Abstract
Anthocyanins contribute to the quality and flavour of fruits. They are produced through the phenylpropanoid pathway, which is regulated by specific key genes that have been identified in many species. The dominant anthocyanin forms are reversibly transformed at different pH states, thus forming different colours in aqueous solutions. In plants, anthocyanins are controlled by specific factors of the biosynthetic pathway: light, temperature, phytohormones and transcription factors. Although great progress in research on anthocyanin structures and the regulation of anthocyanin biosynthesis has been made, the molecular regulatory mechanisms of anthocyanin biosynthesis in different plants remain less clear. In addition, the co-regulation of anthocyanin biosynthesis is poorly understood. In this review, we summarise previous findings on anthocyanin biosynthesis, including the biochemical and biological features of anthocyanins; differences in anthocyanin biosynthesis among fruit species, i.e., apple, red pear, and the model plant Arabidopsis thaliana; and the developmental and environmental regulation of anthocyanin accumulation. This review reveals the molecular mechanisms underlying anthocyanin biosynthesis in different plant species and provides valuable information for the development of anthocyanin-rich red-skinned and red-fleshed apple and pear varieties.
1. Introduction
The accumulation of different pigmentations affects the growth, development and reproduction of plants under various conditions, which, similar to the diversification of coat hair colour in mammals, is appropriate for different environments [1]. Anthocyanins are water-soluble flavonoid pigments widely distributed in the petals, fruits, stems and leaves of plants. [2,3,4]. In addition, anthocyanins are natural antioxidants that can strongly scavenge free radicals and reactive oxygen species (ROS) [5]. Anthocyanins have a pivotal role in the rendering of red, purple and blue colours of plant tissues and organs, and contribute to the dispersal of pollen and seeds by attracting animals such as bees and birds [6,7]. Moreover, anthocyanins are also important indicators of fruit ripening and influence the quality of fruits and their derived products (e.g., wine and fruit juices), which affects the purchasing behaviour of consumers to a certain extent [8].
In the past few decades, great progress in the understanding of anthocyanin metabolism in model plant species has been made, especially with respect to the biosynthetic pathways of these compounds. In plants, anthocyanin biosynthesis is affected by various biotic and abiotic stresses, including UV irradiation, insect attack, drought, and low temperature [9,10,11]. Additionally, the accumulation of anthocyanins is also determined by enzyme-coding structural genes, transcription factors, plant hormones and microRNAs [12,13,14,15].
Anthocyanins participate in the formation of fruit quality, affecting taste, colour and lustre, whilst even affecting human health. The biosynthesis of anthocyanins in fruit crops has attracted large amounts of attention from academics. With the development of molecular biology and bioinformatics, an increasing number of studies have revealed that there are obvious differences in the regulatory mechanisms of anthocyanin biosynthesis between different plant species. Many key players in the complex regulatory network remain to be identified. Therefore, this article aims to provide a detailed overview of the known regulatory mechanisms of anthocyanin biosynthesis in apples and pears and provide a theoretical foundation for the genetic improvement and breeding of crops, fruits and ornamental plant species [16].
2. Basic Information of Anthocyanins
2.1. Classification and Chemical Structure of Anthocyanins
Anthocyanins are important secondary metabolites belonging to the flavonoids (polyphenols). At present, more than twenty kinds of anthocyanins have been identified in nature, and approximately 90% of more than 550 kinds of anthocyanins found in nature are derived from the six most common anthocyanins: cyanidin (Cy), peonidin (Pn), pelargonidin (Pg), malvidin (Mv), delphinidin (Dp) and petunidin (Pt). Anthocyanins exist alongside a variety of monosaccharides, including glucose, rhamnose, galactose and xylose, and disaccharides consisting of rhamnose, gentian disaccharide and sophora disaccharide to form glycosides. Additionally, anthocyanins consist of α-phenylbenzopyran cations and are mainly composed of C6(A)-C3(C)-C6(B) carbon skeleton structures (Figure 1). At the A and C rings of these six anthocyanins, the 3, 5 and 7 positions connect to the hydroxyl group, while at the B ring, the hydroxyl group is connected to position 4′. Whether positions 3′ and 5′ have hydroxyl groups and are methoxylated is considered the standard for distinguishing the six anthocyanins. Due to the methylation and hydroxylation modifications at different positions of the ring, different colours of anthocyanins are formed (Table 1).
