Bovine Satellite Cells Isolated after 2 and 5 Days of Tissue Storage Maintain the Proliferative and Myogenic Capacity Needed for Cultured Meat Production
Abstract
:1. Introduction
2. Result
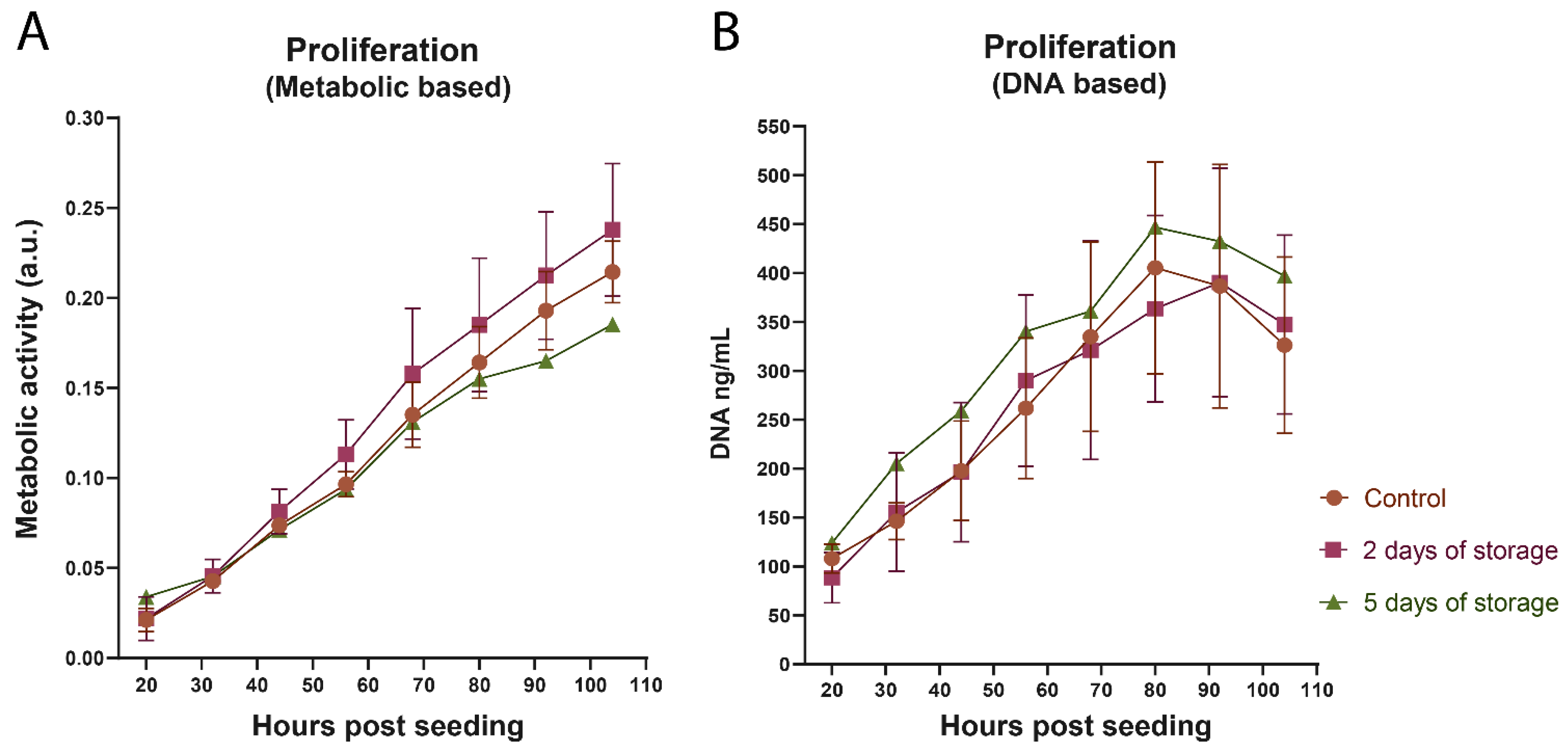
2.1. Proliferation of Bovine Satellite Cells Isolated from Fresh and Stored Muscle Tissue
