Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon
Abstract
:1. Introduction
2. Results
2.1. Identification, Annotation, and Uneven Chromosomal Distribution of Watermelon SWEET Genes
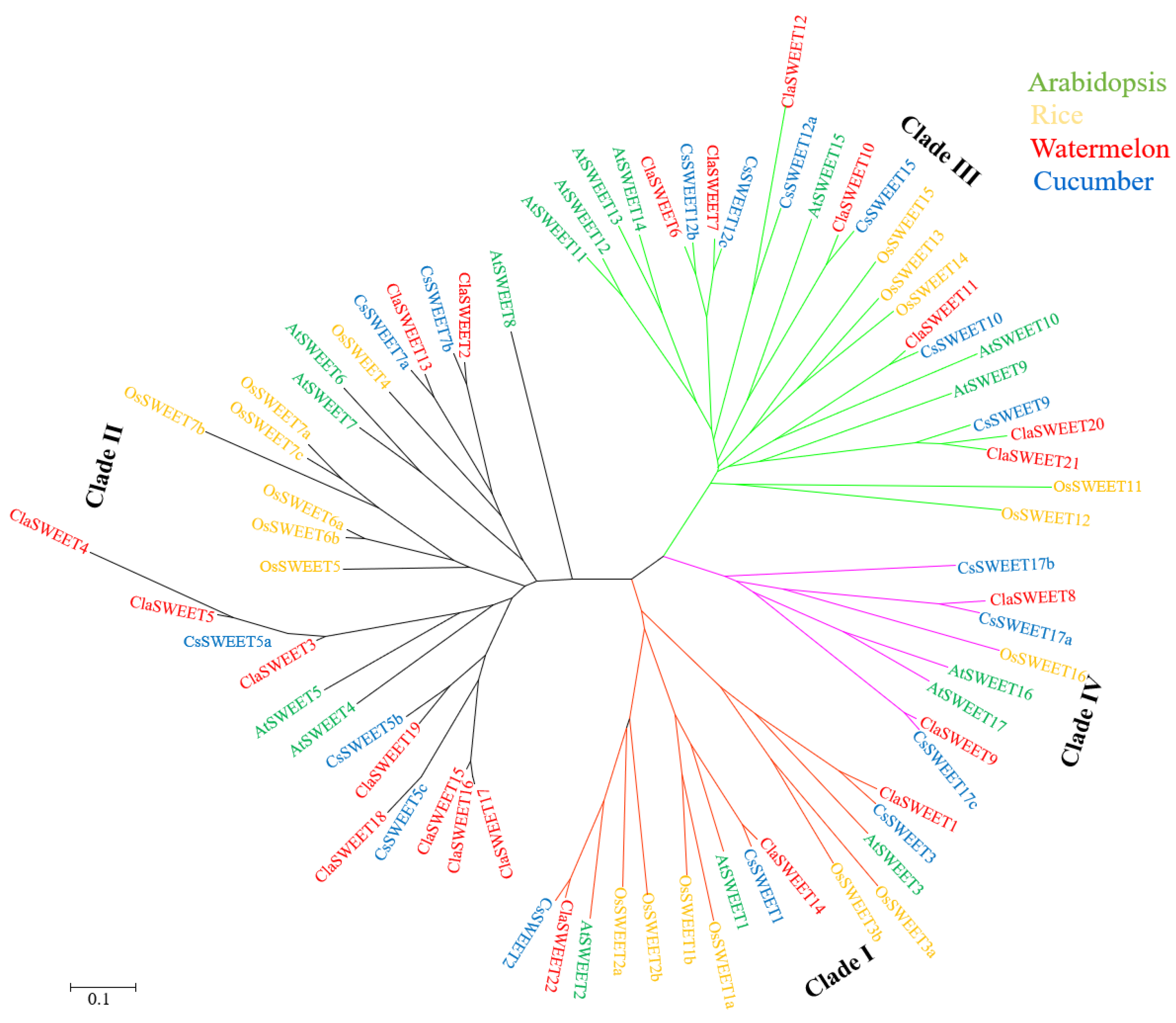
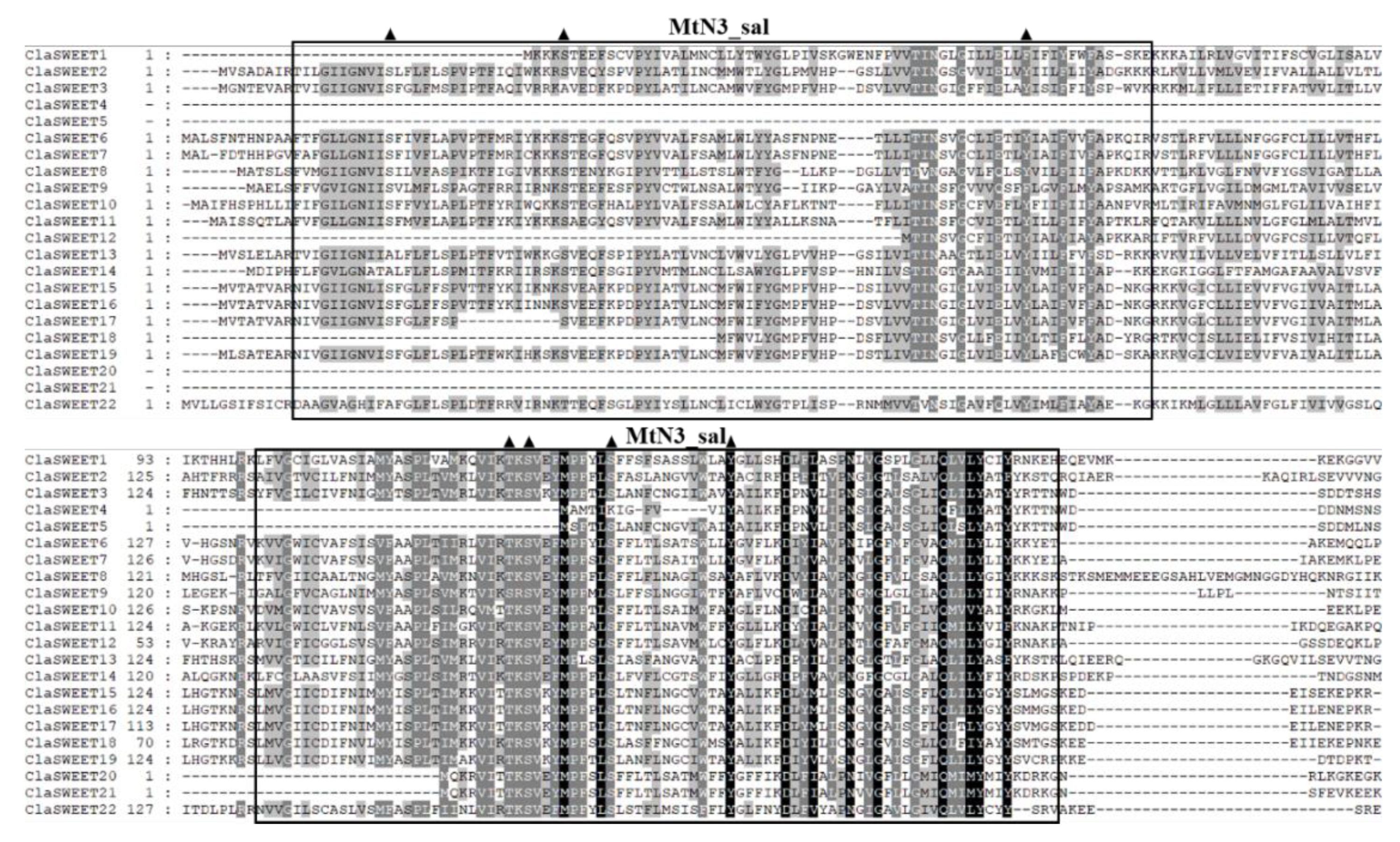
2.2. Phylogenetic and Conserved Domain Analysis of ClaSWEET Genes
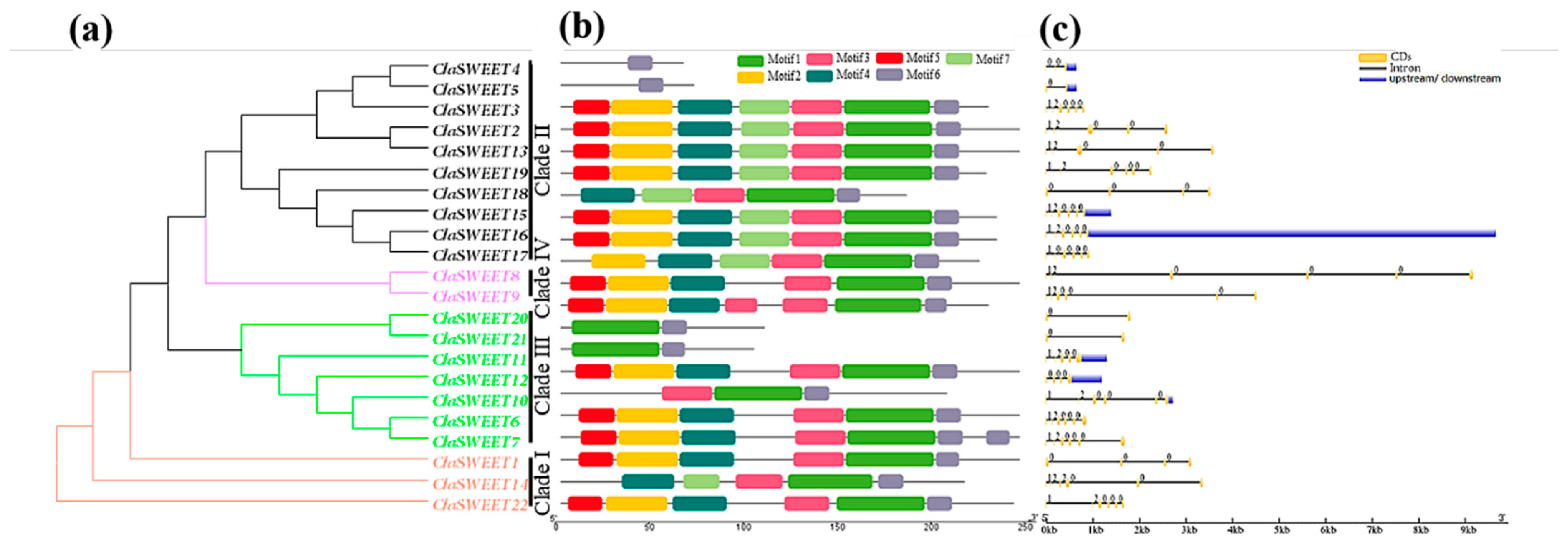
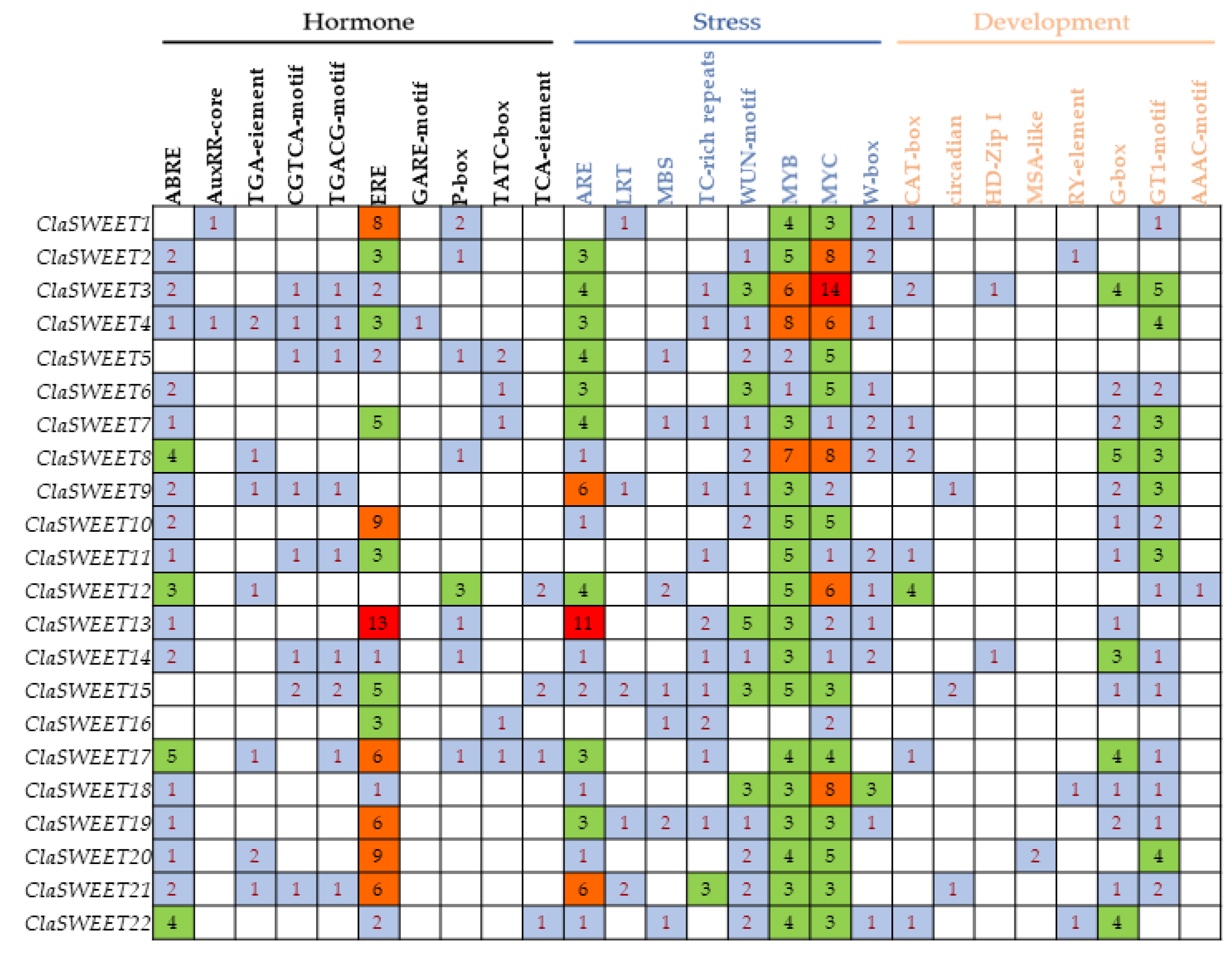
2.3. Gene Structure and Cis-Regulatory Elements Prediction of ClaSWEET Genes
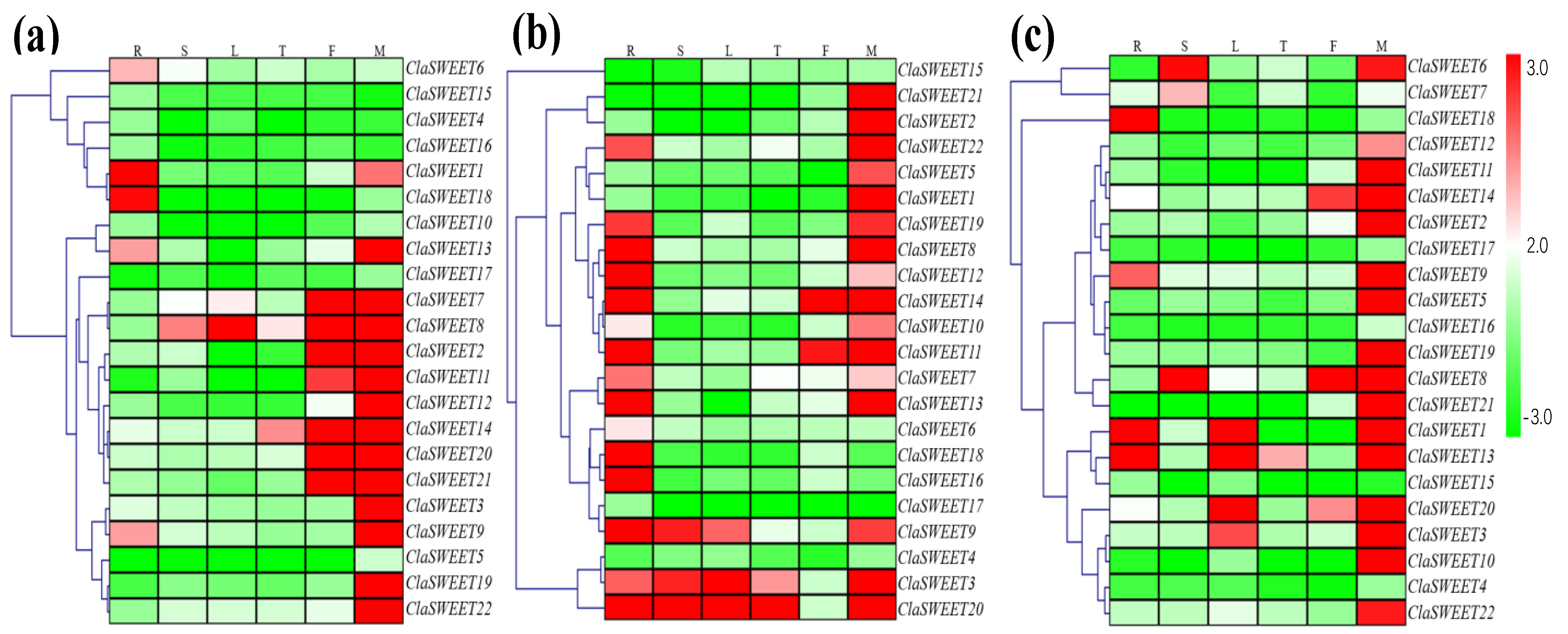
2.4. Expression Patterns of ClaSWEET Genes in Cultivars and Wild Varieties of Watermelon
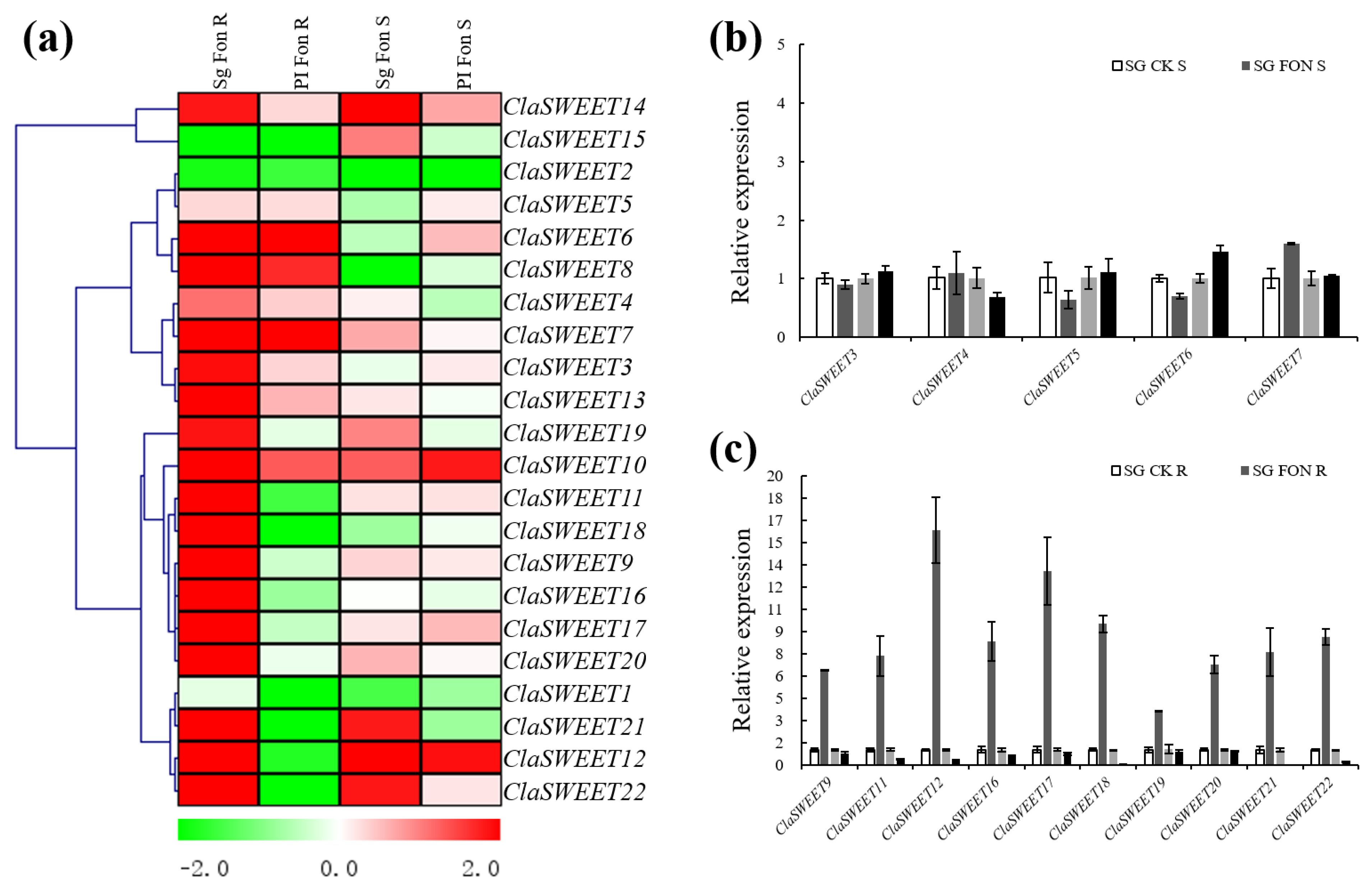
2.5. Expression Patterns of SWEET Genes in Response to Stress Treatment with Fon Infection
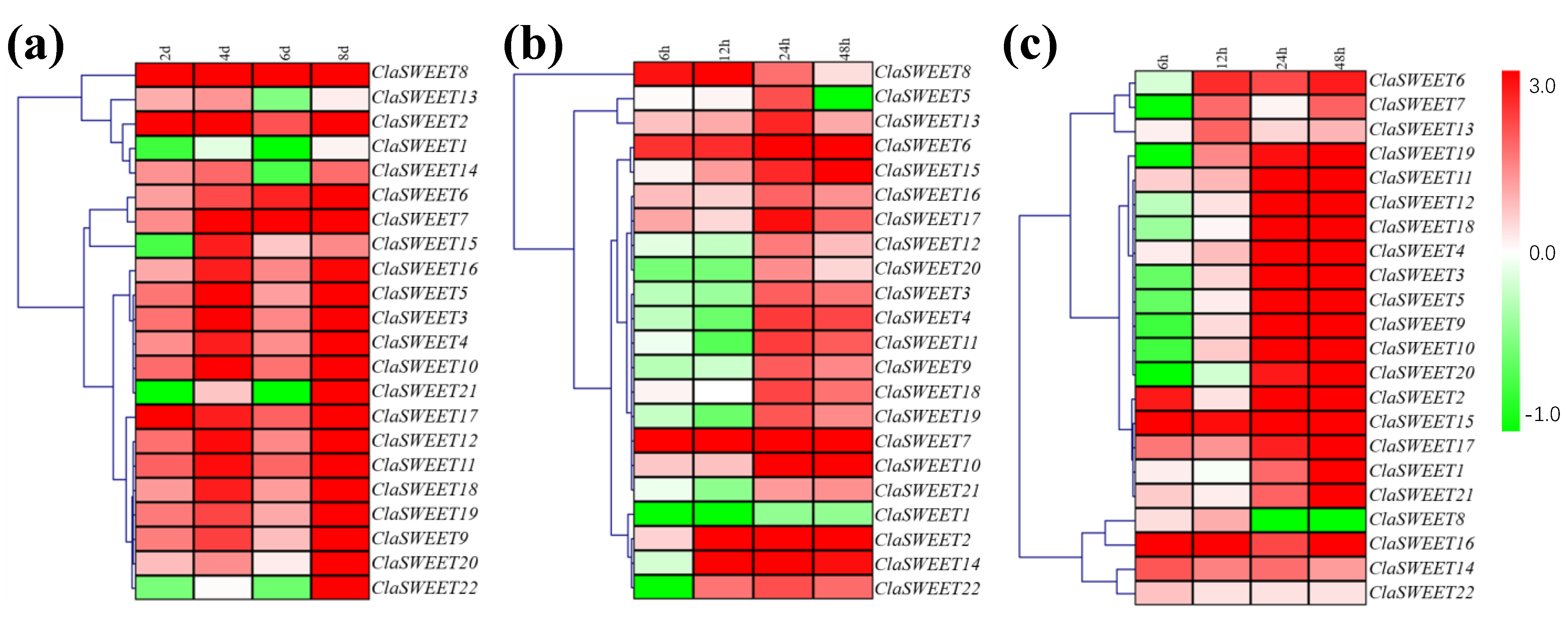
2.6. Induced Expression of SWEET Genes in Response to Low-Temperature, Salt, and Drought Stress
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and Domain Confirmation of Watermelon SWEET genes
4.2. Gene Structure, Cis-Regulatory Element, Protein Properties, and Phylogenetic Analysis
4.3. Plant Cultivation and Treatments
4.4. RNA Extraction and Quantitative RT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SWEET | Sugars Will Eventually be Exported Transporter |
| SUTs | sucrose transporters |
| MSTs | monosaccharide transporters |
| TMDs | transmembrane domains |
| MFS | major facilitator superfamily |
| GA | gibberellin |
| BR | brassinosteroid |
| ABA | abscisic acid |
| htp | hour post treament |
| dtp | day post treament |
References