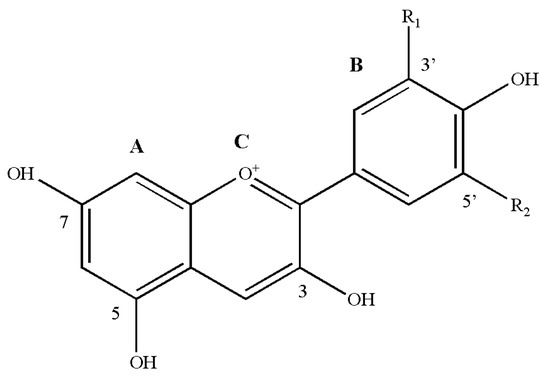
Figure 1.
Basic chemical structures of anthocyanidins. A, C6; B, C6; C, C3, anthocyanins consisting of α-phenylbenzopyran cations are mainly composed of C6-C3-C6 carbon skeleton structures. R1 and R2, structural groups at different positions (3′ and 5′) on the B ring. Detailed information on R1 and R2 substitutions is listed in Table 1.

Table 1.
Basic information on the six common anthocyanidins.
2.2. The Influence of pH on Anthocyanin Chemical Structure
The corresponding four dominant anthocyanin forms, flavylium cations, carbinol pseudobases, neutral quinonoidal bases and chalcone, are reversibly transformed at different pH states (Figure 2), and so different colours will be produced concomitantly in aqueous solutions [17]. Under acidic conditions, three chemical equilibria, including acid-base equilibrium, hydration equilibrium and ring-chain tautomeric equilibrium, were found to coexist. When the pH < 2, anthocyanin exists in the form of red flavylium cations; as the pH increases, the flavylium cation rapidly continues to hydrate to produce a colourless carbinol pseudobase at a pH from 4 to 6. Under alkaline conditions, acid-base equilibrium becomes dominant, and proton transfer reactions swiftly occur to generate products that are unstable and that easily degrade into other products. As the solution pH value varies from 6 to 8, the colours vary from purple to violet. When the pH > 8, anthocyanins exist in the form of chalcone, and a colourless solution emerges. Under the same external conditions, the higher the pH, the faster the degradation rate of anthocyanins [18]. In addition, anthocyanins are unstable in nature, approximately 65% of anthocyanins are acylated, and glycosylation and methylation promote the polymorphism of anthocyanins. The presence of metal ions, oxidative degradation, vitamin C, sugars, temperature and light also affects the stability of the anthocyanin chemical structure [19,20].
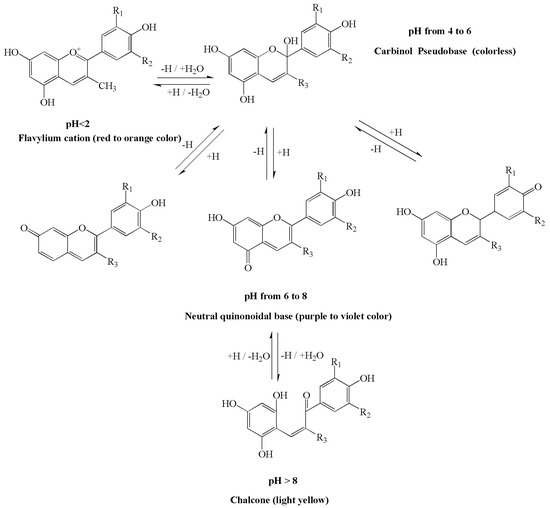
Figure 2.
Scheme of the pH-dependent structural equilibrium interconversion between the dominant anthocyanin forms. Four dominant anthocyanin forms, flavylium cation, carbinol pseudobase, neutral quinonoidal base and chalcone, can reversibly transform at different pH states and then produce different colours. pH < 2, the colour ranges from red to orange; 4 < pH < 6, shows mainly a colourless solution; 6 < pH < 8, the colours vary from purple to violet; pH > 8 a light yellow and colourless solution appears.
