Interest of Homodialkyl Neamine Derivatives against Resistant P. aeruginosa, E. coli, and β-Lactamases-Producing Bacteria—Effect of Alkyl Chain Length on the Interaction with LPS
Abstract
1. Introduction
2. Experimental Procedures
2.1. Bacterial Strain and Growth Conditions
2.2. MIC (Minimal Inhibitory Concentration) Determination
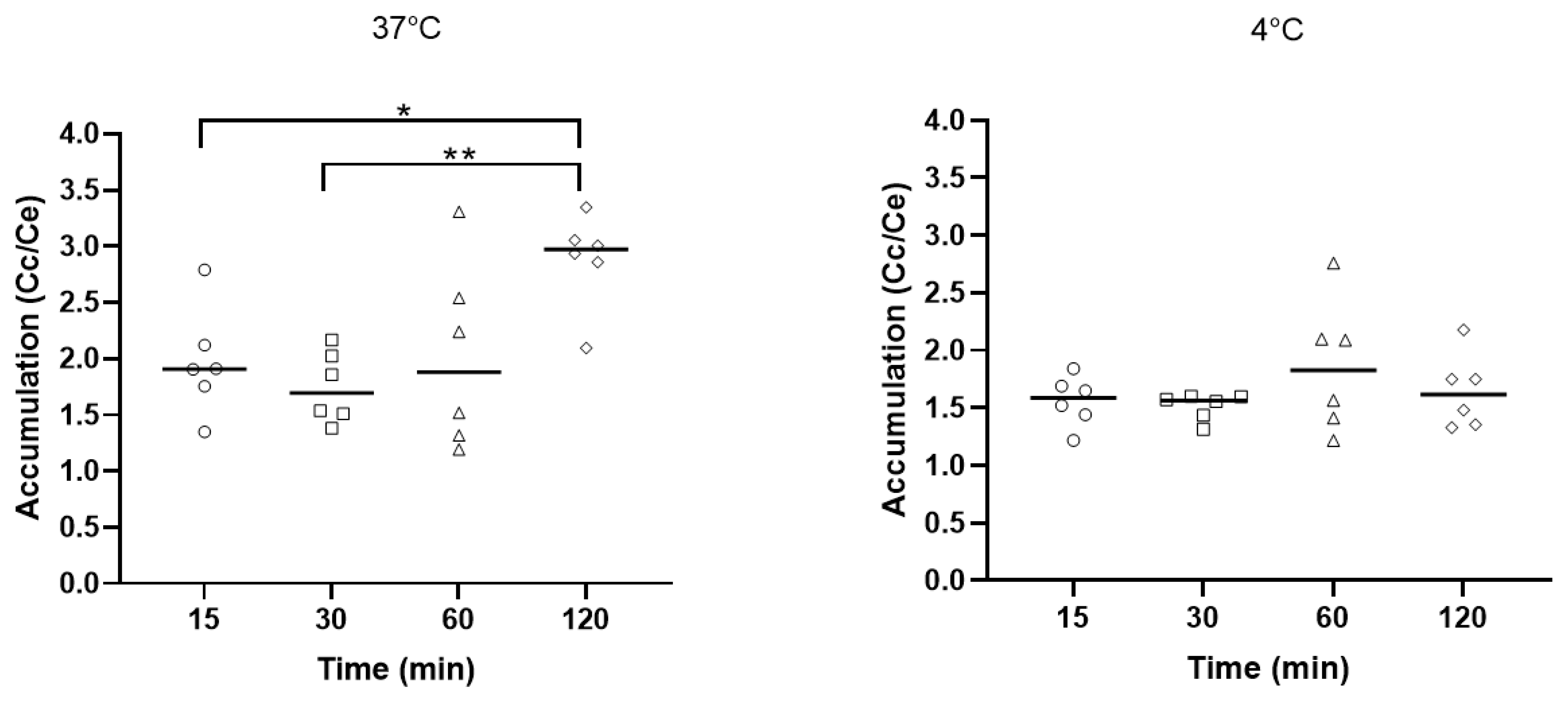
2.3. Accumulation of Fluorescent Amphiphilic Neamine Derivative in P. aeruginosa
2.4. Evaluation of Protein Synthesis—Luciferase Assay
2.5. Interaction with LPS—BODIPY-TR-Cadaverine Displacement Assay
2.6. Bacterial Membrane Permeabilization—Propidium Iodide Assay
2.7. Statistics
3. Results
3.1. 3′,6-Homodialkyl Neamine Derivatives Are Active against Clinical Strains of P. aeruginosa and E. coli
3.2. 3′,6-Homodialkyl Neamine Derivatives Are Active against β-Lactamases-Resistant Strains
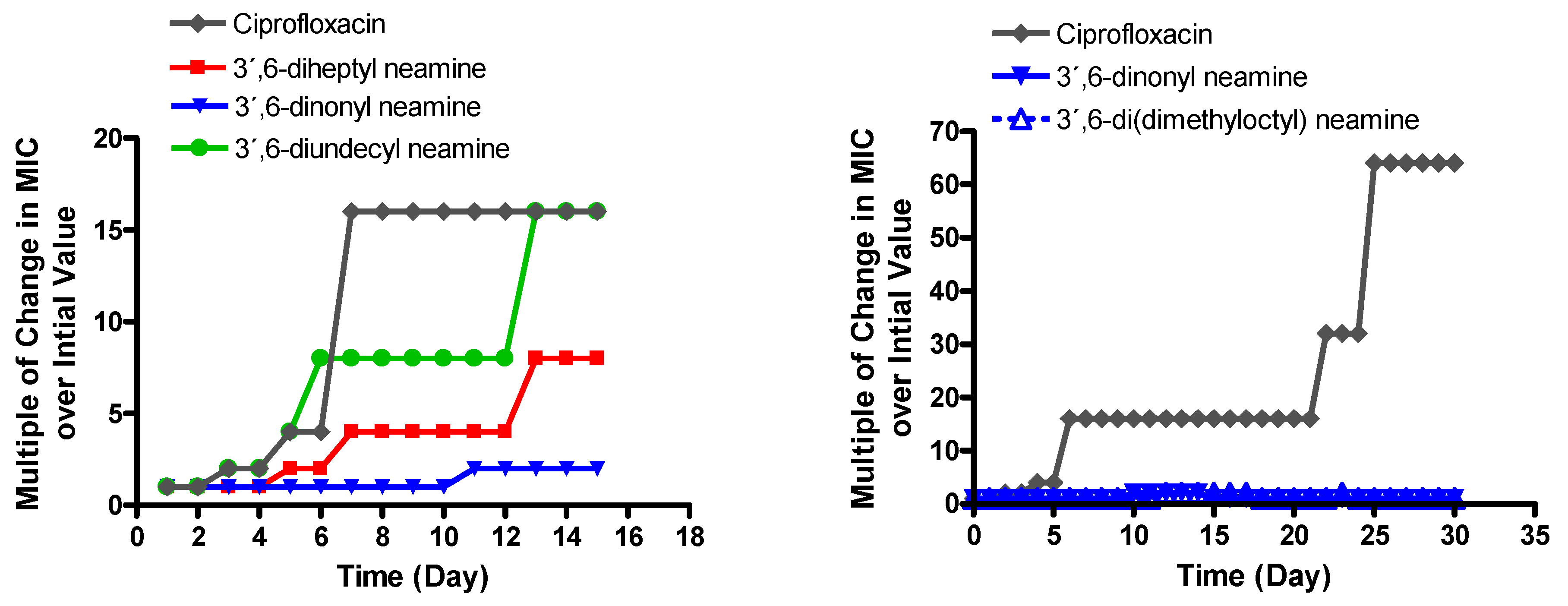
3.3. MIC against P. aeruginosa of 3′,6-Dinonyl Neamine Derivative Increases Slightly upon Long Exposure in Contrast to the Other Antibacterial 3′,6-Dialkyl Neamine Derivatives
3.4. 3′,6-Homodialkyl Neamine Derivatives Are Not Accumulated in Bacteria and Did Not Inhibit Protein Synthesis
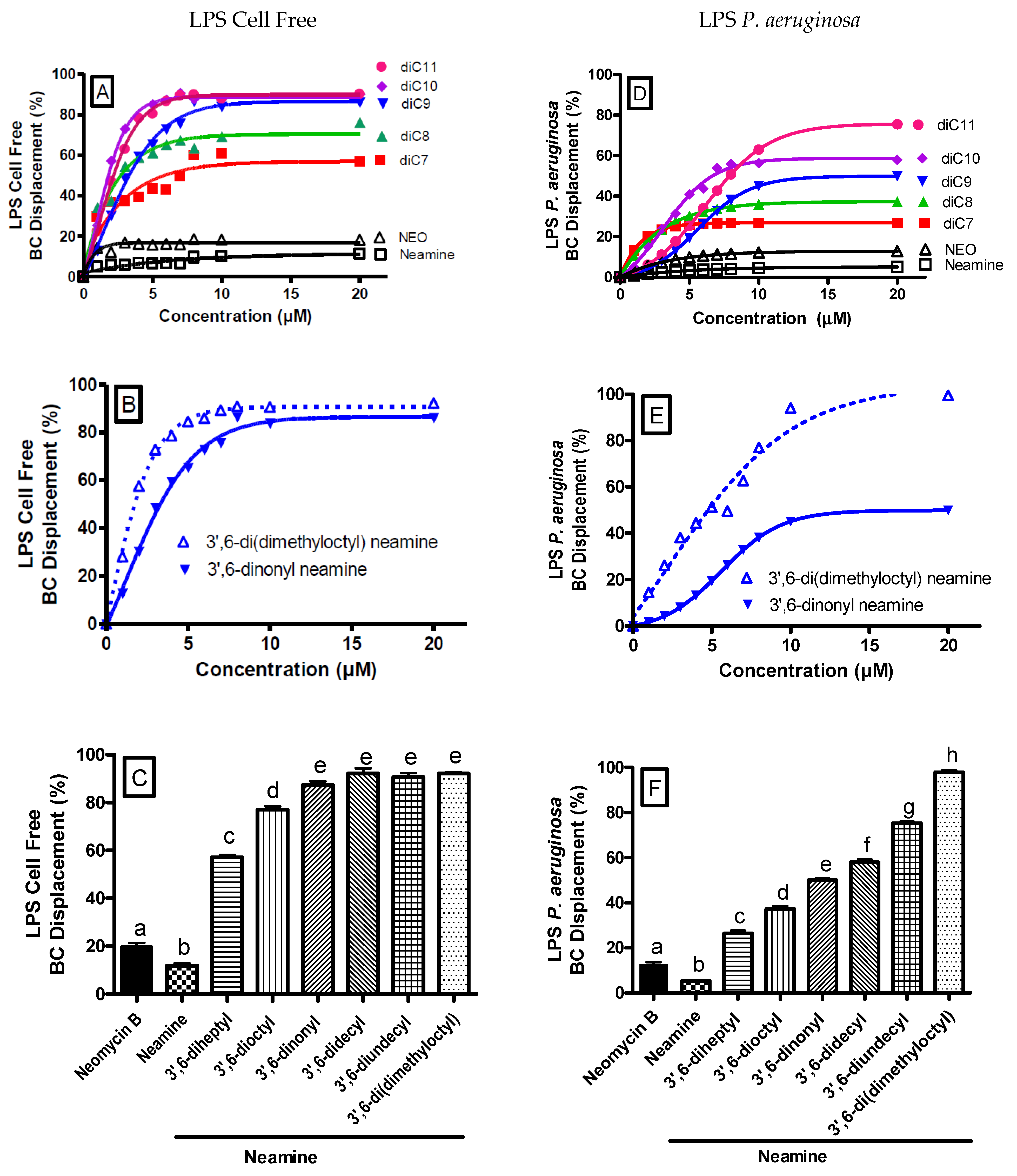
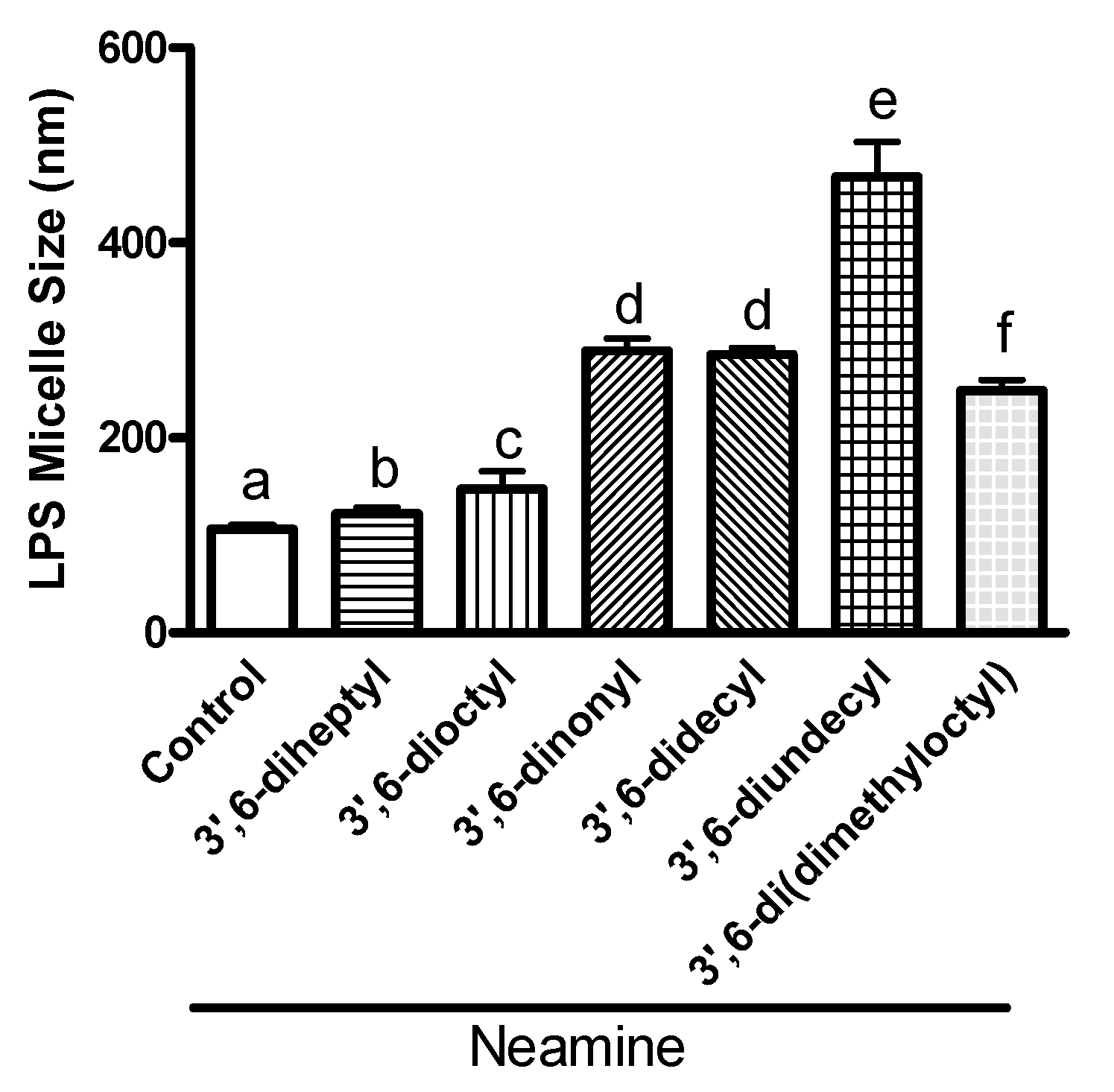
3.5. 3′,6-Homodialkyl Neamine Derivatives Interact with Lipopolysaccharides
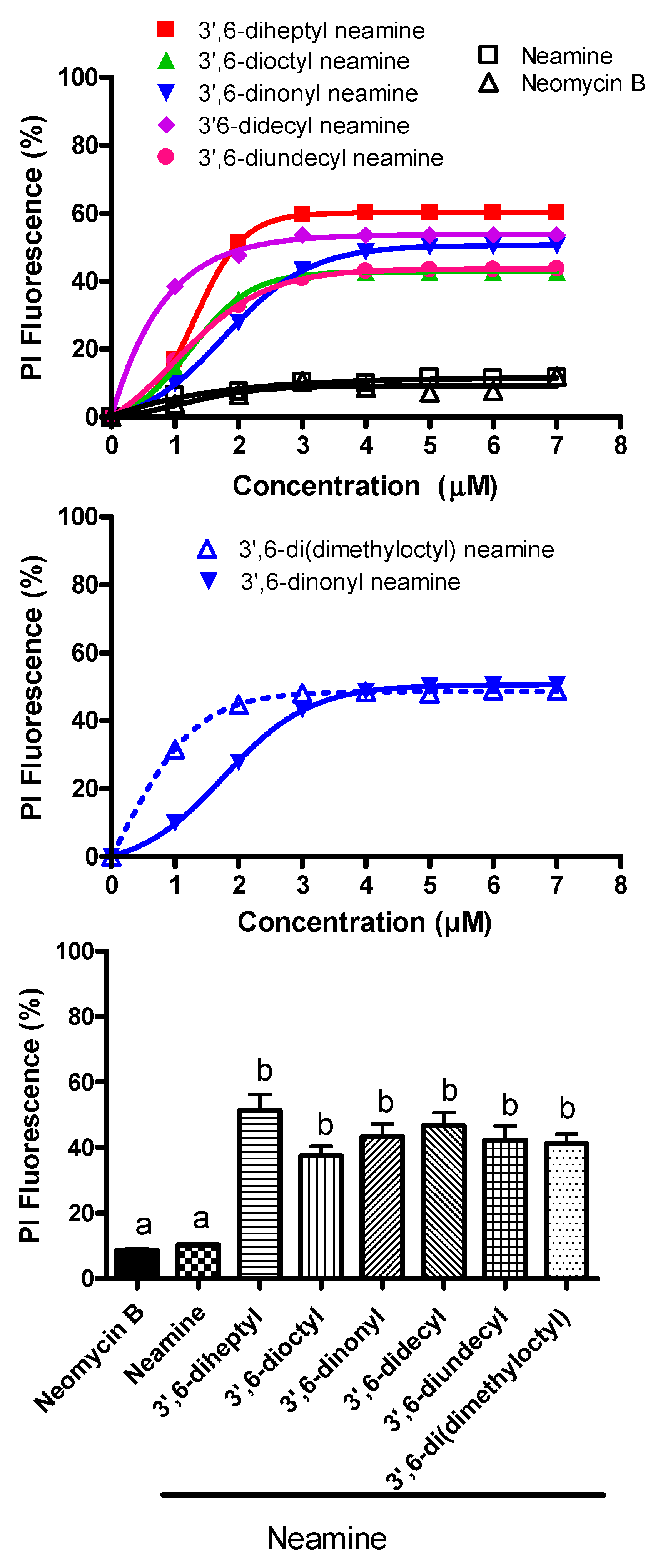
3.6. 3′,6-Homodialkyl Neamine Derivatives Induce Membrane Permeabilization
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rice, L.B. Progress and Challenges in Implementing the Research on ESKAPE Pathogens. Infect. Control Hosp. Epidemiol. 2010, 31, S7–S10. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control 2019, 8, 1–23. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Opperman, T.J.; Nguyen, S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015, 6, 421. [Google Scholar] [CrossRef] [PubMed]
- Housseini, B.I.; Phan, G.; Broutin, I. Functional Mechanism of the Efflux Pumps Transcription Regulators from Pseudomonas aeruginosa Based on 3D Structures. Front. Mol. Biosci. 2018, 5, 57. [Google Scholar] [CrossRef]
- Karampatakis, T.; Antachopoulos, C.; Tsakris, A.; Roilides, E. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa in an endemic area: Comparison with global data. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1211–1220. [Google Scholar] [CrossRef]
- Fourmy, D.; Recht, M.I.; Blanchard, S.C.; Puglisi, J.D. Structure of the A Site of Escherichia coli 16S Ribosomal RNA Complexed with an Aminoglycoside Antibiotic. Science 1996, 274, 1367–1371. [Google Scholar] [CrossRef]