- Tao, Y.; Cheung, L.S.; Li, S.; Eom, J.S.; Chen, L.Q.; Xu, Y.; Perry, K.; Frommer, W.B.; Feng, L. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 2015, 527, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Niu, K.; Ma, H. Identification and expression analysis of the SWEET gene family from Poa pratensis under abiotic stresses. DNA Cell Biol. 2020, 39, 1606–1620. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Xiao, D.; Zhang, C.; Hou, X. The expanded SWEET gene family following whole genome triplication in Brassica rapa. Genes 2019, 10, 722. [Google Scholar] [CrossRef] [Green Version]
- Bisson, L.F.; Fan, Q.W.; Walker, G.A. Sugar and glycerol transport in Saccharomyces cerevisiae. In Yeast Membrane Transport; Ramos, J., Sychrová, H., Kschischo, M., Eds.; Springer: Cham, Switzerland, 2016; Volume 892, pp. 125–168. [Google Scholar] [CrossRef]
- Fan, R.C.; Peng, C.C.; Xu, Y.H.; Wang, X.F.; Li, Y.; Shang, Y.; Du, S.Y.; Zhao, R.; Zhang, X.Y.; Zhang, L.Y.; et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 2009, 150, 1880–1901. [Google Scholar] [CrossRef] [Green Version]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, R.A.; Frommer, B.A. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 3536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, M.; Wang, S.P. Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, D.P.; Zhu, X.F.; Li, T.Y.; Jia, B.L.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.P.; Zhang, F.; Song, S.H.; Tang, X.W.; Xu, H.; Liu, G.M.; Wang, Y.G.; He, H.J. Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J. Integr. Agric. 2017, 16, 1486–1501. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Sui, J.L.; Xiao, X.H.; Qi, J.Y.; Fang, Y.J.; Tang, C.R. The SWEET gene family in Hevea brasiliensis—Its evolution and expression compared with four other plant species. FEBS Open Bio 2017, 7, 1943–1959. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 127. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Piron, M.C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Liu, F.; Chen, C.; Ma, F.; Li, M. The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [Green Version]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Geros, H.; Granell, A. Plant SWEETs: From sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Hu, B.; Wu, H.; Huang, W.; Song, J.; Zhou, Y.; Lin, Y. SWEET gene family in Medicago truncatula: Genome-wide identification, expression and substrate specificity analysis. Plants 2019, 8, 338. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londono, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.L.; Ma, J.F.; Shou, H. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.D.; Liu, S.L.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.J.; Hogan, P.; Sundaresan, V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 2005, 139, 1853–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyama, E.N.; Strope, P.K.; Opiyo, S.O.; Chen, Z.; Jones, A.M. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006, 7, R96. [Google Scholar] [CrossRef] [Green Version]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Andres, F.; Kinoshita, A.; Kalluri, N.; Fernandez, V.; Falavigna, V.S.; Cruz, T.M.D.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef] [Green Version]
- Verdier, V.; Triplett, L.R.; Hummel, A.W.; Corral, R.; Cernadas, R.A.; Schmidt, C.L.; Bogdanove, A.J.; Leach, J.E. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 2012, 196, 1197–1207. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Zhang, H.; Zhang, Q.; Zhai, H.; Liu, Q.; He, S. The plasma membrane-localized sucrose transporter IbSWEET10 contributes to the resistance of sweet potato to Fusarium oxysporum. Front. Plant Sci. 2017, 8, 197. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmulling, T.; Parniske, M.; Ludwig-Muller, J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant-Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, S.; Galletti, R.; Denoux, C.; De Lorenzo, G.; Ausubel, F.M.; Dewdney, J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007, 144, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Morii, M.; Sugihara, A.; Takehara, S.; Kanno, Y.; Kawai, K.; Hobo, T.; Hattori, M.; Yoshimura, H.; Seo, M.; Ueguchi-Tanaka, M. The dual function of OsSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice. Plant Cell Physiol. 2020, 61, 1935–1945. [Google Scholar] [CrossRef]
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1464. [Google Scholar] [CrossRef] [Green Version]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef] [PubMed]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2020, 171, 620–637. [Google Scholar] [CrossRef]
- Le Hir, R.; Spinner, L.; Klemens, P.A.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Gery, C.; et al. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Park, J.M.; Kang, S.K.; Kim, S.G.; Park, C.M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef]