3. Biosynthesis Pathway of Anthocyanins
Anthocyanins are synthesised from a common phenolic precursor through the phenylpropanoid pathway on the cytoplasmic face of the endoplasmic reticulum in plants. Stable anthocyanins formed by different modifications, such as glycosylation, methylation and acylation, are transported into the vacuole, where they accumulate. As one of the end-products of the flavonoid biosynthetic pathway, proanthocyanidins are synthesised from epicatechin, which is catalysed by anthocyanidin reductase (ANR) and subsequently transported and polymerised [21]. The correlative enzymes and central regulatory factors involved in the anthocyanin biosynthetic pathway are shown in schematic models (Figure 3). In the anthocyanin biosynthetic pathway, multiple enzyme-encoding structural genes are involved as follows: upstream structural genes, such as phenylalanine ammonialyase (PAL) and 4-coumarate–CoA ligase (4CL); early biosynthetic genes (EBGs), such as chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), and flavonoid 3′5′-hydroxylase (F3′5′H); late biosynthetic genes (LBGs), such as dihydroflavonol 4-reductase (DFR), anthocyanidin synthase/leucoanthocyanidin dioxygenase (ANS/LDOX), and UDP-glucose flavonoid glucosyl transferase (UFGT); and modifying genes, such methyltransferase (MT), O-methyltransferase (OMT) and anthocyanin transferase (AT). In addition, four structural genes, transparent taste 10 (tt10), transparent taste 12 (tt12), transparent taste 13 (tt13) and transparent taste 19 (tt19), encode polyphenol oxidase (PPO), a secondary transport factor of the MATE (multidrug and toxic compound extrusion) family, H (+)-ATPase and glutathione S-transferase (GST), respectively. These proteins play significant roles in the modification, transport and oxidation of anthocyanidins [22,23,24].
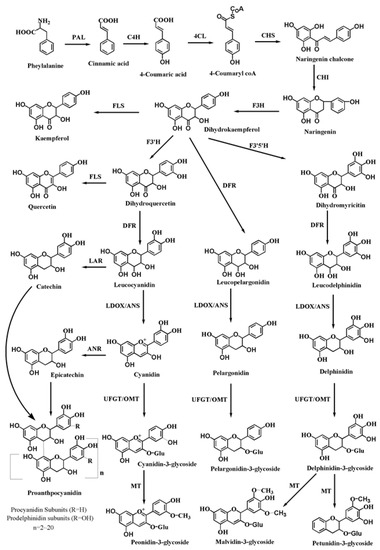
Figure 3.
The biosynthetic pathway of anthocyanins/proanthocyanidins. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′ hydroxylase; F3′5′H, flavonoid 3′5′hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; ANS, anthocyanidin synthase; LDOX, leucoanthocyanidin dioxygenase; UFGT, UDP-galactose flavonoid 3-O-galactosyltransferase; OMT, O-methyl transferase; MT, methyltransferase.
The synthesis of anthocyanins in plants is controlled by regulatory genes. At present, the identified genes involved in the regulation of the anthocyanin biosynthetic pathway consist of the myeloblastosis family (MYB) TFs, the basic helix-loop-helix (bHLH) TFs and the tryptophan-aspartic acid repeat (WDR) TFs [25,26]. Generally, the members of these three families of regulatory factors mainly depend on the MYB-bHLH-WD40 (MBW) complex to exert their effects [27,28].
4. Regulatory Mechanism of Anthocyanin Biosynthesis
The biosynthesis of anthocyanins is affected by many factors, and therefore the regulatory mechanism is complicated in plants. To date, the main factors include environmental factors, phytohormones, transcription factors and epigenetic modification. A schematic of the regulatory mechanism of the activation of anthocyanin biosynthesis is summarised briefly (Figure 4). In plants, MYB TFs are characterised by the highly conserved MYB domain, with MYB proteins usually interacting with bHLH factors and WD-repeat proteins in order to regulate anthocyanin biosynthesis [29]. Internal factors such as plant hormones, and external factors such as light, temperature, fertility, sucrose and drought, can both highly affect the transcriptional activation of target genes and the biosynthesis, accumulation and transport of anthocyanins [30,31]. Anthocyanin metabolic pathways and related gene transcription in different plant species respond to changes under different conditions.
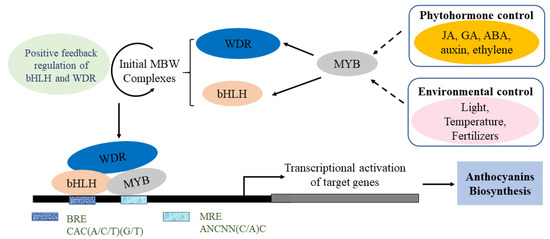
Figure 4.
Simplified model of the regulatory mechanisms of anthocyanin biosynthesis. Developmental and environmental factors could induce MYB, which then activates WDR and bHLH to form the MBW complex. The MBW complex consists of MRE (MYB recognition elements) and BRE (bHLH recognition elements), which bind to the promoter of the target gene. The transcriptional activation of target genes promotes anthocyanin biosynthesis.