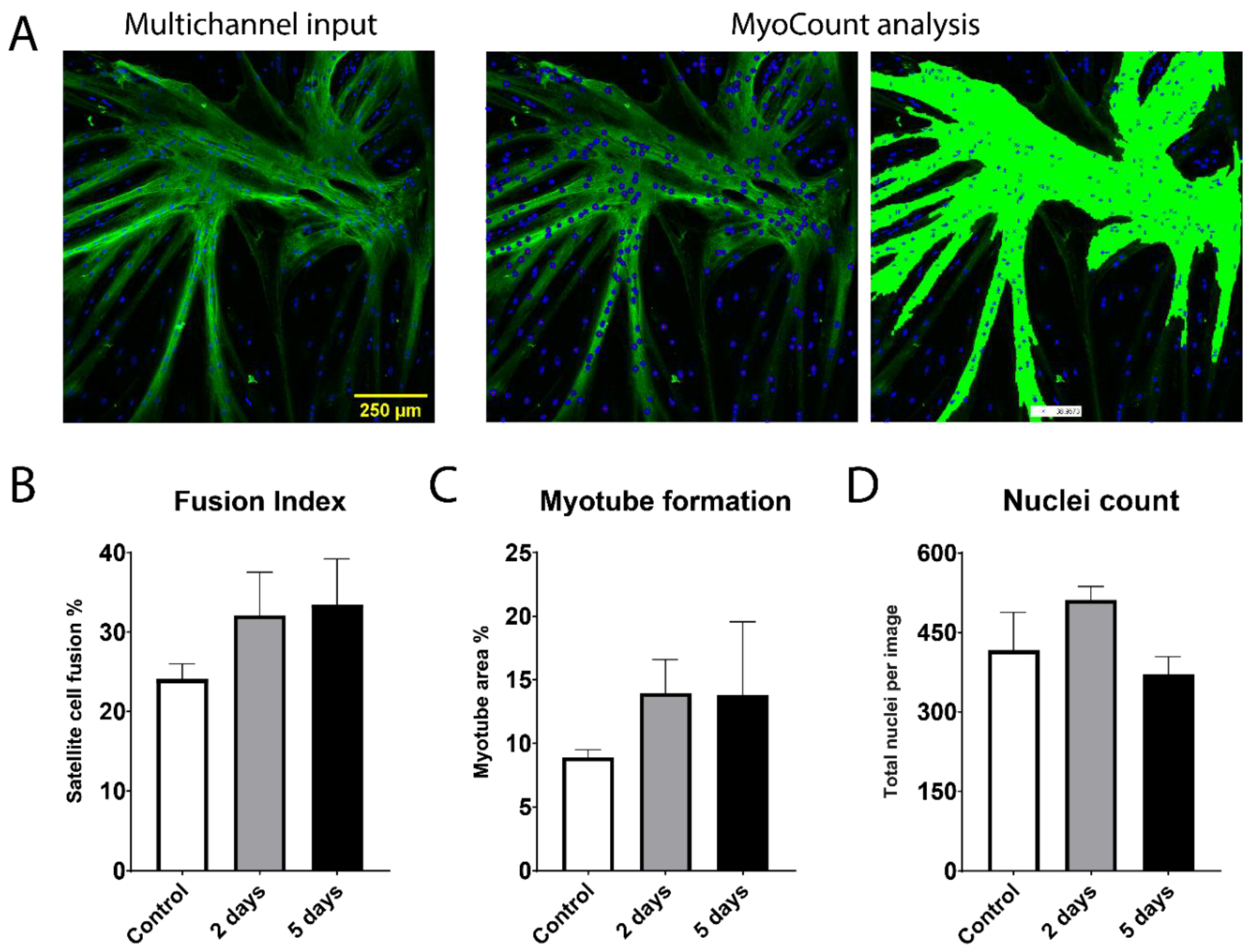
2.2. Fusion Potential of Bovine Satellite Cells Isolated from Fresh and Stored Muscle Tissue
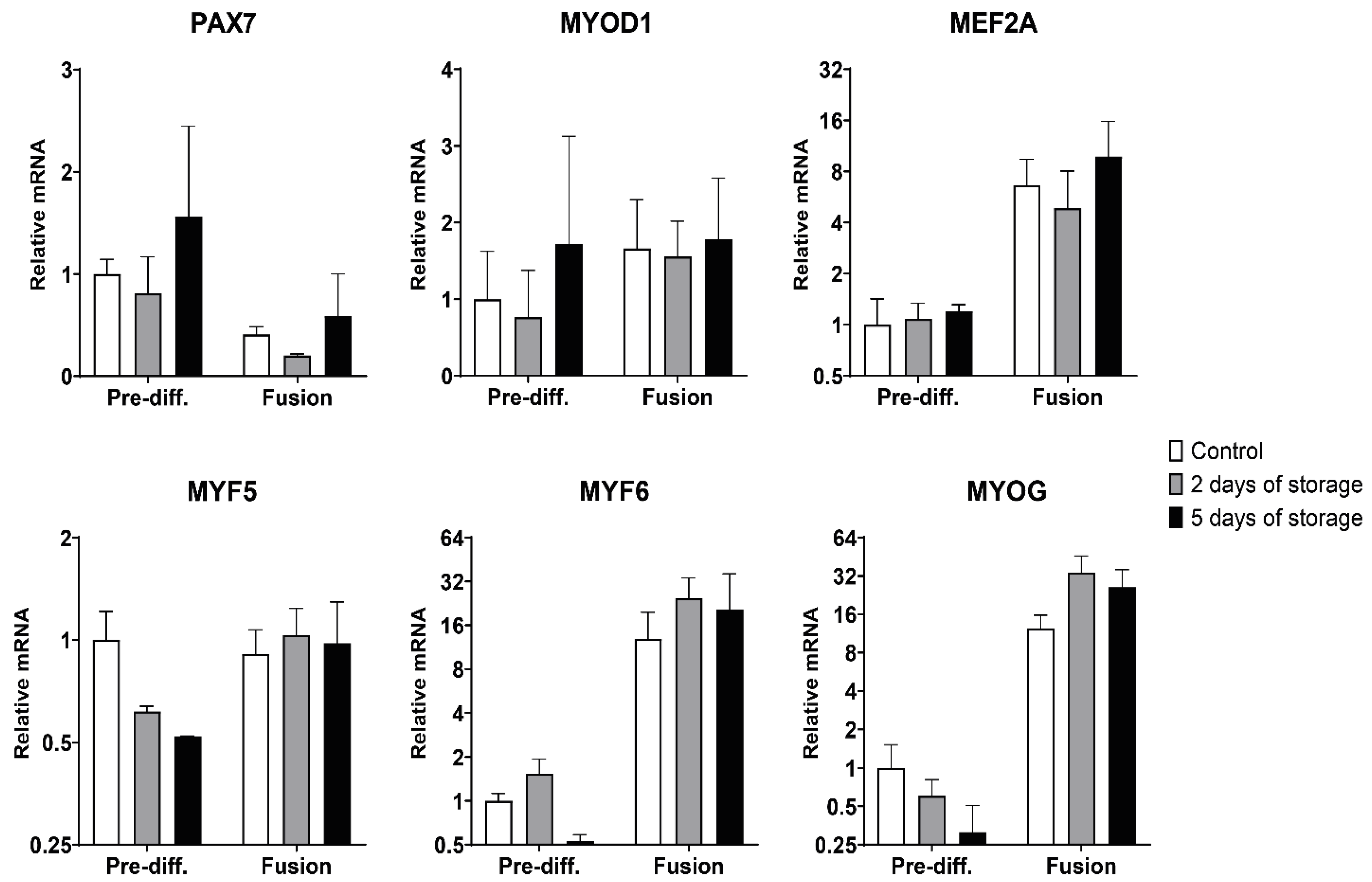
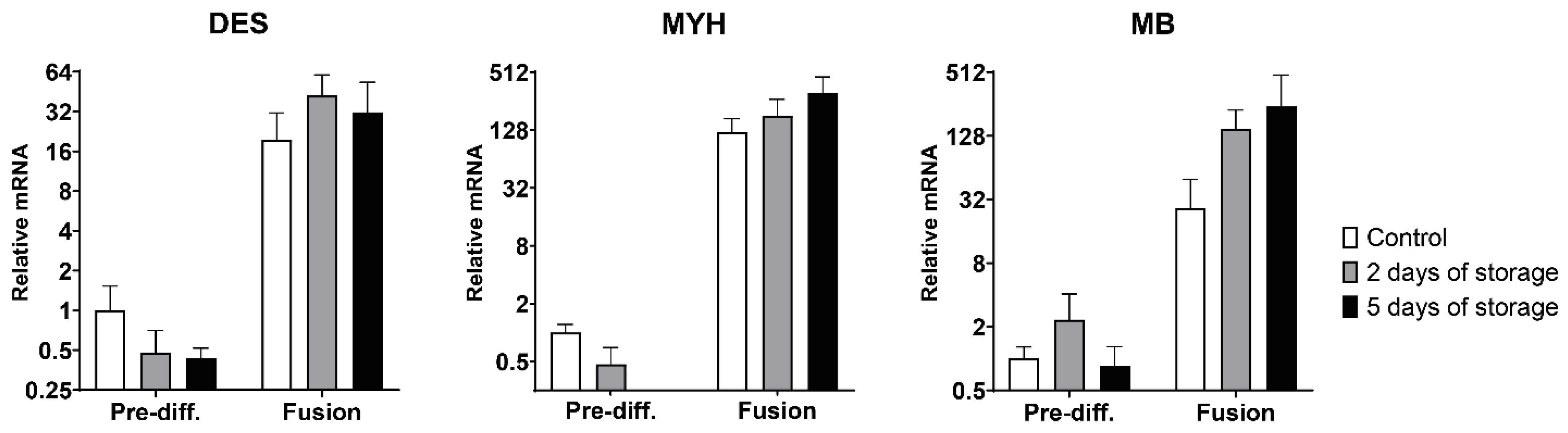
2.3. Myogenic and Muscle-Specific mRNA Expression Profiles of Bovine Satellite Cells Isolated from Fresh and Stored Muscle Tissue
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Satellite Cell Isolation
4.3. Cell Culture and Population Doubling Level (PDL)
4.4. WST-1 and PicoGreen Assays
4.5. Fluorescence Microscopy
4.6. Quantitative RT-PCR
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. In vitro meat: A future animal-free harvest. Crit. Rev. Food Sci. Nutr. 2017, 57, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Tuomisto, H.L.; Teixeira De Mattos, M.J. Environmental impacts of cultured meat production. Environ. Sci. Technol. 2011, 45, 6117–6123. [Google Scholar] [CrossRef] [PubMed]
- Our Story—Mosa Meat. Available online: https://www.mosameat.com/our-story (accessed on 3 December 2020).