- Yao, L.N.; Ding, C.Q.; Hao, X.Y.; Zeng, J.M.; Yang, Y.J.; Wang, X.C.; Wang, L. CsSWEET1a and CsSWEET17 mediate growth and freezing tolerance by promoting sugar transport across the plasma membrane. Plant Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Pfrunder, S.; Yu, Y.C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef] [Green Version]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, 3685–3694. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ren, Z.; Wang, Z.; Sun, K.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Song, C.; Zhou, X.; et al. Evolution and stress responses of Gossypium hirsutum SWEET genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef] [Green Version]
- Li, J.M.; Qin, M.F.; Qiao, X.; Cheng, Y.S.; Li, X.L.; Zhang, H.P.; Wu, J. A new insight into the evolution and functional divergence of SWEET transporters in chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W.; Honys, D.; Ward, J.M.; Padmanaban, S.; Nawrocki, E.P.; Hirschi, K.D.; Twell, D.; Sze, H. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 2006, 140, 1151–1168. [Google Scholar] [CrossRef] [Green Version]
- Branham, S.E.; Levi, A.; Farnham, M.W.; Patrick Wechter, W. A GBS-SNP-based linkage map and quantitative trait loci (QTL) associated with resistance to Fusarium oxysporum f. sp. niveum race 2 identified in Citrullus lanatus var. citroides. Theor. Appl. Genet. 2017, 130, 319–330. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020, 39, 589–595. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755. [Google Scholar] [CrossRef]