4.1. Light and Temperature
Light is essential for plant growth and development, and especially affects anthocyanin biosynthesis. After photostimulation, the photoreceptors activate a range of signal transduction pathways involving photoreactions and corresponding gene expression [11]. In this process, some central light signal elements, such as constitutive photomorphogenic 1 (COP1), suppressor of PYHA (SPA) and elongated hypocotyl 5 (HY5), are involved. In general, HY5 itself can target but not activate the promoters of anthocyanin biosynthesis-related genes [32,33]. Once HY5 accumulates in abundance, it will be degraded by COP1 because of the photoinhibition of COP1 activity [34,35]. Some BBX proteins interact with HY5 and further induce anthocyanin biosynthesis [36,37]. At present, most of the research on the regulation of anthocyanin biosynthesis by light focuses on apple and pear.
In apples (Figure 5A), light negatively regulates anthocyanin biosynthesis mainly by inhibiting the expression of MdBBX37. MdBBX37 belongs to the zinc finger transcription factor family, whose members contain at least one conserved B-box motif in the N-terminal region [38,39]. MdBBX37 reduces the expression level of MdHY5 by directly targeting the promoter of MdHY5. Conversely, MdWRKY72 and MdWRKY11 transcription factors bind to the W-box cis-element of MdHY5 to activate its regulation of anthocyanin biosynthesis [20,40]. Thus, the function of BBX proteins in response to light-induced anthocyanin accumulation requires the participation of HY5 [36,37]. Moreover, as a bZIP TF and positive regulator of light signalling, MdHY5 can enhance anthocyanin biosynthesis by directly activating the expression of MdMYB1/10 [34]. However, MdBBX37 hinders the binding of MdMYB1 and MdMYB9 to their target genes and further reduces anthocyanin accumulation [41,42,43].
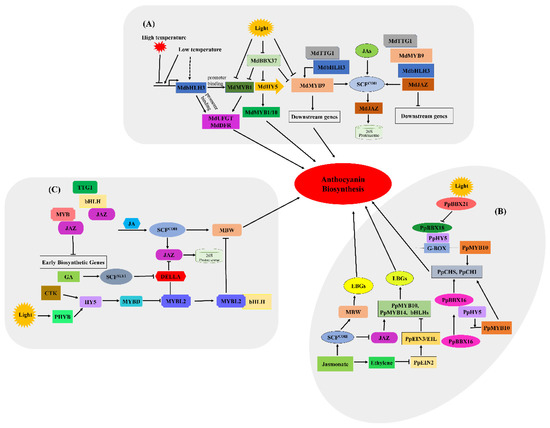
Figure 5.
Simplified model for light, temperature, phytohormones and transcription factors involved in regulating anthocyanin biosynthesis [41]. (A) Light, temperature and transcription factors involved in regulating anthocyanin biosynthesis in red-fleshed apples. (B) Phytohormones and light are involved in regulating anthocyanin biosynthesis in red pears. (C) Light and phytohormone-induced regulation of anthocyanin biosynthesis in A. thaliana.
In red pears, PpBBX16, a positive regulator of light-induced anthocyanin accumulation, cannot combine directly with the promoter of either PpMYB10 or PpCHS, but can interact with PpHY5 physically to form the PpBBX16-PpHY5 complex (Figure 5B). PpHY5 directly binds to the PpMYB10 promoter, but whether PpHY5 can activate gene expression has not been proven [44]. The PpBBX16-PpHY5 complex stimulates the promoter activity of PpMYB10 and subsequently strongly enhances light-induced anthocyanin accumulation, and the overexpression of PpBBX16 also promotes anthocyanin accumulation in the peel of pear fruits [45]. As a positive regulator, PpBBX18 directly interacts with PpHY5, and the heterodimer PpBBX18-pHY5 regulates anthocyanin accumulation by inducing PpMYB10 transcription [46]; conversely, as a negative regulator, PpBBX21 can physically interact with PpBBX18 and PpHY5, repressing anthocyanin biosynthesis by hindering the formation of the PpHY5-PpBBX18 complex [44,47]. In addition, HY5 binds to the MYBD promoter directly in A. thaliana, which positively accelerates the expression of downstream genes in light- or cytokinin-stimulated signalling pathways [40,48,49].
A large number of studies have also revealed that ambient temperature is a key factor regulating the accumulation of anthocyanins. In general, high temperature represses while low temperature promotes the biosynthesis of anthocyanin [50,51,52]. It is likely that the COP1-HY5 module may also play an important role in this process. When the temperature is elevated, COP1 is imported to nucleus and further decreases the biosynthesis of anthocyanins by destabilising HY5 [53]. In contrast, cold temperature depletes COP1 from the nucleus, resulting in HY5 stabilisation and an increase in anthocyanin production [54]. Moreover, low temperature enhances the transcription of MdbHLH3 and promotes the activity of structural genes involved in anthocyanin biosynthesis by phosphorylating MdbHLH3 [31]. Detailed regulatory mechanisms of temperature on anthocyanin biosynthesis still need to be explored.
