Molecular Basis of Interactions between the Antibiotic Nitrofurantoin and Human Serum Albumin: A Mechanism for the Rapid Drug Blood Transportation
Abstract
:1. Introduction
2. Results
2.1. UV-VIS Absorption Spectra of the Nitrofurantoin−HSA Complex
2.2. Fluorescence Characterization of Nitrofurantoin-HSA Complex
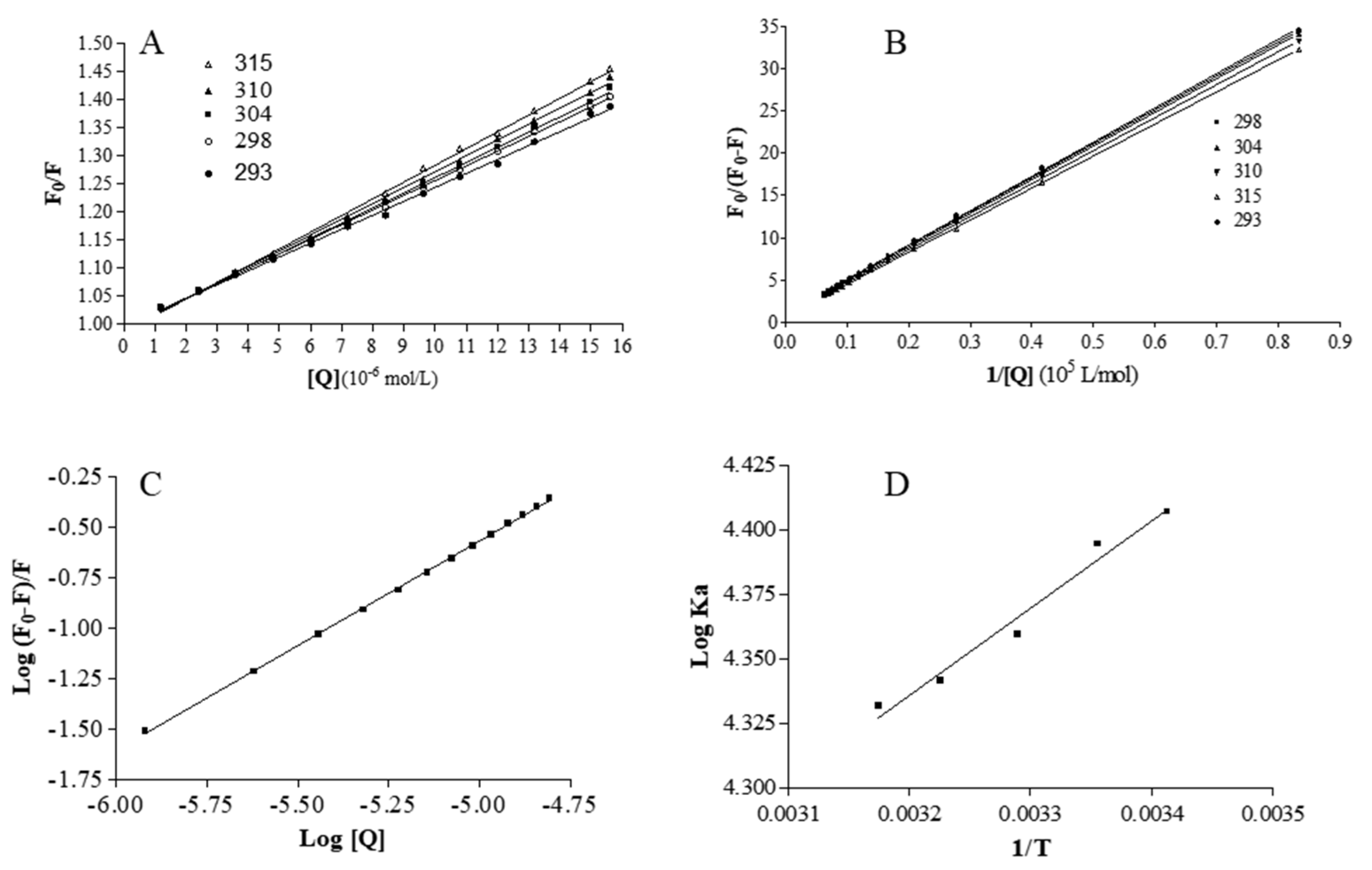
2.3. Analysis of Binding Equilibrium
2.4. Thermodynamics and Acting Forces
2.5. Fluorescence Displacement Binding Experiments
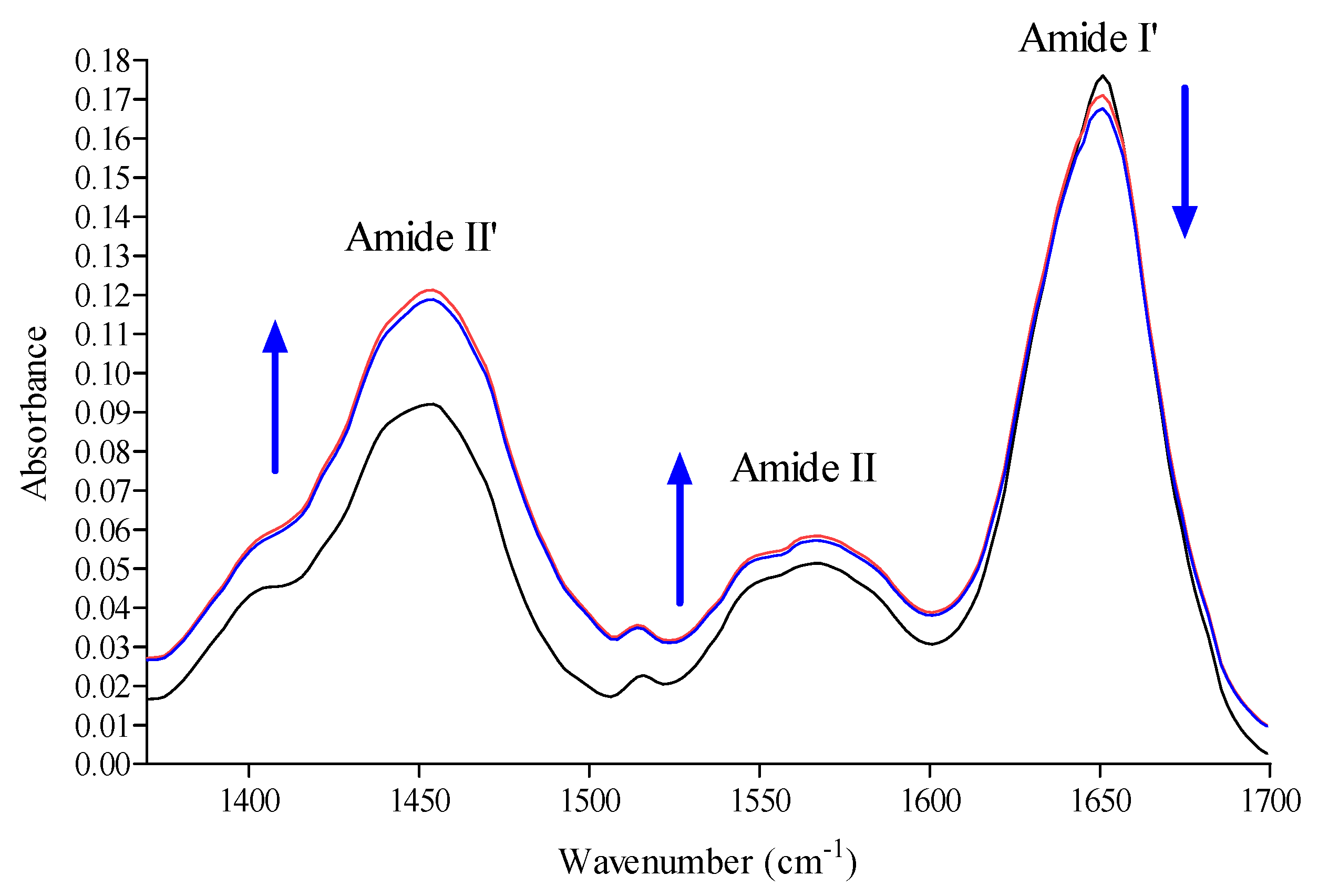
2.6. Conformation Investigation by Fourier Transform Infrared Spectroscopy (FTIR) Spectroscopy
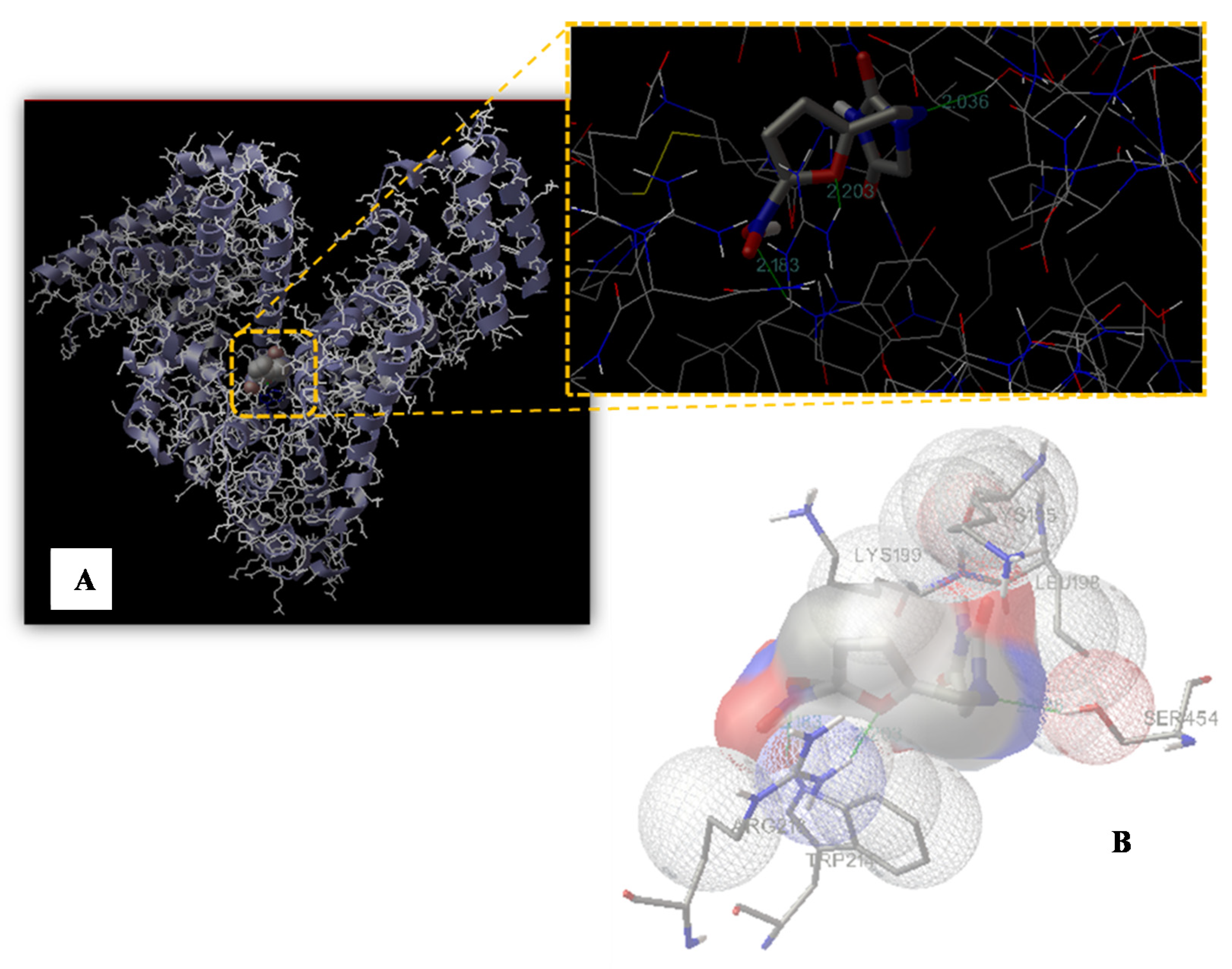
2.7. Molecular Modeling Study
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. UV-VIS Spectra
4.3. FTIR Spectra
4.4. Data Analysis of FTIR Spectra
4.5. Fluorescence Spectra
4.6. Molecular Modeling Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.J.; McLeod, M.; McNulty, C.A.M.; Lecky, D.M.; Holmes, A.H.; Ahmad, R. Trends in Antibiotic Prescribing in Out-of-Hours Primary Care in England from January 2016 to June 2020 to Understand Behaviours during the First Wave of COVID-19. Antibiotics 2021, 10, 32. [Google Scholar] [CrossRef]
- Konwar, M.; Gogtay, N.J.; Ravi, R.; Thatte, U.M.; Bose, D. Evaluation of efficacy and safety of fosfomycin versus nitrofurantoin for the treatment of uncomplicated lower urinary tract infection (UTI) in women—A systematic review and meta-analysis. J. Chemother. 2021, 3, 1–10. [Google Scholar] [CrossRef]
- Ten Doesschate, T.; Hendriks, K.; van Werkhoven, C.H.; van der Hout, E.C.; Platteel, T.N.; Groenewegen, I.A.M.; Muller, A.E.; Hoepelman, A.I.M.; Bonten, M.J.M.; Geerlings, S.E. Nitrofurantoin 100 mg versus 50 mg prophylaxis for urinary tract infections, a cohort study. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 5, 100. [Google Scholar] [CrossRef]
