Cytoskeleton and Associated Proteins: Pleiotropic JNK Substrates and Regulators
Abstract
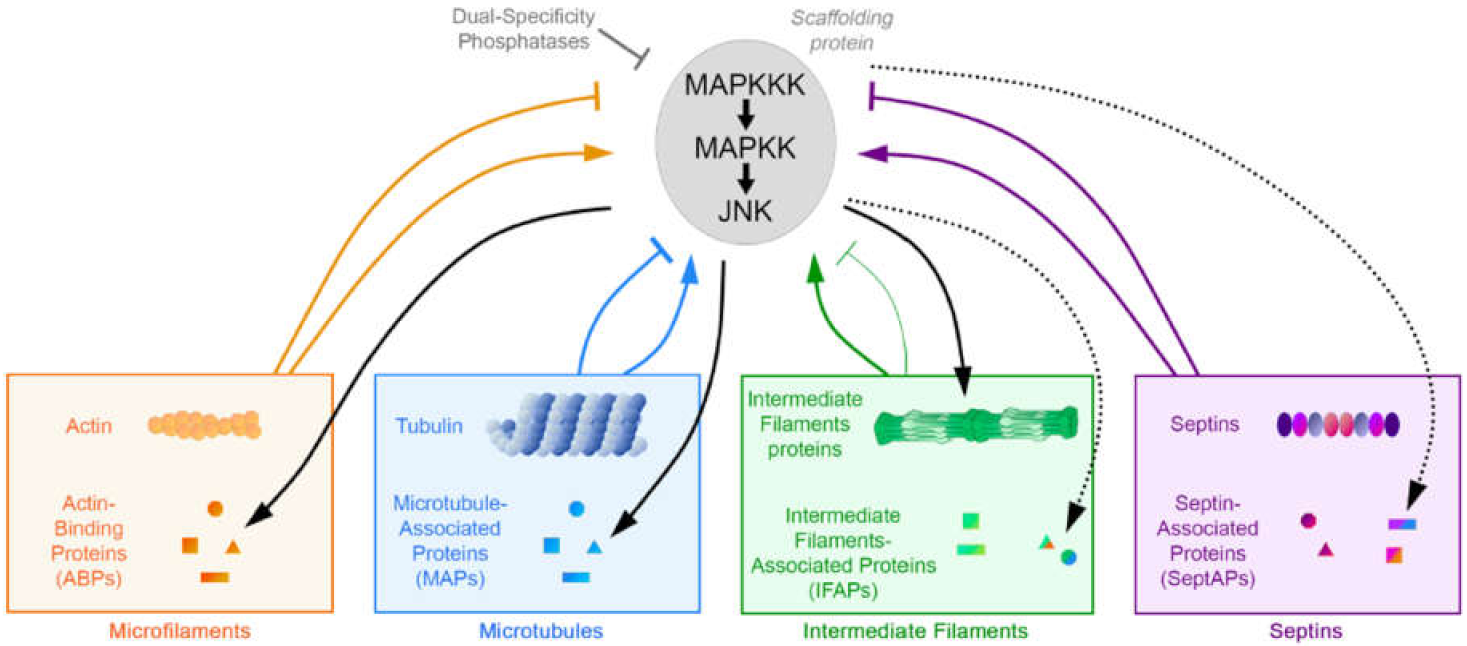
1. Introduction
2. JNK and Polarized Cytoskeletons: Microfilaments and Microtubules
2.1. JNK and Actin Microfilaments
2.1.1. Control of JNK by F-Actin and ABPs
2.1.2. Phosphorylation of Actin Proteins and ABPs by JNK
2.2. JNK and Microtubules
2.2.1. Control of JNK by MTs and MAPs
2.2.2. Phosphorylation of Tubulin Proteins and MAPs by JNK
3. JNK and Non-Polarized Cytoskeletons: Intermediate Filaments and Septins
3.1. JNK and Intermediate Filaments
3.1.1. Control of JNK by IFs
3.1.2. Phosphorylation of IF Proteins and IFAPs by JNK
3.2. JNK and Septins
4. JNK and Newly Considered Cytoskeletons: Spectrin and ESCRT-III
4.1. JNK and Spectrins
4.2. JNK and ESCRT-III
5. Cytoskeletal Localization of Upstream JNK Regulators and Scaffolds
6. JNK and Cytoskeleton Regulations in Time and Space: The Technical Challenge of Looking for Needles in a Haystack
7. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cytoplasmic Intermediate Filaments in Cell Biology. Annu. Rev. Cell Dev. Biol. 2018, 34, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Woods, B.L.; Gladfelter, A.S. The State of the Septin Cytoskeleton from Assembly to Function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Dogterom, M.; Koenderink, G.H. Actin–Microtubule Crosstalk in Cell Biology. Nat. Rev. Mol. Cell Biol. 2019, 20, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, A.J.; Calvo, F. The Borg Family of Cdc42 Effector Proteins Cdc42EP1–5. Biochem. Soc. Trans. 2016, 44, 1709–1716. [Google Scholar] [CrossRef]
- Hu, L.; Huang, Z.; Wu, Z.; Ali, A.; Qian, A. Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. Int. J. Mol. Sci. 2018, 19, 974. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Shanahan, C.M. Nesprins: From the Nuclear Envelope and Beyond. Expert Rev. Mol. Med. 2013, 15, e5. [Google Scholar] [CrossRef]
- Liem, R.K.H. Cytoskeletal Integrators: The Spectrin Superfamily. Cold Spring Harb. Perspect. Biol. 2016, 8, a018259. [Google Scholar] [CrossRef] [PubMed]
- Leite, S.C.; Sousa, M.M. The Neuronal and Actin Commitment: Why Do Neurons Need Rings? Cytoskeleton 2016, 73, 424–434. [Google Scholar] [CrossRef]
- Karasmanis, E.P.; Hwang, D.; Nakos, K.; Bowen, J.R.; Angelis, D.; Spiliotis, E.T. A Septin Double Ring Controls the Spatiotemporal Organization of the ESCRT Machinery in Cytokinetic Abscission. Curr. Biol. 2019, 29, 2174–2182.e7. [Google Scholar] [CrossRef]
- McCullough, J.; Sundquist, W.I. Membrane Remodeling: ESCRT-III Filaments as Molecular Garrotes. Curr. Biol. 2020, 30, R1425–R1428. [Google Scholar] [CrossRef]