4.2. Phytohormones
Phytohormones are indispensable for the processes of plant growth and development, and known plant hormones, such as JA (jasmonate), ABA (abscisic acid), ethylene and auxin, could stimulate the ethylene response factor family (ERF) and be involved in the regulation of anthocyanin biosynthesis [55,56,57]. In red pear and A. thaliana, JAZ (jasmonate ZIM-domain) proteins are substrates of the SCFCOI1 complex, which interferes with the transcriptional levels of WD/bHLH/MYB complexes by interacting directly with the C-terminus of both bHLH and MYB members, in turn repressing anthocyanin biosynthesis [58]. Moreover, JAZs act as negative regulatory factors in apples [59]. The direct interaction of MdJAZ with MdbHLH3 interferes with the recruitment of MdbHLH3 to the MdMYB9 promoter, which represses the transcription of the MBW complexes and further decreases anthocyanin biosynthesis [44]. In addition, ABI5 promotes ABA-induced anthocyanin biosynthesis by regulating the MYB1-bHLH3 complex in apples [60]. Ethylene inhibits anthocyanin biosynthesis by downregulating the expression of R2R3-MYBs (including PpMYB10 and PpMYB114) and LBGs (late biosynthetic genes) in red pears, and ethylene signal transduction occurs via ethylene response factors (ERFs) and EIN3/EILs (ethylene-insensitive 3, EIN3; EIN3-like, EIL) [57]. Usually, high levels of auxin can inhibit anthocyanin biosynthesis by suppressing regulatory genes and structural genes [45,61]. Cytokinin plays a positive role in photomorphogenesis in Arabidopsis thaliana, while GA signalling curbs anthocyanin biosynthesis via DELLA proteins [62,63]. DELLAs may also be essential for the function of gibberellic acids (GA) in regulating the biosynthesis of anthocyanins. In the presence of GA, DELLAs are polyubiquitinated and then degraded by the 26S proteasome [64].
4.3. Transcription Factors
The expression of structural genes during anthocyanin biosynthesis is directly controlled by the MYB-bHLH-WDR complex. R2R3-MYB transcription factors play a critical role in this regulatory pathway, which can directly regulate the expression of related genes and lead to tissue-specific anthocyanin accumulation [65,66]. BHLH transcription factors are indispensable for the activity of R2R3-MYBs, mainly by stabilising the MYB complex or promoting its transcription [29,67]. For example, some MYB transcription factors, such as MdMYB1, MdMYB9, MdMYB10 and MdMYB114, can promote apple fruit colouration by means of interaction with bHLH3 and WD40 [42,68]. In fleshy fruit skin, MdMYB1 is initially found in apple skin, and its product participates in photoinduction by activating the transcription activity of MdDFR and MdUFGT promoters. The transcript level of MdMYB1 is positively correlated with the accumulation of anthocyanin and the expression of structural genes because the MdDFR and MdUFGT promoters include light-response elements, such as ACGTs and MRE or MRE-like sequences [43,69]. Members of the WD40 protein family have 4–10 random WD repeat domains, which consist of 40 amino acid sequences ending in tryptophan (W) and aspartic acid (D). MdTTG1 was the first WD40 protein isolated from apple, similar to AtTTG1 in Arabidopsis, MdTTG1 can interact with MdbHLH3 and MdMYB9 to control the expression of downstream structural genes [70]. Similarly, PpMYB10 and PpMYB114 also contribute to the colouration of red pears. However, some MYB TFs, such as Arabidopsis MYBL2, apple MYB16, peach MYB17-20, and strawberry MYB1, negatively regulate anthocyanin biosynthesis [71,72,73]. The interaction of MYBL2 with bHLH hinders the formation of the MBW complex, and MYBL2 competitively interacts with the MYB/bHLH and bHLH subunits. Once the expression level of MYBL2 is reduced, the expression of early biosynthetic genes and the accumulation of anthocyanin increase (Figure 5C). Given that TFs from the same family have different functions in the regulation of anthocyanin biosynthesis, the functional elucidation of more TFs will be a fertile research area in the coming years.
4.4. Other Regulatory Factors
Demethylation and methylation of DNA also have important roles in regulating anthocyanin accumulation, and studies have shown that the DNA methylation inhibitor 5-azacytidine can induce red pigmentation in apple and peach fruits [74,75]. Coincidentally, in red-fleshed radish, DNA methylation of the RsMYB1 promoter inhibits anthocyanin biosynthesis [76]. In addition, microRNAs also play vital roles in the biosynthesis of anthocyanins. For example, mdm-miR828 inhibits anthocyanin accumulation in response to high temperature in apple [77], and miR156 regulates anthocyanin biosynthesis by targeting SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) in poplar and Arabidopsis [78,79,80]. Moreover, phosphorylated MYB75 is essential for light-induced Arabidopsis anthocyanin accumulation [13].