- Crosser, N.; Bushnell, C.; Derbes, E.; Friedrich, B.; Lamy, J.; Manu, N.; Swartz, E. 2019 State of the Industry Report Cultivated Meat. Good Food Inst. 2020, 3, 24–35. [Google Scholar]
- Ben-Arye, T.; Levenberg, S. Tissue Engineering for Clean Meat Production. Front. Sustain. Food Syst. 2019, 3, 1–19. [Google Scholar] [CrossRef]
- Fraeye, I.; Kratka, M.; Vandenburgh, H.; Thorrez, L. Sensorial and Nutritional Aspects of Cultured Meat in Comparison to Traditional Meat: Much to Be Inferred. Front. Nutr. 2020, 7, 35. [Google Scholar] [CrossRef] [Green Version]
- Bogliotti, Y.S.; Wu, J.; Vilarino, M.; Okamura, D.; Soto, D.A.; Zhong, C.; Sakurai, M.; Sampaio, R.V.; Suzuki, K.; Izpisua Belmonte, J.C.; et al. Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. USA 2018, 115, 2090–2095. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y. Capturing bovine pluripotency. Proc. Natl. Acad. Sci. USA 2018, 115, 1962–1963. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Yin, J.; Zhu, M.J. Cellular signaling pathways regulating the initial stage of adipogenesis and marbling of skeletal muscle. Meat Sci. 2010, 86, 103–109. [Google Scholar] [CrossRef]
- Okamura, L.H.; Cordero, P.; Palomino, J.; Parraguez, V.H.; Torres, C.G.; Peralta, O.A. Myogenic Differentiation Potential of Mesenchymal Stem Cells Derived from Fetal Bovine Bone Marrow. Anim. Biotechnol. 2018, 29, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Espinosa, J.J.; González-Dávalos, L.; Shimada, A.; Pinã, E.; Varela-Echavarria, A.; Mora, O. Bovine (Bos taurus) Bone Marrow Mesenchymal Cell Differentiation to Adipogenic and Myogenic Lineages. Cells Tissues Organs 2015, 201, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Specht, E.A.; Welch, D.R.; Rees Clayton, E.M.; Lagally, C.D. Opportunities for applying biomedical production and manufacturing methods to the development of the clean meat industry. Biochem. Eng. J. 2018, 132, 161–168. [Google Scholar] [CrossRef]
- Snijders, T.; Nederveen, J.P.; McKay, B.R.; Joanisse, S.; Verdijk, L.B.; van Loon, L.J.C.; Parise, G. Satellite cells in human skeletal muscle plasticity. Front. Physiol. 2015, 6, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summerbell, D.; Halai, C.; Rigby, P.W.J. Expression of the myogenic regulatory factor Mrf4 precedes or is contemporaneous with that of Myf5 in the somitic bud. Mech. Dev. 2002, 117, 331–335. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef] [Green Version]
- Yablonka-Reuveni, Z.; Day, K.; Vine, A.; Shefer, G. Defining the transcriptional signature of skeletal muscle stem cells. J. Anim. Sci. 2008, 86, E207. [Google Scholar] [CrossRef] [Green Version]
- Blais, A.; Tsikitis, M.; Acosta-Alvear, D.; Sharan, R.; Kluger, Y.; Dynlacht, B.D. An initial blueprint for myogenic differentiation. Genes Dev. 2005, 19, 553–569. [Google Scholar] [CrossRef] [Green Version]
- Molkentin, J.D.; Olson, E.N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 1996, 93, 9366–9373. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Nelson, B.R.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 4109–4114. [Google Scholar] [CrossRef] [Green Version]
- Kassar-Duchossoy, L.; Gayraud-Morel, B.; Gomès, D.; Rocancourt, D.; Buckingham, M.; Shinin, V.; Tajbakhsh, S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 2004, 431, 466–471. [Google Scholar] [CrossRef]
- Hinterberger, T.J.; Sassoon, D.A.; Rhodes, S.J.; Konieczny, S.F. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev. Biol. 1991, 147, 144–156. [Google Scholar] [CrossRef]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the muscle cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar] [CrossRef] [Green Version]
- Hnia, K.; Ramspacher, C.; Vermot, J.; Laporte, J. Desmin in muscle and associated diseases: Beyond the structural function. Cell Tissue Res. 2015, 360, 591–608. [Google Scholar] [CrossRef]
- Weiss, A.; Leinwand, L.A. The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 1996, 12, 417–439. [Google Scholar] [CrossRef]