- Zhang, G.; Ding, Q.; Wei, B. Genome-wide identification of superoxide dismutase gene families and their expression patterns under low-temperature, salt and osmotic stresses in watermelon and melon. 3 Biotech 2021, 11, 194. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, R.; Yang, X.; Zhu, C.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Comparative analysis of calcium-dependent protein kinase in cucurbitaceae and expression studies in watermelon. Int. J. Mol. Sci. 2019, 20, 2527. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative Analysis, characterization and evolutionary study of dirigent gene family in cucurbitaceae and expression of novel dirigent peptide against powdery mildew stress. Genes 2021, 12, 326. [Google Scholar] [CrossRef] [PubMed]
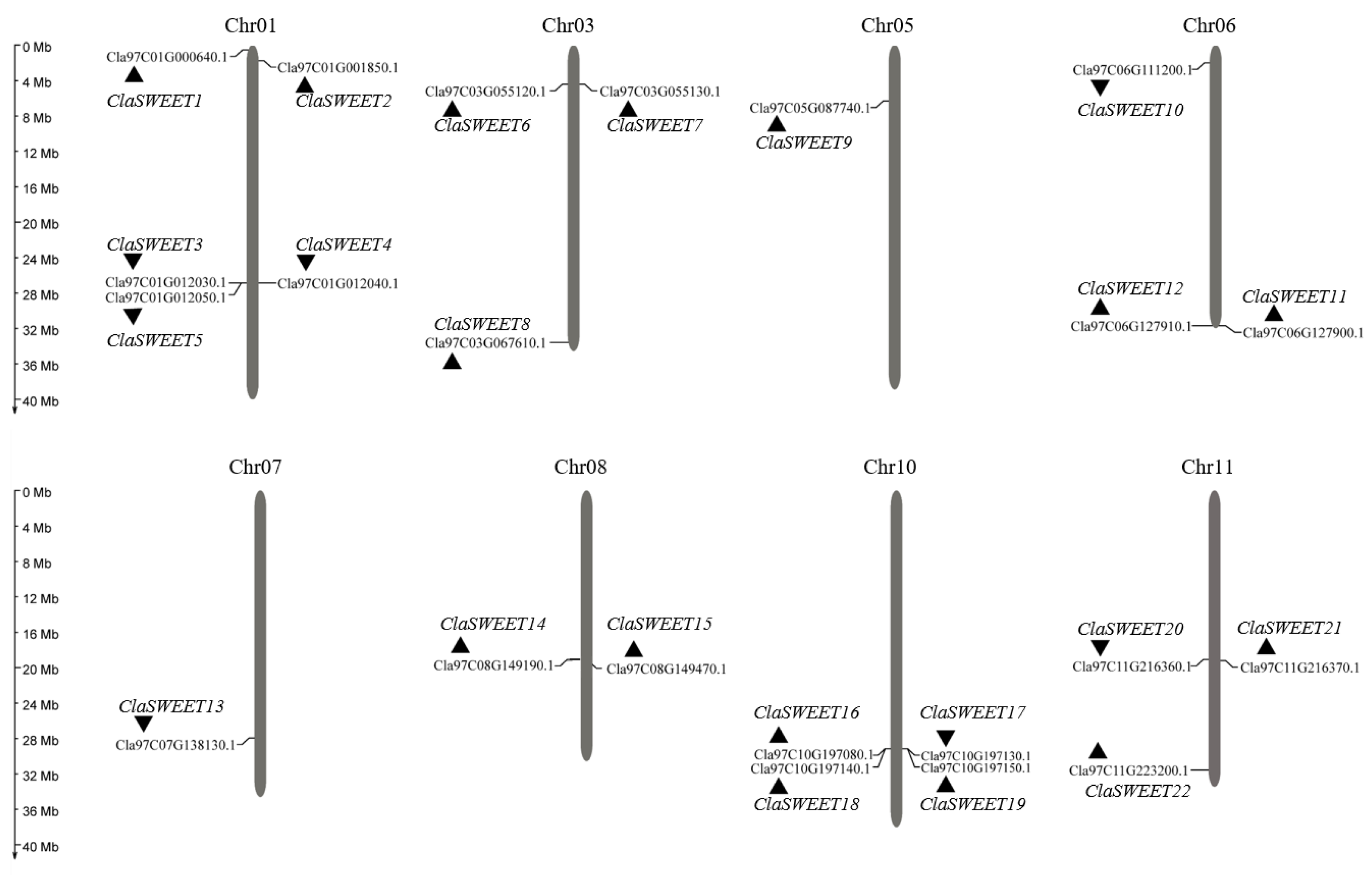
| Arabidopsis Homologous | Gene Name | Gene ID | Location (Strand) | CDS (bp) 1 | TMDs 2 | Protein Length (aa) | pI 3 | MWs (kDa) 3 | E-Value | Subcellular Localization 4 |
|---|---|---|---|---|---|---|---|---|---|---|
| AtSWEET3/AT5G53190.1 | ClaSWEET1 | Cla97C01G000640.1 | Chr:01 436923-440033 (−) | 675 | 6 | 224 | 9.12 | 25.23 | 6.17 × 10−83 | PM |
| AtSWEET7/AT4G10850.1 | ClaSWEET2 | Cla97C01G001850.1 | Chr:01 1665964-1668736 (−) | 795 | 6 | 264 | 9.83 | 29.1 | 3.96 × 10−92 | PM |
| AtSWEET5/AT5G62850.1 | ClaSWEET3 | Cla97C01G012030.1 | Chr:01 24835894-24837119 (+) | 714 | 7 | 237 | 9.11 | 26.89 | 1.15 × 10−105 | PM |
| AtSWEET5/AT5G62850.1 | ClaSWEET4 | Cla97C01G012040.1 | Chr:01 24846574-24847102 (+) | 207 | 1 | 68 | 4.85 | 7.66 | 5.33 × 10−18 | EX |
| AtSWEET5/AT5G62850.1 | ClaSWEET5 | Cla97C01G012050.1 | Chr:01 24854037-24854591 (+) | 225 | 2 | 74 | 4.46 | 8.29 | 9.12 × 10−26 | CY |
| AtSWEET12/AT5G23660.1 | ClaSWEET6 | Cla97C03G055120.1 | Chr:03 4110250-4112328 (−) | 885 | 7 | 294 | 8.36 | 33.02 | 6.10 × 10−105 | PM |