- Fourmy, D.I.; Recht, M.; Puglisi, J.D. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 s rRNA. J. Mol. Biol. 1998, 277, 347–362. [Google Scholar] [CrossRef]
- Fourmy, D.; Yoshizawa, S.; Puglisi, J.D. Paromomycin binding induces a local conformational change in the A-site of 16 s rRNA. J. Mol. Biol. 1998, 277, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Mingeot-Leclercq, M.-P.; Glupczynski, Y.; Tulkens, P.M. Aminoglycosides: Activity and Resistance. Antimicrob. Agents Chemother. 1999, 43, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Bolla, J.M.; Alibert, S.; Handzlik, J.; Chevalier, J.; Mahamoud, A.; Boyer, G.; Kiec-Kononowicz, K.; Pagès, J.-M. Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Lett. 2011, 585, 1682–1690. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.; Bell, A. Antibiotic uptake into gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 1988, 7, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Domalaon, R.; Idowu, T.; Zhanel, G.G.; Schweizer, F. Antibiotic Hybrids: The Next Generation of Agents and Adjuvants against Gram-Negative Pathogens? Clin. Microbiol. Rev. 2018, 31, e00077-17. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.W.; Deshayes, S.; Hawker, S.; Blacker, A.; Kasko, A.M.; Wong, G.C.L. Engineering Persister-Specific Antibiotics with Synergistic Antimicrobial Functions. ACS Nano 2014, 8, 8786–8793. [Google Scholar] [CrossRef]
- Dezanet, C.; Kempf, J.; Mingeot-Leclercq, M.-P.; Décout, J.-L. Amphiphilic Aminoglycosides as Medicinal Agents. Int. J. Mol. Sci. 2020, 21, 7411. [Google Scholar] [CrossRef]
- Cooper, C.J.; Krishnamoorthy, G.; Wolloscheck, D.; Walker, J.K.; Rybenkov, V.V.; Parks, J.M.; Zgurskaya, H.I. Molecular Properties That Define the Activities of Antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 2018, 4, 1223–1234. [Google Scholar] [CrossRef]
- Zimmermann, L.; Bussière, A.; Ouberai, M.; Baussanne, I.; Jolivalt, C.; Mingeot-Leclercq, M.-P.; Décout, J.-L. Tuning the Antibacterial Activity of Amphiphilic Neamine Derivatives and Comparison to Paromamine Homologues. J. Med. Chem. 2013, 56, 7691–7705. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Das, I.; Desire, J.; Sautrey, G.; Barros, R.S.V.; El, K.M.; Mingeot-Leclercq, M.P.; Decout, J.L. New Broad-Spectrum Antibacterial Amphiphilic Aminoglycosides Active against Resistant Bacteria: From Neamine Derivatives to Smaller Neosamine Analogues. J. Med. Chem. 2016, 59, 9350–9369. [Google Scholar] [CrossRef]
