.; Chesney, L.; Evans, C.; Phillips, R.J. Vagal innervation of the stomach reassessed: Brain-gut connectome uses smart terminals. Ann. N. Y. Acad. Sci. 2019, 1454, 14–30. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Food, immunity, and the microbiome. Gastroenterology 2015, 148, 1107–1119. [Google Scholar] [CrossRef]
- Neurath, M.F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 2019, 20, 970–979. [Google Scholar] [CrossRef]
- Korn, T.; Kallies, A. T cell responses in the central nervous system. Nat. Rev. Immunol. 2017, 17, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Haghikia, A.; Jorg, S.; Duscha, A.; Berg, J.; Manzel, A.; Waschbisch, A.; Hammer, A.; Lee, D.H.; May, C.; Wilck, N.; et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 2015, 43, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Yissachar, N.; Zhou, Y.; Ung, L.; Lai, N.Y.; Mohan, J.F.; Ehrlicher, A.; Weitz, D.A.; Kasper, D.L.; Chiu, I.M.; Mathis, D.; et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell 2017, 168, 1135–1148.e12. [Google Scholar] [CrossRef]
- Powell, N.; Walker, M.M.; Talley, N.J. The mucosal immune system: Master regulator of bidirectional gut-brain communications. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; Kim, S.W.; Suda, W.; Awasumi, M.; Onawa, S.; Taguchi-Atarashi, N.; Morita, H.; Taylor, T.D.; Hattori, M.; Ohno, H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 2020, 585, 102–106. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, J.; Li, J.; Kim, G.; Stewart, A.; Urban Jr, J.F.; Huang, Y.; Chen, S.; Wu, L.; Chesler, A.; et al. Interleukin-33 Promotes Serotonin Release from Enterochromaffin Cells for Intestinal Homeostasis. Immunity 2021, 54, 151–163.e6. [Google Scholar] [CrossRef]
- Flamar, A.L.; Klose, C.S.N.; Moeller, J.B.; Mahlakoiv, T.; Bessman, N.J.; Zhang, W.; Moriyama, S.; Stokic-Trtica, V.; Rankin, L.C.; Putzel, G.C.; et al. Interleukin-33 Induces the Enzyme Tryptophan Hydroxylase 1 to Promote Inflammatory Group 2 Innate Lymphoid Cell-Mediated Immunity. Immunity 2020, 52, 606–619.e6. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.C.; Wernig, M.; Duncan, I.D.; Brustle, O.; Thomson, J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem. Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y. Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture. Cell Stem. Cell 2013, 12, 520–530. [Google Scholar] [CrossRef]
- Mariani, J.; Simonini, M.V.; Palejev, D.; Tomasini, L.; Coppola, G.; Szekely, A.M.; Horvath, T.L.; Vaccarino, F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 12770–12775. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–202849. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Brauninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef]
- Pasca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.D.; Goke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem. Cell 2016, 19, 248–257. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef]
- Pellegrini, L.; Bonfio, C.; Chadwick, J.; Begum, F.; Skehel, M.; Lancaster, M.A. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 2020, 369, 6500. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Levi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef]
- Barber, K.; Studer, L.; Fattahi, F. Derivation of enteric neuron lineages from human pluripotent stem cells. Nat. Protoc. 2019, 14, 1261–1279. [Google Scholar] [CrossRef]
- Fattahi, F.; Steinbeck, J.A.; Kriks, S.; Tchieu, J.; Zimmer, B.; Kishinevsky, S.; Zeltner, N.; Mica, Y.; El-Nachef, W.; Zhao, H.; et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 2016, 531, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Zorn, A.M.; Wells, J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 221–251. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yan, L.; Wang, R.; Li, J.; Yong, J.; Zhou, X.; Wei, Y.; Wu, X.; Wang, X.; Fan, X.; et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat. Cell Biol. 2018, 20, 721–734. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Huch, M. Disease modelling in human organoids. Dis. Model. Mech. 2019, 12, 039347. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Mithal, A.; Capilla, A.; Heinze, D.; Berical, A.; Villacorta-Martin, C.; Vedaie, M.; Jacob, A.; Abo, K.; Szymaniak, A.; Peasley, M.; et al. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat. Commun. 2020, 11, 215. [Google Scholar] [CrossRef]
- Forster, R.; Chiba, K.; Schaeffer, L.; Regalado, S.G.; Lai, C.S.; Gao, Q.; Kiani, S.; Farin, H.F.; Clevers, H.; Cost, G.J.; et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem. Cell Rep. 2014, 2, 838–852. [Google Scholar] [CrossRef]
- McCracken, K.W.; Howell, J.C.; Wells, J.M.; Spence, J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011, 6, 1920–1928. [Google Scholar] [CrossRef]