- Addi, C.; Bai, J.; Echard, A. Actin, Microtubule, Septin and ESCRT Filament Remodeling during Late Steps of Cytokinesis. Curr. Opin. Cell Biol. 2018, 50, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Ong, M.S.; Deng, S.; Halim, C.E.; Cai, W.; Tan, T.Z.; Huang, R.Y.-J.; Sethi, G.; Hooi, S.C.; Kumar, A.P.; Yap, C.T. Cytoskeletal Proteins in Cancer and Intracellular Stress: A Therapeutic Perspective. Cancers 2020, 12, 238. [Google Scholar] [CrossRef]
- Musi, C.A.; Agrò, G.; Santarella, F.; Iervasi, E.; Borsello, T. JNK3 as Therapeutic Target and Biomarker in Neurodegenerative and Neurodevelopmental Brain Diseases. Cells 2020, 9, 2190. [Google Scholar] [CrossRef]
- Zeke, A.; Misheva, M.; Reményi, A.; Bogoyevitch, M.A. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiol. Mol. Biol. Rev. 2016, 80, 793–835. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.M.; Davis, R.J. Targeting JNK for Therapeutic Benefit: From Junk to Gold? Nat. Rev. Drug Discov. 2003, 2, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.K.; Davis, R.J. Regulation of Map Kinase Signaling Modules by Scaffold Proteins in Mammals. Annu. Rev. Cell Dev. Biol. 2003, 19, 91–118. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Kashef, K.; Lee, C.M.; Xu, H.; Reddy, E.P. Scaffold Proteins of MAP-Kinase Modules. Oncogene 2007, 26, 3185–3202. [Google Scholar] [CrossRef]
- Ha, J.; Kang, E.; Seo, J.; Cho, S. Phosphorylation Dynamics of JNK Signaling: Effects of Dual-Specificity Phosphatases (DUSPs) on the JNK Pathway. Int. J. Mol. Sci. 2019, 20, 6157. [Google Scholar] [CrossRef]
- Bogoyevitch, M.A.; Kobe, B. Uses for JNK: The Many and Varied Substrates of the c-Jun N-Terminal Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 1061–1095. [Google Scholar] [CrossRef]
- Barlan, K.; Gelfand, V.I. Microtubule-Based Transport and the Distribution, Tethering, and Organization of Organelles. Cold Spring Harb. Perspect. Biol. 2017, 9, a025817. [Google Scholar] [CrossRef] [PubMed]
- Titus, M.A. Myosin-Driven Intracellular Transport. Cold Spring Harb. Perspect. Biol. 2018, 10, a021972. [Google Scholar] [CrossRef]
- Rottner, K.; Faix, J.; Bogdan, S.; Linder, S.; Kerkhoff, E. Actin Assembly Mechanisms at a Glance. J. Cell Sci. 2017, 130, 3427–3435. [Google Scholar] [CrossRef] [PubMed]
- Møller, L.L.V.; Klip, A.; Sylow, L. Rho GTPases—Emerging Regulators of Glucose Homeostasis and Metabolic Health. Cells 2019, 8, 434. [Google Scholar] [CrossRef] [PubMed]
- Izdebska, M.; Zielińska, W.; Hałas-Wiśniewska, M.; Grzanka, A. Involvement of Actin and Actin-Binding Proteins in Carcinogenesis. Cells 2020, 9, 2245. [Google Scholar] [CrossRef]
- Yang, C.; Patel, K.; Harding, P.; Sorokin, A.; Glass, W.F. Regulation of TGF-Β1/MAPK-Mediated PAI-1 Gene Expression by the Actin Cytoskeleton in Human Mesangial Cells. Exp. Cell Res. 2007, 313, 1240–1250. [Google Scholar] [CrossRef]
- Christerson, L.B.; Vanderbilt, C.A.; Cobb, M.H. MEKK1 Interacts with A-actinin and Localizes to Stress Fibers and Focal Adhesions. Cell Motil. Cytoskel. 1999, 43, 186–198. [Google Scholar] [CrossRef]