- Zimmermann, L.; Kempf, J.; Briée, F.; Swain, J.; Mingeot-Leclercq, M.-P.; Décout, J.-L. Broad-spectrum antibacterial amphiphilic aminoglycosides: A new focus on the structure of the lipophilic groups extends the series of active dialkyl neamines. Eur. J. Med. Chem. 2018, 157, 1512–1525. [Google Scholar] [CrossRef]
- Baussanne, I.; Bussiere, A.; Halder, S.; Ganem-Elbaz, C.; Ouberai, M.; Riou, M.; Paris, J.M.; Ennifar, E.; Mingeot-Leclercq, M.P.; Decout, J.L. Synthesis and antimicrobial evaluation of amphiphilic neamine derivatives. J. Med. Chem. 2010, 53, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Sautrey, G.; Zimmermann, L.; Deleu, M.; Delbar, A.; Souza, M.L.; Jeannot, K.; Van, B.F.; Buyck, J.M.; Decout, J.L.; Mingeot-Leclercq, M.P. New amphiphilic neamine derivatives active against resistant Pseudomonas aeruginosa and their interactions with lipopolysaccharides. Antimicrob. Agents Chemother. 2014, 58, 4420–4430. [Google Scholar] [CrossRef]
- El Khoury, M.; Swain, J.; Sautrey, G.; Zimmermann, L.; Van Der Smissen, P.; Décout, J.-L.; Mingeot-Leclercq, M.-P. Targeting Bacterial Cardiolipin Enriched Microdomains: An Antimicrobial Strategy Used by Amphiphilic Aminoglycoside Antibiotics. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.J.; Miller, K.A.; David, S.A. Anti-Endotoxin Agents. Development of a Fluorescent Probe Displacement Method Optimized for the Rapid Identification of Lipopolysaccharide-Binding Agents. 1. Comb. Chem. High Throughput Screen. 2004, 7, 239–249. [Google Scholar] [CrossRef]
- Wood, S.J.; Miller, K.A.; David, S.A. Anti-endotoxin agents. 2. Pilot high-throughput screening for novel lipopolysaccharide-recognizing motifs in small molecules. Comb. Chem. High Throughput Screen. 2004, 7, 733–747. [Google Scholar] [CrossRef]
- Niven, G.W.; Mulholland, F. Cell membrane integrity and lysis in Lactococcus lactis: The detection of a population of permeable cells in post-logarithmic phase cultures. J. Appl. Microbiol. 1998, 84, 90–96. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef]
- Ouberai, M.; El Garch, F.; Bussiere, A.; Riou, M.; Alsteens, D.; Lins, L.; Baussanne, I.; Dufrêne, Y.; Brasseur, R.; Decout, J.-L.; et al. The Pseudomonas aeruginosa membranes: A target for a new amphiphilic aminoglycoside derivative? Biochim. Biophys. Acta BBA Biomembr. 2011, 1808, 1716–1727. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Inácio, R.G.; Raimundo, M.; Martins, M.V.; Castanho, M.; Santos, N. Biophysical characterization of polymyxin b interaction with LPS aggregates and membrane model systems. Biopolymers 2012, 98, 338–344. [Google Scholar] [CrossRef]