- Workman, M.J.; Mahe, M.M.; Trisno, S.; Poling, H.M.; Watson, C.L.; Sundaram, N.; Chang, C.F.; Schiesser, J.; Aubert, P.; Stanley, E.G.; et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017, 23, 49–59. [Google Scholar] [CrossRef]
- Sarvestani, S.K.; Signs, S.; Hu, B.; Yeu, Y.; Feng, H.; Ni, Y.; Hill, D.R.; Fisher, R.C.; Ferrandon, S.; DeHaan, R.K.; et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 2021, 12, 262. [Google Scholar] [CrossRef]
- Yoshida, S.; Miwa, H.; Kawachi, T.; Kume, S.; Takahashi, K. Generation of intestinal organoids derived from human pluripotent stem cells for drug testing. Sci. Rep. 2020, 10, 5989. [Google Scholar] [CrossRef]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef]
- Williams, J.M.; Duckworth, C.A.; Burkitt, M.D.; Watson, A.J.; Campbell, B.J.; Pritchard, D.M. Epithelial cell shedding and barrier function: A matter of life and death at the small intestinal villus tip. Vet. Pathol. 2015, 52, 445–455. [Google Scholar] [CrossRef]
- Alison, M.R.; Lin, W.R. Hepatocyte turnover and regeneration: Virtually a virtuoso performance. Hepatology 2011, 53, 1393–1396. [Google Scholar] [CrossRef]
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef]
- Gunther, C.; Brevini, T.; Sampaziotis, F.; Neurath, M.F. What gastroenterologists and hepatologists should know about organoids in 2019. Dig. Liver Dis. 2019, 51, 753–760. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Min, S.; Kim, S.; Cho, S.W. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp. Mol. Med. 2020, 52, 227–237. [Google Scholar] [CrossRef]
- Hefele, M.; Stolzer, I.; Ruder, B.; He, G.W.; Mahapatro, M.; Wirtz, S.; Neurath, M.F.; Gunther, C. Intestinal epithelial Caspase-8 signaling is essential to prevent necroptosis during Salmonella Typhimurium induced enteritis. Mucosal. Immunol. 2018, 11, 1191–1202. [Google Scholar] [CrossRef]
- Gunther, C.; Ruder, B.; Stolzer, I.; Dorner, H.; He, G.W.; Chiriac, M.T.; Aden, K.; Strigli, A.; Bittel, M.; Zeissig, S.; et al. Interferon Lambda Promotes Paneth Cell Death Via STAT1 Signaling in Mice and Is Increased in Inflamed Ileal Tissues of Patients With Crohn’s Disease. Gastroenterology 2019, 157, 1310–1322.e13. [Google Scholar] [CrossRef]
- Bittel, M.; Kremer, A.E.; Sturzl, M.; Wirtz, S.; Stolzer, I.; Neurath, M.F.; Ballon, G.; Gunther, C. Modulation of the extrinsic cell death signaling pathway by viral Flip induces acute-death mediated liver failure. Cell Death Dis. 2019, 10, 878. [Google Scholar] [CrossRef]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef]
- Boccellato, F.; Woelffling, S.; Imai-Matsushima, A.; Sanchez, G.; Goosmann, C.; Schmid, M.; Berger, H.; Morey, P.; Denecke, C.; Ordemann, J.; et al. Polarised epithelial monolayers of the gastric mucosa reveal insights into mucosal homeostasis and defence against infection. Gut 2019, 68, 400–413. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Piazuelo, M.B.; Chaturvedi, R.; Schumacher, M.; Aihara, E.; Feng, R.; Noto, J.M.; Delgado, A.; Israel, D.A.; Zavros, Y.; et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 2015, 64, 720–730. [Google Scholar] [CrossRef]
- Shaffiey, S.A.; Jia, H.; Keane, T.; Costello, C.; Wasserman, D.; Quidgley, M.; Dziki, J.; Badylak, S.; Sodhi, C.P.; March, J.C.; et al. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen. Med. 2016, 11, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.R.; Yu, Q. Correction: Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2021, 28, 2025–2027. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015, 64, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef]
- Shi, Y.; Sun, L.; Wang, M.; Liu, J.; Zhong, S.; Li, R.; Li, P.; Guo, L.; Fang, A.; Chen, R.; et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020, 18, e3000705. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Goncalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Günther, C.; Rothhammer, V.; Karow, M.; Neurath, M.F.; Winner, B. The Gut-Brain Axis in Inflammatory Bowel Disease—Current and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 8870. https://doi.org/10.3390/ijms22168870
Günther C, Rothhammer V, Karow M, Neurath MF, Winner B. The Gut-Brain Axis in Inflammatory Bowel Disease—Current and Future Perspectives. International Journal of Molecular Sciences. 2021; 22(16):8870. https://doi.org/10.3390/ijms22168870
Chicago/Turabian StyleGünther, Claudia, Veit Rothhammer, Marisa Karow, Markus F. Neurath, and Beate Winner. 2021. "The Gut-Brain Axis in Inflammatory Bowel Disease—Current and Future Perspectives" International Journal of Molecular Sciences 22, no. 16: 8870. https://doi.org/10.3390/ijms22168870
APA StyleGünther, C., Rothhammer, V., Karow, M., Neurath, M. F., & Winner, B. (2021). The Gut-Brain Axis in Inflammatory Bowel Disease—Current and Future Perspectives. International Journal of Molecular Sciences, 22(16), 8870. https://doi.org/10.3390/ijms22168870