- Gajdacs, M.; Abrok, M.; Lazar, A.; Burian, K. Comparative Epidemiology and Resistance Trends of Common Urinary Pathogens in a Tertiary-Care Hospital: A 10-Year Surveillance Study. Medicina 2019, 55, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garau, J. Other antimicrobials of interest in the era of extended-spectrum beta-lactamases: Fosfomycin, nitrofurantoin and tigecycline. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2008, 14 (Suppl. 1), 198–202. [Google Scholar] [CrossRef] [Green Version]
- Hooper, D.C. Urinary tract agents: Nitrofurantoin and methenamine. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 6th ed.; Churchill Livingstone: Philadelphia, PA, USA, 2005; pp. 423–428. [Google Scholar]
- Munsimbwe, L.; Seetsi, A.; Namangala, B.; N’Da, D.D.; Inoue, N.; Suganuma, K. In Vitro and In Vivo Trypanocidal Efficacy of Synthesized Nitrofurantoin Analogs. Molecules 2021, 26, 3372. [Google Scholar] [CrossRef] [PubMed]
- Manin, A.N.; Drozd, K.V.; Voronin, A.P.; Churakov, A.V.; Perlovich, G.L. A Combined Experimental and Theoretical Study of Nitrofuran Antibiotics: Crystal Structures, DFT Computations, Sublimation and Solution Thermodynamics. Molecules 2021, 26, 3444. [Google Scholar] [CrossRef] [PubMed]
- Day, M.A.; Jarrom, D.; Christofferson, A.J.; Graziano, A.E.; Anderson, J.L.R.; Searle, P.F.; Hyde, E.I.; White, S.A. The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN. Biochem. J. 2021, 478, 2601–2617. [Google Scholar] [CrossRef]
- Edowik, Y.; Caspari, T.; Williams, H.M. The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections. Microorganisms 2020, 8, 1983. [Google Scholar] [CrossRef]
- Gajdacs, M.; Urban, E. Comparative Epidemiology and Resistance Trends of Proteae in Urinary Tract Infections of Inpatients and Outpatients: A 10-Year Retrospective Study. Antibiotics 2019, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Kragh-Hansen, U.; Chuang, V.T.; Otagiri, M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol. Pharm. Bull. 2002, 25, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Barreca, D.; Lagana, G.; Toscano, G.; Calandra, P.; Kiselev, M.A.; Lombardo, D.; Bellocco, E. The interaction and binding of flavonoids to human serum albumin modify its conformation, stability and resistance against aggregation and oxidative injuries. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3531–3539. [Google Scholar] [CrossRef]