- Roes, S.; Seydel, U.; Gutsmann, T. Probing the Properties of Lipopolysaccharide Monolayers and Their Interaction with the Antimicrobial Peptide Polymyxin B by Atomic Force Microscopy. Langmuir 2005, 21, 6970–6978. [Google Scholar] [CrossRef]
- Moore, R.A.; Bates, N.C.; Hancock, R. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 1986, 29, 496–500. [Google Scholar] [CrossRef]
- Sousa, M.C. New antibiotics target the outer membrane of bacteria. Nature 2019, 576, 389–390. [Google Scholar] [CrossRef]
- Robinson, J.A. Folded Synthetic Peptides and Other Molecules Targeting Outer Membrane Protein Complexes in Gram-Negative Bacteria. Front. Chem. 2019, 7, 45. [Google Scholar] [CrossRef]
- Lehman, K.M.; Grabowicz, M. Countering Gram-Negative Antibiotic Resistance: Recent Progress in Disrupting the Outer Membrane with Novel Therapeutics. Antibiotics 2019, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Fosso, M.Y.; Shrestha, S.K.; Chandrika, N.T.; Dennis, E.K.; Green, K.D.; Garneau-Tsodikova, S. Differential Effects of Linkers on the Activity of Amphiphilic Tobramycin Antifungals. Molecules 2018, 23, 899. [Google Scholar] [CrossRef]
- Steinbuch, K.B.; Benhamou, R.; Levin, L.; Stein, R.; Fridman, M. Increased Degree of Unsaturation in the Lipid of Antifungal Cationic Amphiphiles Facilitates Selective Fungal Cell Disruption. ACS Infect. Dis. 2018, 4, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.K.; Fosso, M.Y.; Green, K.D.; Garneau-Tsodikova, S. Amphiphilic Tobramycin Analogues as Antibacterial and Antifungal Agents. Antimicrob. Agents Chemother. 2015, 59, 4861–4869. [Google Scholar] [CrossRef]
- Lyu, Y.; Yang, X.; Goswami, S.; Gorityala, B.K.; Idowu, T.; Domalaon, R.; Zhanel, G.G.; Shan, A.; Schweizer, F. Amphiphilic Tobramycin–Lysine Conjugates Sensitize Multidrug Resistant Gram-Negative Bacteria to Rifampicin and Minocycline. J. Med. Chem. 2017, 60, 3684–3702. [Google Scholar] [CrossRef]
- Dhondikubeer, R.; Bera, S.; Zhanel, G.G.; Schweizer, F. Antibacterial activity of amphiphilic tobramycin. J. Antibiot. 2012, 65, 495–498. [Google Scholar] [CrossRef][Green Version]
- Berkov-Zrihen, Y.; Herzog, I.M.; Benhamou, R.I.; Feldman, M.; Steinbuch, K.B.; Shaul, P.; Lerer, S.; Eldar, A.; Fridman, M. Tobramycin and Nebramine as Pseudo-oligosaccharide Scaffolds for the Development of Antimicrobial Cationic Amphiphiles. Chem. A Eur. J. 2015, 21, 4340–4349. [Google Scholar] [CrossRef]
- Yang, X.; Ammeter, D.; Idowu, T.; Domalaon, R.; Brizuela, M.; Okunnu, O.; Bi, L.; Guerrero, Y.A.; Zhanel, G.G.; Kumar, A.; et al. Amphiphilic nebramine-based hybrids Rescue legacy antibiotics from intrinsic resistance in multidrug-resistant Gram-negative bacilli. Eur. J. Med. Chem. 2019, 175, 187–200. [Google Scholar] [CrossRef]