5. Conclusions and Perspectives
Anthocyanins protect against abiotic and biotic stresses in plants, and optimising anthocyanin content is regarded as the goal of breeding programmes in apples and pears. Many factors, such as light, transcription factors, phytohormones, affect the anthocyanin accumulation in plant tissues. Although HY5 and the MBW complex play core roles in anthocyanin biosynthesis in apples, pears and A. thaliana, there are still obvious differences in the regulatory mechanism of anthocyanin biosynthesis between different plants. For example, BBX proteins are a link between light signals and HY5 in apples and pears, but PHYTOCHROMES B (PHYB) functions as a “bridge” connecting light stimulus and HY5 in A. thaliana. Therefore, it is necessary to elucidate the regulatory mechanisms of anthocyanin biosynthesis in different plant species.
In addition, as follows, there are other problems that also remain to be studied: (1) although the biosynthesis and accumulation of anthocyanins are reportedly regulated by environmental factors, plant hormones, structural genes and regulatory genes, their interrelationships in this pathway need to be further explored. (2) What kind of transcription factor modifications exist in the dynamic balance of anthocyanin synthesis and degradation? Four dominant anthocyanin forms are reversibly transformed at different pH states and affect the colour of aqueous solution. However, what is the mechanism by which pH affects anthocyanin transformation? (3) Current research is limited to the molecular level, but as science and technology progresses, we can strengthen integrated genomic, phenotypic genomics, proteomic, and metabolomic research and use a variety of means to explore unknown parts of anthocyanin regulatory mechanisms and metabolic pathways in apple and pear, and then reveal relationships among anthocyanin and colour, sugar content, and other flavour attributes. In addition, to put this information in perspective, the mechanisms involved in the function of the MBW complex are highly conserved during the regulation of anthocyanin biosynthesis in higher plants, but mechanisms related to environmental and developmental factors affecting anthocyanin biosynthesis are complicated, even the information based on these mechanisms from different species is very fragmented. Thus, additional regulatory pathways and key genes and proteins during the process of anthocyanin biosynthesis in crop, ornamental and other fruit species would also be worthwhile for further investigation.
Overall, with the deepening of research, we will have a clearer understanding of the mechanism of anthocyanin synthesis and accumulation, therefore establishing a stable genetic transformation system using gene engineering technology to improve plant colour, which will provide a solid technology and information basis for crop breeding and mechanised intensive planting in the future.
Author Contributions
H.L. and Z.L. collected the literature and wrote the manuscript; G.Z. amended the manuscript; Y.W. and L.Z. took part in the literature collection. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under grant numbers 31872672 and 31470399.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Galliot, C.; Stuurman, J.; Kuhlemeier, C. The genetic dissection of floral pollination syndromes. Curr. Opin. Plant Biol. 2006, 9, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Knekt, P.; Kumpulainen, J.; Järvinen, R.; Rissanen, H.; Heliövaara, M.; Reunanen, A.; Hakulinen, T.; Aromaa, A. Flavonoid intake and risk of chronic diseases. Am. J. Clin. Nutr. 2002, 76, 560–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar]
- Harborne, J.B.; Williams, C. Anthocyanins and other flavonoids. Nat. Prod. Rep. 1998, 21, 539–573. [Google Scholar] [CrossRef]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: Chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef]
- Schaefer, H.M.; Schaefer, V.; Levey, D.J. How plant–animal interactions signal new insights in communication. Trends Ecol. Evol. 2004, 19, 577–584. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.T.; Goto-Yamamoto, N.; Hashizume, K.; Esaka, M. Expression of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci. 2006, 170, 61–69. [Google Scholar] [CrossRef]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W.; et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2019, 101, 573–589. [Google Scholar] [CrossRef]
- Ilk, N.; Ding, J.; Ihnatowicz, A.; Koornneef, M.; Reymond, M. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG-4 2 (HUA 2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP 1) and PAP 2. New Phytol. 2014, 206, 422–435. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Cheng, Y.; Peng, Z.; Han, J. Transcriptome profiling of anthocyanin-related genes reveals effects of light intensity on anthocyanin biosynthesis in red leaf lettuce. PeerJ 2018, 6, e4607. [Google Scholar] [CrossRef]