- Barreca, D.; Lagana, G.; Ficarra, S.; Tellone, E.; Leuzzi, U.; Galtieri, A.; Bellocco, E. Ornithine carbamoyltransferase unfolding states in the presence of urea and guanidine hydrochloride. Appl. Biochem. Biotechnol. 2014, 172, 854–866. [Google Scholar] [CrossRef]
- Barreca, D.; Lagana, G.; Bruno, G.; Magazu, S.; Bellocco, E. Diosmin binding to human serum albumin and its preventive action against degradation due to oxidative injuries. Biochimie 2013, 95, 2042–2049. [Google Scholar] [CrossRef]
- Di Salvo, C.; Barreca, D.; Lagana, G.; di Bella, M.; Tellone, E.; Ficarra, S.; Bellocco, E. Myelin basic protein: Structural characterization of spherulites formation and preventive action of trehalose. Int. J. Biol. Macromol. 2013, 57, 63–68. [Google Scholar] [CrossRef]
- Barreca, D.; Lagana, G.; Ficarra, S.; Gattuso, G.; Magazu, S.; La Torre, R.; Tellone, E.; Bellocco, E. Spectroscopic determination of lysozyme conformational changes in the presence of trehalose and guanidine. Cell Biochem. Biophys. 2013, 66, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Barreca, D.; Lagana, G.; Ficarra, S.; Tellone, E.; Leuzzi, U.; Magazu, S.; Galtieri, A.; Bellocco, E. Anti-aggregation properties of trehalose on heat-induced secondary structure and conformation changes of bovine serum albumin. Biophys. Chem. 2010, 147, 146–152. [Google Scholar] [CrossRef]
- Curry, S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metab. Pharmacokinet. 2009, 24, 342–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreca, D.; Laganà, G.; Magazù, S.; Migliardo, F.; Bellocco, E. Glycerol, trehalose and glycerol–trehalose mixture effects on thermal stabilization of OCT. Chem. Phys. 2013, 424, 100–104. [Google Scholar] [CrossRef]
- Mandeville, J.S.; Tajmir-Riahi, H.A. Complexes of dendrimers with bovine serum albumin. Biomacromolecules 2010, 11, 465–472. [Google Scholar] [CrossRef]
- Ascenzi, P.; Fasano, M. Allostery in a monomeric protein: The case of human serum albumin. Biophys. Chem. 2010, 148, 16–22. [Google Scholar] [CrossRef]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef]