- Mingeot-Leclercq, M.-P.; Décout, J.-L. Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides. MedChemComm 2016, 7, 586–611. [Google Scholar] [CrossRef]
- Chen, F.; Chen, G.; Liu, Y.; Jin, Y.; Cheng, Z.; Liu, Y.; Yang, L.; Jin, S.; Wu, W. Pseudomonas aeruginosa Oligoribonuclease Contributes to Tolerance to Ciprofloxacin by Regulating Pyocin Biosynthesis. Antimicrob. Agents Chemother. 2017, 61, e02256-16. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Chen, H.; Li, M.; Pan, X.; Fu, W.; Ren, H.; Chen, R.; Bai, F.; Jin, Y.; Cheng, Z.; et al. Pseudomonas aeruginosa Polynucleotide Phosphorylase Contributes to Ciprofloxacin Resistance by Regulating PrtR. Front. Microbiol. 2019, 10, 1762. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.; Wong, P.G. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 1984, 26, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Rutten, L.; Geurtsen, J.; Lambert, W.; Smolenaers, J.J.M.; Bonvin, A.M.; de Haan, A.; van der Ley, P.; Egmond, M.R.; Gros, P.; Tommassen, J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 7071–7076. [Google Scholar] [CrossRef] [PubMed]
- Simpson, B.W.; Trent, M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Genet. 2019, 17, 403–416. [Google Scholar] [CrossRef]
- Brandenburg, K.; Richter, W.; Koch, M.; Meyer, H.W.; Seydel, U. Characterization of the nonlamellar cubic and HII structures of lipid A from Salmonella enterica serovar Minnesota by X-ray diffraction and freeze-fracture electron microscopy. Chem. Phys. Lipids 1998, 91, 53–69. [Google Scholar] [CrossRef]
- Schromm, A.B.; Brandenburg, K.; Loppnow, H.; Moran, A.P.; Koch, M.H.J.; Rietschel, E.T.; Seydel, U. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. JBIC J. Biol. Inorg. Chem. 2000, 267, 2008–2013. [Google Scholar] [CrossRef]
- Seydel, U.; Hawkins, L.; Schromm, A.B.; Heine, H.; Scheel, O.; Koch, M.H.J.; Brandenburg, K. The generalized endotoxic principle. Eur. J. Immunol. 2003, 33, 1586–1592. [Google Scholar] [CrossRef]
- Sciuto, A.L.; Martorana, A.M.; Fernández-Piñar, R.; Mancone, C.; Polissi, A.; Imperi, F. Pseudomonas aeruginosa LptE is crucial for LptD assembly, cell envelope integrity, antibiotic resistance and virulence. Virulence 2018, 9, 1718–1733. [Google Scholar] [CrossRef]
- Mashburn-Warren, L.; McLean, R.; Whiteley, M. Gram-negative outer membrane vesicles: Beyond the cell surface. Geobiology 2008, 6, 214–219. [Google Scholar] [CrossRef]