- Simsa, R.; Yuen, J.; Stout, A.; Rubio, N.; Fogelstrand, P.; Kaplan, D.L. Extracellular heme proteins influence bovine myosatellite cell proliferation and the color of cell-based meat. Foods 2019, 8, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Post, M.J. Cultured meat from stem cells: Challenges and prospects. Meat Sci. 2012, 92, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Melzener, L.; Verzijden, K.E.; Buijs, A.J.; Post, M.J.; Flack, J.E. Cultured beef: From small biopsy to substantial quantity. J. Sci. Food Agric. 2021, 101, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Zhao, X.; Zhou, J.; Du, G.; Chen, J. A conceptual air-lift reactor design for large scale animal cell cultivation in the context of in vitro meat production. Chem. Eng. Sci. 2020, 211, 115269. [Google Scholar] [CrossRef]
- Erker, L.; Azuma, H.; Lee, A.Y.; Guo, C.; Orloff, S.; Eaton, L.; Benedetti, E.; Jensen, B.; Finegold, M.; Willenbring, H.; et al. Therapeutic liver reconstitution with murine cells isolated long after death. Gastroenterology 2010, 139, 1019–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhu, Y.; Gao, W. Isolation of neural stem cells from the spinal cords of low temperature preserved abortuses. J. Neurosci. Methods 2006, 157, 64–70. [Google Scholar] [CrossRef]
- Celikkan, F.T.; Mungan, C.; Sucu, M.; Ulus, A.T.; Cinar, O.; Ili, E.G.; Can, A.L.P. Optimizing the transport and storage conditions of current Good Manufacturing Practice–grade human umbilical cord mesenchymal stromal cells for transplantation (HUC-HEART Trial). Cytotherapy 2019, 21, 64–75. [Google Scholar] [CrossRef]
- Latil, M.; Rocheteau, P.; Châtre, L.; Sanulli, S.; Mémet, S.; Ricchetti, M.; Tajbakhsh, S.; Chrétien, F. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat. Commun. 2012, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mahipal Singh, H.S.A. Recovery of fibroblast-like cells after 160 days of postmortem storage of goat skin tissues in refrigerated media. J. Vet. Sci. Technol. 2015, 06. [Google Scholar] [CrossRef] [Green Version]
- Chaillou1, T.; Lanner, J.T. Regulation of myogenesis and skeletal muscle regeneration: Effects of oxygen levels on satellite cell activity. FASEB J. 2016, 30, 3929–3941. [Google Scholar] [CrossRef] [Green Version]
- Csete, M. Oxygen in the cultivation of stem cells. Ann. N. Y. Acad. Sci. 2005, 1049, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bürgers, H.F.; Schelshorn, D.W.; Wagner, W.; Kuschinsky, W.; Maurer, M.H. Acute anoxia stimulates proliferation in adult neural stem cells from the rat brain. Exp. Brain Res. 2008, 188, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wen, Y.; Bi, P.; Lai, X.; Liu, X.S.; Liu, X.; Kuang, S. Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development 2012, 139, 2857–2865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csete, M.; Walikonis, J.; Slawny, N.; Wei, Y.; Korsnes, S.; Doyle, J.C.; Wold, B. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. J. Cell. Physiol. 2001, 189, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Baquero-Perez, B.; Kuchipudi, S.V.; Nelli, R.K.; Chang, K.C. A simplified but robust method for the isolation of avian and mammalian muscle satellite cells. BMC Cell Biol. 2012, 13. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Swennen, G.N.M.; Messmer, T.; Gagliardi, M.; Molin, D.G.M.; Li, C.; Zhou, G.; Post, M.J. Maintaining bovine satellite cells stemness through p38 pathway. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ishii, K.; Sakurai, H.; Suzuki, N.; Mabuchi, Y.; Sekiya, I.; Sekiguchi, K.; Akazawa, C. Recapitulation of Extracellular LAMININ Environment Maintains Stemness of Satellite Cells In Vitro. Stem Cell Rep. 2018, 10, 568–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahini, A.; Vydiam, K.; Choudhury, D.; Rajabian, N.; Nguyen, T.; Lei, P.; Andreadis, S.T. Efficient and high yield isolation of myoblasts from skeletal muscle. Stem Cell Res. 2018, 30, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Balci-Hayta, B.; Bekircan-Kurt, C.E.; Aksu, E.; Dayangac-Erden, D.; Tan, E.; Erdem-Ozdamar, S. Establishment of primary myoblast cell cultures from cryopreserved skeletal muscle biopsies to serve as a tool in related research & development studies. J. Neurol. Sci. 2018, 393, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.O.M.; Toner, M. Long-term storage of tissues by cryopreservation: Critical issues. Biomaterials 1996, 17, 243–256. [Google Scholar] [CrossRef]