- Curry, S. Beyond expansion: Structural studies on the transport roles of human serum albumin. Vox Sang. 2002, 83 (Suppl. 1), 315–319. [Google Scholar] [CrossRef]
- Peters, T. All about Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Colmenarejo, G.; Alvarez-Pedraglio, A.; Lavandera, J.L. Cheminformatic models to predict binding affinities to human serum albumin. J. Med. Chem. 2001, 44, 4370–4378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufour, C.; Dangles, O. Flavonoid-serum albumin complexation: Determination of binding constants and binding sites by fluorescence spectroscopy. Biochim. Biophys. Acta 2005, 1721, 164–173. [Google Scholar] [CrossRef]
- Timaseff, S.N. Thermodynamics of Protein Interactions. In Proteins of Biological Fluids; Peeters, H., Ed.; Pergamon Press: Oxford, UK, 1972; pp. 511–519. [Google Scholar]
- Barreca, D.; Lagana, G.; Magazu, S.; Migliardo, F.; Gattuso, G.; Bellocco, E. FTIR, ESI-MS, VT-NMR and SANS study of trehalose thermal stabilization of lysozyme. Int. J. Biol. Macromol. 2014, 63, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Ou-Yang, Y.; Dai, C.M.; Liu, Y.; Xiao, X.H. Site-selective binding of human serum albumin by palmatine: Spectroscopic approach. Biomacromolecules 2010, 11, 106–112. [Google Scholar] [CrossRef]
- Barreca, D.; Bellocco, E.; Galli, G.; Lagana, G.; Leuzzi, U.; Magazu, S.; Migliardo, F.; Galtieri, A.; Telling, M.T. Stabilization effects of kosmotrope systems on ornithine carbamoyltransferase. Int. J. Biol. Macromol. 2009, 45, 120–128. [Google Scholar] [CrossRef]
- Hu, Y.J.; Liu, Y.; Xiao, X.H. Investigation of the interaction between Berberine and human serum albumin. Biomacromolecules 2009, 10, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Bellocco, E.; Barreca, D.; Lagana, G.; Tellone, E.; Ficarra, S.; Migliardo, F.; Leuzzi, U.; Magazu, S.; Galtieri, A. Influences of temperature and threshold effect of NaCl concentration on Alpias vulpinus OCT. Int. J. Biol. Macromol. 2008, 43, 474–480. [Google Scholar] [CrossRef]