- Li, A.; Schertzer, J.W.; Yong, X. Molecular conformation affects the interaction of the Pseudomonas quinolone signal with the bacterial outer membrane. J. Biol. Chem. 2019, 294, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Tegaga, G.; Sanchez-Gomez, S.; Razquin-Olazaran, I.; Kowalski, I.; Kaconis, Y.; Heinbockel, L.; Andra, J.; Schurholz, T.; Hornef, M.; Dupont, A.; et al. Bacterial cell wall compounds as promising targets of antimicrobial agents I. Antimicrobial peptides and lipopolyamines. Curr. Drug Targets 2012, 13, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Boll, J.; Tucker, A.T.; Klein, D.R.; Beltran, A.M.; Brodbelt, J.S.; Davies, B.W.; Trent, M.S. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. mBio 2015, 6, e00478-15. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Snyder, D.S.; McIntosh, T.J. The Lipopolysaccharide Barrier: Correlation of Antibiotic Susceptibility with Antibiotic Permeability and Fluorescent Probe Binding Kinetics. Biochemistry 2000, 39, 11777–11787. [Google Scholar] [CrossRef]
- Snyder, S.; Kim, D.; McIntosh, T.J. Lipopolysaccharide Bilayer Structure: Effect of Chemotype, Core Mutations, Divalent Cations, and Temperature. Biochemistry 1999, 38, 10758–10767. [Google Scholar] [CrossRef]
- Santos, D.; Pol-Fachin, L.; Lins, R.D.; Soares, T.A. Polymyxin Binding to the Bacterial Outer Membrane Reveals Cation Displacement and Increasing Membrane Curvature in Susceptible but Not in Resistant Lipopolysaccharide Chemotypes. J. Chem. Inf. Model. 2017, 57, 2181–2193. [Google Scholar] [CrossRef]
- Li, X.-Z.; Nikaido, H. Efflux-Mediated Drug Resistance in Bacteria. Drugs 2004, 64, 159–204. [Google Scholar] [CrossRef]
- Cama, J.; Henney, A.M.; Winterhalter, M. Breaching the Barrier: Quantifying Antibiotic Permeability across Gram-negative Bacterial Membranes. J. Mol. Biol. 2019, 431, 3531–3546. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayanan, R.; Tan, W.X.; Aung, T.T.; Goh, E.T.; Muruganantham, N.; Li, J.; Chang, J.Y.; Dikshit, N.; Saraswathi, P.; Lim, R.R.; et al. Branched Peptide, B2088, Disrupts the Supramolecular Organization of Lipopolysaccharides and Sensitizes the Gram-negative Bacteria. Sci. Rep. 2016, 6, 25905. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vaara, M. Polymyxin Derivatives that Sensitize Gram-Negative Bacteria to Other Antibiotics. Molecules 2019, 24, 249. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; MacNair, C.R.; Ilyas, B.; French, S.; Côté, J.-P.; Bouwman, C.; Farha, M.A.; Sieron, A.O.; Whitfield, C.; Coombes, B.K.; et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Corbett, D.; Wise, A.; Langley, T.; Skinner, K.; Trimby, E.; Birchall, S.; Dorali, A.; Sandiford, S.; Williams, J.; Warn, P.; et al. Potentiation of Antibiotic Activity by a Novel Cationic Peptide: Potency and Spectrum of Activity of SPR741. Antimicrob. Agents Chemother. 2017, 61, 00200-17. [Google Scholar] [CrossRef] [PubMed]