| AtSWEET12/AT5G23660.1 | ClaSWEET7 | Cla97C03G055130.1 | Chr:03 4123823-4125227 (−) | 900 | 7 | 299 | 6.5 | 33.68 | 1.28 × 10−108 | ER |
| AtSWEET17/AT4G15920.1 | ClaSWEET8 | Cla97C03G067610.1 | Chr:03 31033031-31041340 (−) | 879 | 7 | 292 | 9.3 | 32.13 | 2.66 × 10−62 | PM |
| AtSWEET17/AT4G15920.1 | ClaSWEET9 | Cla97C05G087740.1 | Chr:09 5868309-5872614 (−) | 714 | 7 | 237 | 8.96 | 26.13 | 1.13 × 10−73 | TM |
| AtSWEET15/AT5G13170.1 | ClaSWEET10 | Cla97C06G111200.1 | Chr:06 1891143-1893938 (+) | 804 | 7 | 267 | 9.02 | 30.26 | 8.57 × 10−96 | PM |
| AtSWEET12/AT5G23660.1 | ClaSWEET11 | Cla97C06G127900.1 | Chr:06 29274606-29275898 (−) | 870 | 7 | 289 | 9.14 | 32.51 | 3.20 × 10−95 | PM |
| AtSWEET11/AT3G48740.1 | ClaSWEET12 | Cla97C06G127910.1 | Chr:06 29284604-29285549 (−) | 645 | 5 | 214 | 8.65 | 23.91 | 2.97 × 10−62 | PM |
| AtSWEET7/AT4G10850.1 | ClaSWEET13 | Cla97C07G138130.1 | Chr:07 25774648-25778233 (+) | 774 | 7 | 257 | 9.13 | 28.45 | 6.01 × 10−95 | PM |
| AtSWEET1/AT1G21460.1 | ClaSWEET14 | Cla97C08G149190.1 | Chr:08 17567657-17571032 (−) | 756 | 7 | 251 | 9.25 | 27.629 | 3.61 × 10−123 | PM |
| AtSWEET4/AT3G28007.1 | ClaSWEET15 | Cla97C08G149470.1 | Chr:08 17854068-17855319 (−) | 729 | 7 | 242 | 9.27 | 26.94 | 7.34 × 10−95 | PM |
| AtSWEET4/AT3G28007.1 | ClaSWEET16 | Cla97C10G197080.1 | Chr:10 26877106-26878414 (−) | 729 | 7 | 242 | 8.95 | 27.05 | 1.07 × 10−94 | PM |
| AtSWEET4/AT3G28007.1 | ClaSWEET17 | Cla97C10G197130.1 | Chr:10 26899001-26900306 (+) | 699 | 7 | 232 | 8.28 | 25.77 | 2.19 × 10−88 | PM |
| AtSWEET5/AT5G62850.1 | ClaSWEET18 | Cla97C10G197140.1 | Chr:10 26903294-26906670 (−) | 579 | 5 | 192 | 8.53 | 21.99 | 9.81 × 10−60 | PM |