- Gonzalez, A.; Brown, M.; Hatlestad, G.; Akhavan, N.; Smith, T.; Hembd, A.; Moore, J.A.; Montes, D.; Mosley, T.; Resendez, J.; et al. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 2016, 419, 54–63. [Google Scholar] [CrossRef]
- Li, S.; Wang, W.; Gao, J.; Yin, K.; Wang, R.; Wang, C.; Petersen, M.; Mundy, J.; Qiu, J.-L. MYB75 Phosphorylation by MPK4 Is Required for Light-Induced Anthocyanin Accumulation in Arabidopsis. Plant Cell 2016, 28, 2866–2883. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Lai, B.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Identification of MicroRNAs and Their Target Genes Related to the Accumulation of Anthocyanins in Litchi chinensis by High-Throughput Sequencing and Degradome Analysis. Front. Plant Sci. 2017, 7, 758. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, T. Dietary anthocyanin-rich plants: Biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 2011, 56, 159–170. [Google Scholar] [CrossRef]
- Osawa, Y. Copigmentation of Anthocyanins. In Anthocyanins as Food Colors; Academic Press: New York, NY, USA, 1982; pp. 41–68. [Google Scholar]
- Reyes, L.F.; Cisneros-Zevallos, L. Degradation kinetics and colour of anthocyanins in aqueous extracts of purple- and red-flesh potatoes (Solanum tuberosum L.). Food Chem. 2007, 100, 885–894. [Google Scholar] [CrossRef]
- Irani, N.G.; Grotewold, E. Light-induced morphological alteration in anthocyanin-accumulating vacuoles of maize cells. BMC Plant Biol. 2005, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, Y.; Yu, L.; Jiang, H.; Guo, Z.; Xu, H.; Jiang, S.; Fang, H.; Zhang, J.; Su, M.; et al. MdWRKY11 Participates in Anthocyanin Accumulation in Red-Fleshed Apples by Affecting MYB Transcription Factors and the Photoresponse Factor MdHY5. J. Agric. Food Chem. 2019, 67, 8783–8793. [Google Scholar] [CrossRef] [PubMed]
- Villegas, D.; Handford, M.; Alcalde, J.A.; Perez-Donoso, A. Exogenous application of pectin-derived oligosaccharides to grape berries modifies anthocyanin accumulation, composition and gene expression. Plant Physiol. Biochem. 2016, 104, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, I.; Young, J.C.; Armstrong, G.; Foster, N.; Bogenschutz, N.; Cordova, T.; Peer, W.; Hazen, S.; Murphy, A.S.; Harper, J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 2649–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, S.; Shikazono, N.; Tanaka, A. Transparent Testa 19is involved in the accumulation of both anthocyanins and proanthocyanidins inArabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Routaboul, J.-M.; Dubos, C.; Beck, G.; Marquis, C.; Bidzinski, P.; Loudet, O.; Lepiniec, L. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 2012, 63, 3749–3764. [Google Scholar] [CrossRef] [Green Version]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Goswami, G.; Nath, U.K.; Park, J.-I.; Hossain, M.R.; Biswas, M.K.; Kim, H.-T.; Kim, H.R.; Nou, I.-S. Transcriptional regulation of anthocyanin biosynthesis in a high-anthocyanin resynthesized Brassica napus cultivar. J. Biol. Res. 2018, 25, 19. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Xie, X.-B.; Li, S.; Zhang, R.-F.; Zhao, J.; Chen, Y.-C.; Zhao, Q.; Yao, Y.-X.; You, C.-X.; Zhang, X.-S.; Hao, Y.-J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.-B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.-P.; Wang, X.-F.; Espley, R.; Lin-Wang, K.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An Apple B-Box Protein MdBBX37 Modulates Anthocyanin Biosynthesis and Hypocotyl Elongation Synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2019, 61, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Hoecker, U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 2017, 37, 63–69. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-Box Proteins Regulate Photomorphogenesis by Oppositely Modulating HY5 through their Diverse C-Terminal Domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef]
- Wei, C.-Q.; Chien, C.-W.; Ai, L.-F.; Zhao, J.; Zhang, Z.; Li, K.H.; Burlingame, A.L.; Sun, Y.; Wang, Z.-Y. The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J. Genet. Genom. 2016, 43, 555–563. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef] [Green Version]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Hu, J.; Fang, H.; Wang, J.; Yue, X.; Su, M.; Mao, Z.; Zou, Q.; Jiang, H.; Guo, Z.; Yu, L.; et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 2020, 292, 110377. [Google Scholar] [CrossRef]