- Belloco, E.; Lagana, G.; Barreca, D.; Ficarra, S.; Tellone, E.; Magazu, S.; Branca, C.; Kotyk, A.; Galtieri, A.; Leuzzi, U. Role of polyols in thermal inactivation of shark ornithine transcarbamoylase. Physiol. Res. 2005, 54, 395–402. [Google Scholar] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Fabian, H.M.W. Infrared spectroscopy of proteins. In Handbook of Vibrational Spectroscopy; John Wiley & Sons: Chichester, UK, 2002; Volume 5. [Google Scholar]
- Mantsch, H.H.; Chapman, D. Infrared Spectroscopy of Biomolecules; Wiley-Liss: New York, NY, USA, 1996. [Google Scholar]
- Havel, H.A. Spectroscopic Methods for Determining Protein Structure in Solution; John Wiley & Sons: Chichester, UK, 1995. [Google Scholar]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal structure analysis of warfarin binding to human serum albumin: Anatomy of drug site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [Green Version]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Rimac, H.; Dufour, C.; Debeljak, Z.; Zorc, B.; Bojic, M. Warfarin and Flavonoids Do Not Share the Same Binding Region in Binding to the IIA Subdomain of Human Serum Albumin. Molecules 2017, 22, 1153. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; The International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Gajdacs, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreca, D.; Trombetta, D.; Smeriglio, A.; Mandalari, G.; Romeo, O.; Felice, M.R.; Gattuso, G.; Nabavi, S.M. Food flavonols: Nutraceuticals with complex health benefits and functionalities. Trends Food Sci. Technol. 2021, 3, 26. [Google Scholar] [CrossRef]
- Schmidt, S.; Rock, K.; Sahre, M.; Burkhardt, O.; Brunner, M.; Lobmeyer, M.T.; Derendorf, H. Effect of protein binding on the pharmacological activity of highly bound antibiotics. Antimicrob. Agents Chemother. 2008, 52, 3994–4000. [Google Scholar] [CrossRef] [Green Version]
- Yasmeen, S.; Rabbani, G. Calorimetric and spectroscopic binding studies of amoxicillin with human serum albumin. J. Therm. Anal. Calorim. 2017, 127, 1445–1455. [Google Scholar] [CrossRef]