- Murphy, D.P.; Nicholson, T.; Jones, S.W.; O’Leary, M.F. MyoCount: A software tool for the automated quantification of myotube surface area and nuclear fusion index [version 1; referees: 2 approved]. Wellcome Open Res. 2019, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.K.; Zamaratskaia, G.; Ekstrand, B. Gender-related Differences in Cytochrome P450 in Porcine Liver - Implication for Activity, Expression and Inhibition by Testicular Steroids. Reprod. Domest. Anim. 2011, 46, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.K.; Bertholdt, L.; Gudiksen, A.; Pilegaard, H.; Knudsen, J.G. Impact of fasting followed by short-term exposure to interleukin-6 on cytochrome P450 mRNA in mice. Toxicol. Lett. 2018, 282, 93–99. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Stern-Straeter, J.; Bonaterra, G.A.; Hörmann, K.; Kinscherf, R.; Goessler, U.R. Identification of valid reference genes during the differentiation of human myoblasts. BMC Mol. Biol. 2009, 10, 66. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, M.; Bernard, L.; Bes, S.; Leroux, C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [Green Version]
- Moretti, I.; Ciciliot, S.; Dyar, K.A.; Abraham, R.; Murgia, M.; Agatea, L.; Akimoto, T.; Bicciato, S.; Forcato, M.; Pierre, P.; et al. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Nygaard, A.-B.; Bøttcher, C. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Wu, X.; Guo, X.; Bao, P.; Ding, X.; Chu, M.; Liang, C.; Yan, P. Identification of optimal reference genes for examination of gene expression in different tissues of fetal yaks. Czech J. Anim. Sci. 2017, 62, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Zhou, X.; Ding, X.; Chu, M.; Liang, C.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. Reference gene selection and myosin heavy chain (MyHC) isoform expression in muscle tissues of domestic yak (Bos grunniens). PLoS ONE 2020, 15, e0228493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, R.; Sodhi, M.; Sharma, A.; Sharma, V.L.; Verma, P.; Swami, S.K.; Kumari, P.; Mukesh, M. Selection of suitable reference genes for normalization of quantitative RT-PCR (RT-qPCR) expression data across twelve tissues of riverine buffaloes (Bubalus bubalis). PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skrivergaard, S.; Rasmussen, M.K.; Therkildsen, M.; Young, J.F. Bovine Satellite Cells Isolated after 2 and 5 Days of Tissue Storage Maintain the Proliferative and Myogenic Capacity Needed for Cultured Meat Production. Int. J. Mol. Sci. 2021, 22, 8376. https://doi.org/10.3390/ijms22168376
Skrivergaard S, Rasmussen MK, Therkildsen M, Young JF. Bovine Satellite Cells Isolated after 2 and 5 Days of Tissue Storage Maintain the Proliferative and Myogenic Capacity Needed for Cultured Meat Production. International Journal of Molecular Sciences. 2021; 22(16):8376. https://doi.org/10.3390/ijms22168376
Chicago/Turabian StyleSkrivergaard, Stig, Martin Krøyer Rasmussen, Margrethe Therkildsen, and Jette Feveile Young. 2021. "Bovine Satellite Cells Isolated after 2 and 5 Days of Tissue Storage Maintain the Proliferative and Myogenic Capacity Needed for Cultured Meat Production" International Journal of Molecular Sciences 22, no. 16: 8376. https://doi.org/10.3390/ijms22168376
APA StyleSkrivergaard, S., Rasmussen, M. K., Therkildsen, M., & Young, J. F. (2021). Bovine Satellite Cells Isolated after 2 and 5 Days of Tissue Storage Maintain the Proliferative and Myogenic Capacity Needed for Cultured Meat Production. International Journal of Molecular Sciences, 22(16), 8376. https://doi.org/10.3390/ijms22168376