- Akhoundsadegh, N.; Belanger, C.; Hancock, R.E.W. Outer Membrane Interaction Kinetics of New Polymyxin B Analogs in Gram-Negative Bacilli. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Torrens, G.; Pérez-Gallego, M.; Moyà, B.; Munar-Bestard, M.; Zamorano, L.; Cabot, G.; Blázquez, J.; Ayala, J.; Oliver, A.; Juan, C. Targeting the permeability barrier and peptidoglycan recycling pathways to disarm Pseudomonas aeruginosa against the innate immune system. PLoS ONE 2017, 12, e0181932. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.; Torrens, G.; Gonzalez-Nicolau, M.; Oliver, A. Diversity and regulation of intrinsic beta-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol. Rev. 2017, 41, 781–815. [Google Scholar] [CrossRef]
- Ying, L.; Zhu, H.; Fosso, M.Y.; Garneau-Tsodikova, S.; Fredrick, K. Modified Aminoglycosides Bind Nucleic Acids in High-Molecular-Weight Complexes. Antibiotics 2020, 9, 93. [Google Scholar] [CrossRef]
- Alfindee, M.N.; Subedi, Y.P.; Grilley, M.M.; Takemoto, J.Y.; Chang, C.T. Antifungal Activities of 4″,6″-Disubstituted Amphiphilic Kanamycins. Molecules 2019, 24, 1882. [Google Scholar] [CrossRef] [PubMed]
- Howard, K.C.; Dennis, E.K.; Watt, D.S.; Garneau-Tsodikova, S. A comprehensive overview of the medicinal chemistry of antifungal drugs: Perspectives and promise. Chem. Soc. Rev. 2020, 49, 2426–2480. [Google Scholar] [CrossRef] [PubMed]
- Subedi, Y.; Roberts, P.; Grilley, M.; Takemoto, J.Y.; Chang, C.-W.T. Development of Fungal Selective Amphiphilic Kanamycin: Cost-Effective Synthesis and Use of Fluorescent Analogs for Mode of Action Investigation. ACS Infect. Dis. 2018, 5, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.; Lee, C.-R. Antimicrobial Agents That Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. J. Microbiol. Biotechnol. 2019, 29, 1–10. [Google Scholar] [CrossRef]

| Antibiotics | MICs (µg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | E. coli | |||||||||||
| ATCC 27853 | Psa.F03 a | PA22 b | PA 406 c | PA 238 d | PA 272 d | PA 307 d | PA 313 d | ATCC 25922 | PAZ505H8101 e | L8058.1 f | 06AB003 g | |
| gentamicin | <1 | >128 | 4 | <0.25 | 4 | 32 | 32 | 0.5 | 0.5/1 | 1 | 64 | >128 |
| tobramycin | <0.25 | ND | ND | ND | 1 | 8 | 16 | 0.5 | 1 | 32 | 64 | >128 |