- Nakagawa, K.; Sugahara, M.; Yamasaki, T.; Kajiho, H.; Takahashi, S.; Hirayama, J.; Minami, Y.; Ohta, Y.; Watanabe, T.; Hata, Y.; et al. Filamin Associates with Stress Signalling Kinases MKK7 and MKK4 and Regulates JNK Activation. Biochem. J. 2010, 427, 237–245. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Ka, S.H.; Cho, Y.; Choi, E.-J.; Baek, S.H.; Seol, J.H.; Park, D.; et al. Filamin B Serves as a Molecular Scaffold for Type I Interferon-Induced c-Jun NH2-Terminal Kinase Signaling Pathway. Mol. Biol. Cell 2008, 19, 5116–5130. [Google Scholar] [CrossRef][Green Version]
- Whitmarsh, A.J. Filamin B: A Scaffold for Interferon Signalling. EMBO Rep. 2009, 10, 349–351. [Google Scholar] [CrossRef]
- Chen, L.; Shi, K.; Frary, C.E.; Ditzel, N.; Hu, H.; Qiu, W.; Kassem, M. Inhibiting Actin Depolymerization Enhances Osteoblast Differentiation and Bone Formation in Human Stromal Stem Cells. Stem. Cell Res. 2015, 15, 281–289. [Google Scholar] [CrossRef]
- Lanzardo, S.; Curcio, C.; Forni, G.; Antón, I.M. A Role for WASP Interacting Protein, WIP, in Fibroblast Adhesion, Spreading and Migration. Int. J. Biochem. Cell Biol. 2007, 39, 262–274. [Google Scholar] [CrossRef]
- Mengistu, M.; Brotzman, H.; Ghadiali, S.; Lowe-Krentz, L. Fluid Shear Stress-induced JNK Activity Leads to Actin Remodeling for Cell Alignment. J. Cell. Physiol. 2011, 226, 110–121. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, H.; Jiang, K.; Wang, Y.; Zhang, W.; Chu, Q.; Li, J.; Huang, H.; Cai, T.; Ji, H.; et al. MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Rep. 2016, 16, 1810–1819. [Google Scholar] [CrossRef]
- Yang, G.; Liu, Y.; Yang, K.; Liu, R.; Zhu, S.; Coquinco, A.; Wen, W.; Kojic, L.; Jia, W.; Cynader, M. Isoform-Specific Palmitoylation of JNK Regulates Axonal Development. Cell Death Differ. 2012, 19, 553–561. [Google Scholar] [CrossRef]
- Valakh, V.; Frey, E.; Babetto, E.; Walker, L.J.; DiAntonio, A. Cytoskeletal Disruption Activates the DLK/JNK Pathway, Which Promotes Axonal Regeneration and Mimics a Preconditioning Injury. Neurobiol. Dis. 2015, 77, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Adib, E.A.; Smithson, L.J.; Collins, C.A. An Axonal Stress Response Pathway: Degenerative and Regenerative Signaling by DLK. Curr. Opin. Neurobiol. 2018, 53, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Densham, R.M.; O’Neill, E.; Munro, J.; König, I.; Anderson, K.; Kolch, W.; Olson, M.F. MST Kinases Monitor Actin Cytoskeletal Integrity and Signal via C-Jun N-Terminal Kinase Stress-Activated Kinase To Regulate P21Waf1/Cip1 Stability. Mol. Cell. Biol. 2009, 29, 6380–6390. [Google Scholar] [CrossRef]
- Gross, S.R.; Sin, C.G.T.; Barraclough, R.; Rudland, P.S. Joining S100 Proteins and Migration: For Better or for Worse, in Sickness and in Health. Cell. Mol. Life Sci. 2014, 71, 1551–1579. [Google Scholar] [CrossRef] [PubMed]