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Liu, X.-J.; Liu, D.-D.; Xie, X.-B.; Cheng, C.-G.; Cong, P.-H.; Hao, Y.-J. MdMYB9 and MdMYB11 are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene that is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.; Walker, A.R. Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Bai, S.; Ni, J.; Yang, Q.; Zhao, Y.; Teng, Y. The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 2018, 248, 37–48. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, M.-Z.; Xie, D.-Y. Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins. Planta 2013, 239, 765–781. [Google Scholar] [CrossRef]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-Induced Basic/Helix-Loop-Helix64 Enhances Anthocyanin Biosynthesis and Undergoes Constitutively Photomorphogenic1-Mediated Degradation in Pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef] [PubMed]
- Abbas, N.; Maurya, J.P.; Senapati, D.; Gangappa, S.; Chattopadhyay, S. Arabidopsis CAM7 and HY5 Physically Interact and Directly Bind to the HY5 Promoter to Regulate Its Expression and Thereby Promote Photomorphogenesis. Plant Cell 2014, 26, 1036–1052. [Google Scholar] [CrossRef] [Green Version]
- Cluis, C.P.; Mouchel, C.F.; Hardtke, C.S. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004, 38, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Feng, X.F.; Li, W.; Li, K. High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome 2019, 62, 503–512. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef] [Green Version]
- Sra, B.; Jhh, A.; Jgc, A.; Jhj, A.; Skl, A.; Hjlb, C. High temperature at veraison inhibits anthocyanin biosynthesis in berry skins during ripening in ’Kyoho’ grapevines. Plant Physiol. Biochem. 2020, 157, 219–228. [Google Scholar]
- Park, Y.-J.; Lee, H.; Ha, J.; Kim, J.Y.; Park, C. COP 1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 2017, 215, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Catalá, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16475–16480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Liu, H.-T.; Zhao, G.-P.; Song, J.-X.; Wang, X.-L.; Yang, C.-Q.; Zhai, R.; Wang, Z.-G.; Ma, F.-W.; Xu, L.-F.; et al. Jasmonate and Ethylene-Regulated Ethylene Response Factor 22 Promotes Lanolin-Induced Anthocyanin Biosynthesis in ‘Zaosu’ Pear (Pyrus bretschneideri Rehd.) Fruit. Biomolecules 2020, 10, 278. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM Domain Proteins Interact with the R2R3-MYB Transcription Factors MYB21 and MYB24 to Affect Jasmonate-Regulated Stamen Development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, Å.; Qian, M.; Li, J.; Li, Y.; Shen, J.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007, 19, 2470–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Liu, Y.J.; Wang, X.F.; You, C.X.; Hao, Y.J. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef]
- Ji, X.H.; Wang, Y.T.; Zhang, R. Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f.niedzwetzkyana). Plant Cell Tissue Organ Cult. (PCTOC) 2015, 120, 325–337. [Google Scholar] [CrossRef]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lochmanová, G.; Zdráhal, Z.; Konecná, H.; Koukalová, S.; Malbeck, J.; Soucek, P.; Válková, M.; Kiran, N.S.; Brzobohaty, B. Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: A proteomic analysis. J. Exp. Bot. 2008, 59, 3705–3719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant. 2015, 8, 689–708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Sun, Q.; Zhang, T.; Liu, W.; Wang, N.; Chen, X. MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4-like in apple fruit. J. Plant Physiol. 2021, 257, 153353. [Google Scholar] [CrossRef]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef] [Green Version]
- Brueggemann, J.; Weisshaar, B.; Sagasser, M. A WD40-repeat gene from Malus x domestica is a functional homologue of Arabidopsis thaliana Transparent Testa Glabra1. Plant Cell. Rep. 2010, 29, 285–294. [Google Scholar] [CrossRef]
- Aharoni, A.; De Vos, C.H.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Wang, Y.; Sun, H.; Sun, L.; Zhang, L. Transposon-induced methylation of the RsMYB1 promoter disturbs anthocyanin accumulation in red-fleshed radish. J. Exp. Bot. 2020, 71, 2537–2550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yang, H.J.; Yang, Y.Z.; Zhu, Z.Z.; Li, Y.N.; Qu, D.; Zhao, Z.Y. mdm-miR828 Participates in the Feedback Loop to Regulate Anthocyanin Accumulation in Apple Peel. Front. Plant Sci. 2020, 11, 608109. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Tang, R.; Shi, X.; Wang, W.; Cao, Q.; Liu, X.; Wang, T.; Sun, Y.; Zhang, H.; Li, R.; et al. Uncovering anthocyanin biosynthesis related microRNAs and their target genes by small RNA and degradome sequencing in tuberous roots of sweetpotato. BMC Plant Biol. 2019, 19, 232. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).