| neomycin B | 32–64 | 128 | 32–64 | 2–4 | 64 | >128 | >128 | 4 | 1–2 | 4 | 1 | 1 |
| neamine | ≥64 | >128 | >128 | 64 | >128 | >128 | >128 | 128 | 16–32 | >128 | 32 | 8 |
| 3′,6-dinonyl neamine | 0.5 | 4 | 4 | 2 | 1 | 1 | 1 | 1 | 2–4 | 2–4 | 2–4 | 2 |
| 3′,6-di(dimethyloctyl) neamine | 0.25 | 8 | 4–8 | 2–4 | 1 | 1 | 1 | 1 | 2–4 | 1–2 | 1–2 | 1 |
| Antibiotics | MIC (µg/mL) ESBL | |||||
|---|---|---|---|---|---|---|
| Class B | Class A | Class C | Class D | |||
| P.aeruginosa VIM-2 | E. coli NDM-1 | P. aeruginosa BEL-1 and PER-1 ESBL | ESBL (CTX-M -15) S208/1R2 | P. aeruginosa Amp C | Klebsiella Oxa-48 | |
| gentamicin | >128 | >128 | 4 | 0.5 | <0.25 | <0.25 |
| tobramycin | 128 | >128 | 64 | 16 | <0.25 | 0.5 |
| neomycin B | 4 | 2 | 64 | 1 | 2 | 16 |
| neamine | 128 | 16 | >128 | 16 | 32 | >128 |
| cefotaxime | 128 | >128 | 128 | 32 | 64 | <0.25 |
| meropenem | 128 | 128 | 4 | <0.25 | 2 | 4 |
| 3,6′dinonyl neamine | 1 | 1 | 1 | 2 | 0.5 | 4 |
| 3,6′di(dimethyloctyl) neamine | 1 | 2 | 1 | 1 | 0.5 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swain, J.; Dezanet, C.; Chalhoub, H.; Auquière, M.; Kempf, J.; Décout, J.-L.; Mingeot-Leclercq, M.-P. Interest of Homodialkyl Neamine Derivatives against Resistant P. aeruginosa, E. coli, and β-Lactamases-Producing Bacteria—Effect of Alkyl Chain Length on the Interaction with LPS. Int. J. Mol. Sci. 2021, 22, 8707. https://doi.org/10.3390/ijms22168707
Swain J, Dezanet C, Chalhoub H, Auquière M, Kempf J, Décout J-L, Mingeot-Leclercq M-P. Interest of Homodialkyl Neamine Derivatives against Resistant P. aeruginosa, E. coli, and β-Lactamases-Producing Bacteria—Effect of Alkyl Chain Length on the Interaction with LPS. International Journal of Molecular Sciences. 2021; 22(16):8707. https://doi.org/10.3390/ijms22168707
Chicago/Turabian StyleSwain, Jitendriya, Clément Dezanet, Hussein Chalhoub, Marie Auquière, Julie Kempf, Jean-Luc Décout, and Marie-Paule Mingeot-Leclercq. 2021. "Interest of Homodialkyl Neamine Derivatives against Resistant P. aeruginosa, E. coli, and β-Lactamases-Producing Bacteria—Effect of Alkyl Chain Length on the Interaction with LPS" International Journal of Molecular Sciences 22, no. 16: 8707. https://doi.org/10.3390/ijms22168707
APA StyleSwain, J., Dezanet, C., Chalhoub, H., Auquière, M., Kempf, J., Décout, J.-L., & Mingeot-Leclercq, M.-P. (2021). Interest of Homodialkyl Neamine Derivatives against Resistant P. aeruginosa, E. coli, and β-Lactamases-Producing Bacteria—Effect of Alkyl Chain Length on the Interaction with LPS. International Journal of Molecular Sciences, 22(16), 8707. https://doi.org/10.3390/ijms22168707