- Anfinogenova, N.D.; Quinn, M.T.; Schepetkin, I.A.; Atochin, D.N. Alarmins and C-Jun N-Terminal Kinase (JNK) Signaling in Neuroinflammation. Cells 2020, 9, 2350. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, N.; Enyindah-Asonye, G.; Bagheri, N.; Shah, N.B.; Gupta, N. Spatial Coupling of JNK Activation to the B Cell Antigen Receptor by Tyrosine-Phosphorylated Ezrin. J. Immunol. 2013, 190, 2017–2026. [Google Scholar] [CrossRef] [PubMed]
- Romanov, V.S.; Brichkina, A.I.; Morrison, H.; Pospelova, T.V.; Pospelov, V.A.; Herrlich, P. Novel Mechanism of JNK Pathway Activation by Adenoviral E1A. Oncotarget 2014, 5, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Egge, N.; Arneaud, S.L.B.; Wales, P.; Mihelakis, M.; McClendon, J.; Fonseca, R.S.; Savelle, C.; Gonzalez, I.; Ghorashi, A.; Yadavalli, S.; et al. Age-Onset Phosphorylation of a Minor Actin Variant Promotes Intestinal Barrier Dysfunction. Dev. Cell 2019, 51, 587–601.e7. [Google Scholar] [CrossRef] [PubMed]
- Björkblom, B.; Padzik, A.; Mohammad, H.; Westerlund, N.; Komulainen, E.; Hollos, P.; Parviainen, L.; Papageorgiou, A.C.; Iljin, K.; Kallioniemi, O.; et al. C-Jun N-Terminal Kinase Phosphorylation of MARCKSL1 Determines Actin Stability and Migration in Neurons and in Cancer Cells. Mol. Cell. Biol. 2012, 32, 3513–3526. [Google Scholar] [CrossRef]
- Gordon, E.A.; Whisenant, T.C.; Zeller, M.; Kaake, R.M.; Gordon, W.M.; Krotee, P.; Patel, V.; Huang, L.; Baldi, P.; Bardwell, L. Combining Docking Site and Phosphosite Predictions to Find New Substrates: Identification of Smoothelin-like-2 (SMTNL2) as a c-Jun N-Terminal Kinase (JNK) Substrate. Cell Signal. 2013, 25, 2518–2529. [Google Scholar] [CrossRef]
- Hachimi, M.; Grabowski, C.; Campanario, S.; Herranz, G.; Baonza, G.; Serrador, J.M.; Gomez-Lopez, S.; Barea, M.D.; Bosch-Fortea, M.; Gilmour, D.; et al. Smoothelin-like 2 Inhibits Coronin-1B to Stabilize the Apical Actin Cortex during Epithelial Morphogenesis. Curr. Biol. 2021, 31, 696–706.e9. [Google Scholar] [CrossRef]
- Eyers, C.E.; McNeill, H.; Knebel, A.; Morrice, N.; Arthur, S.J.C.; Cuenda, A.; Cohen, P. The Phosphorylation of CapZ-Interacting Protein (CapZIP) by Stress-Activated Protein Kinases Triggers Its Dissociation from CapZ. Biochem. J. 2005, 389, 127–135. [Google Scholar] [CrossRef]
- Baik, J.; Ok, S.-H.; Cho, H.; Yu, J.; Kim, W.; Nam, I.-K.; Choi, M.-J.; Lee, H.-K.; Sohn, J.-T. Dexmedetomidine-Induced Contraction Involves Phosphorylation of Caldesmon by JNK in Endothelium-Denuded Rat Aortas. Int. J. Biol. Sci. 2014, 10, 1108–1115. [Google Scholar] [CrossRef]
- Ulu, A.; Oh, W.; Zuo, Y.; Frost, J.A. Stress-Activated MAPKs and CRM1 Regulate the Subcellular Localization of Net1A to Control Cell Motility and Invasion. J. Cell Sci. 2017, 131, jcs204644. [Google Scholar] [CrossRef]
- Ulu, A.; Frost, J.A. Regulation of RhoA Activation and Cell Motility by C-Jun N-Terminal Kinases and Net1. Small GTPases 2018, 11, 1–7. [Google Scholar] [CrossRef]