| AtSWEET5/AT5G62850.1 | ClaSWEET19 | Cla97C10G197150.1 | Chr:10 26918854-26921273 (−) | 711 | 7 | 236 | 9.07 | 26.24 | 1.88 × 10−95 | PM |
| AtSWEET15/AT5G13170.1 | ClaSWEET20 | Cla97C11G216360.1 | Chr:11 17582166-17583939 (+) | 342 | 2 | 113 | 9.73 | 13.06 | 2.60 × 10−26 | CTM |
| AtSWEET15/AT5G13170.1 | ClaSWEET21 | Cla97C11G216370.1 | Chr:11 17709586-17711235 (−) | 324 | 2 | 107 | 5.82 | 12.63 | 1.91 × 10−28 | CTM |
| AtSWEET2/AT3G14770.1 | ClaSWEET22 | Cla97C11G223200.1 | Chr:11 29080934-29082856 (−) | 702 | 7 | 233 | 9.22 | 26.02 | 1.07 × 10−115 | PM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xuan, C.; Lan, G.; Si, F.; Zeng, Z.; Wang, C.; Yadav, V.; Wei, C.; Zhang, X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. Int. J. Mol. Sci. 2021, 22, 8407. https://doi.org/10.3390/ijms22168407
Xuan C, Lan G, Si F, Zeng Z, Wang C, Yadav V, Wei C, Zhang X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. International Journal of Molecular Sciences. 2021; 22(16):8407. https://doi.org/10.3390/ijms22168407
Chicago/Turabian StyleXuan, Changqing, Guangpu Lan, Fengfei Si, Zhilong Zeng, Chunxia Wang, Vivek Yadav, Chunhua Wei, and Xian Zhang. 2021. "Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon" International Journal of Molecular Sciences 22, no. 16: 8407. https://doi.org/10.3390/ijms22168407
APA StyleXuan, C., Lan, G., Si, F., Zeng, Z., Wang, C., Yadav, V., Wei, C., & Zhang, X. (2021). Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. International Journal of Molecular Sciences, 22(16), 8407. https://doi.org/10.3390/ijms22168407