- Lehrer, S.S. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry 1971, 10, 3254–3263. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Ferlazzo, N.; de Luca, L.; Gitto, R.; Navarra, M. The link between the AMPK/SIRT1 axis and a flavonoid-rich extract of Citrus bergamia juice: A cell-free, in silico, and in vitro study. Phytother. Res. PTR 2019, 33, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]



| ΔH. | ΔS | ΔG | ||||
|---|---|---|---|---|---|---|
| (kJ mol−1) | (J mol−1K−1) | (kJ mol−1) | ||||
| 293 | 298 | 304 | 310 | 315 | ||
| −3.93 ± 0.26 | 27.05 ± 0.85 | −11.32 ± 1.25 | −11.45 ± 1.11 | −11.62 ± 1.13 | −11.78 ± 1.41 | −11.91 ± 1.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderaro, A.; Maugeri, A.; Magazù, S.; Laganà, G.; Navarra, M.; Barreca, D. Molecular Basis of Interactions between the Antibiotic Nitrofurantoin and Human Serum Albumin: A Mechanism for the Rapid Drug Blood Transportation. Int. J. Mol. Sci. 2021, 22, 8740. https://doi.org/10.3390/ijms22168740
Calderaro A, Maugeri A, Magazù S, Laganà G, Navarra M, Barreca D. Molecular Basis of Interactions between the Antibiotic Nitrofurantoin and Human Serum Albumin: A Mechanism for the Rapid Drug Blood Transportation. International Journal of Molecular Sciences. 2021; 22(16):8740. https://doi.org/10.3390/ijms22168740
Chicago/Turabian StyleCalderaro, Antonella, Alessandro Maugeri, Salvatore Magazù, Giuseppina Laganà, Michele Navarra, and Davide Barreca. 2021. "Molecular Basis of Interactions between the Antibiotic Nitrofurantoin and Human Serum Albumin: A Mechanism for the Rapid Drug Blood Transportation" International Journal of Molecular Sciences 22, no. 16: 8740. https://doi.org/10.3390/ijms22168740
APA StyleCalderaro, A., Maugeri, A., Magazù, S., Laganà, G., Navarra, M., & Barreca, D. (2021). Molecular Basis of Interactions between the Antibiotic Nitrofurantoin and Human Serum Albumin: A Mechanism for the Rapid Drug Blood Transportation. International Journal of Molecular Sciences, 22(16), 8740. https://doi.org/10.3390/ijms22168740