- Slee, J.B.; Lowe-Krentz, L.J. Actin Realignment and Cofilin Regulation Are Essential for Barrier Integrity during Shear Stress. J. Cell. Biochem. 2013, 114, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Kohno, T.; Kikuchi, S.; Shimada, H.; Satohisa, S.; Saito, T.; Kondoh, M.; Kojima, T. Epithelial Barrier Dysfunction and Cell Migration Induction via JNK/Cofilin/Actin by Angubindin-1. Tissue Barriers 2020, 8, 1695475. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Ninomiya, T.; Kohno, T.; Kikuchi, S.; Sawada, N.; Kojima, T. C-Jun N-Terminal Kinase Inhibitor SP600125 Enhances Barrier Function and Elongation of Human Pancreatic Cancer Cell Line HPAC in a Ca-Switch Model. Histochem. Cell Biol. 2015, 143, 471–479. [Google Scholar] [CrossRef]
- Otto, I.M.; Raabe, T.; Rennefahrt, U.E.E.; Bork, P.; Rapp, U.R.; Kerkhoff, E. The P150-Spir Protein Provides a Link between c-Jun N-Terminal Kinase Function and Actin Reorganization. Curr. Biol. 2000, 10, 345–348. [Google Scholar] [CrossRef]
- Kim, M.J.; Futai, K.; Jo, J.; Hayashi, Y.; Cho, K.; Sheng, M. Synaptic Accumulation of PSD-95 and Synaptic Function Regulated by Phosphorylation of Serine-295 of PSD-95. Neuron 2007, 56, 488–502. [Google Scholar] [CrossRef]
- Kunde, S.-A.; Rademacher, N.; Tzschach, A.; Wiedersberg, E.; Ullmann, R.; Kalscheuer, V.M.; Shoichet, S.A. Characterisation of de Novo MAPK10/JNK3 Truncation Mutations Associated with Cognitive Disorders in Two Unrelated Patients. Hum. Genet. 2013, 132, 461–471. [Google Scholar] [CrossRef]
- Tu, W.; Ji, L.; Qian, H.; Zhou, M.; Zhao, J.; Xu, J. Tributyltin Induces Disruption of Microfilament in HL7702 Cells via MAPK-mediated Hyperphosphorylation of VASP. Environ. Toxicol. 2016, 31, 1530–1538. [Google Scholar] [CrossRef]
- Pan, Y.-R.; Tseng, W.-S.; Chang, P.-W.; Chen, H.-C. Phosphorylation of Moesin by Jun N-Terminal Kinase Is Important for Podosome Rosette Formation in Src-Transformed Fibroblasts. J. Cell Sci. 2013, 126, 5670–5680. [Google Scholar] [CrossRef]
- You, H.; Lei, P.; Andreadis, S.T. JNK Is a Novel Regulator of Intercellular Adhesion. Tissue Barriers 2013, 1, e26845. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, C.; Rajfur, Z.; Borchers, C.; Schaller, M.D.; Jacobson, K. JNK Phosphorylates Paxillin and Regulates Cell Migration. Nature 2003, 424, 219–223. [Google Scholar] [CrossRef]
- Chen, J.; Gallo, K.A. MLK3 Regulates Paxillin Phosphorylation in Chemokine-Mediated Breast Cancer Cell Migration and Invasion to Drive Metastasis. Cancer Res. 2012, 72, 4130–4140. [Google Scholar] [CrossRef]
- Ameka, M.; Kahle, M.P.; Perez-Neut, M.; Gentile, S.; Mirza, A.A.; Cuevas, B.D. MEKK2 Regulates Paxillin Ubiquitylation and Localization in MDA-MB 231 Breast Cancer Cells. Biochem. J. 2014, 464, 99–108. [Google Scholar] [CrossRef]
- Png, E.; Tong, L. Transglutaminase-2 in Cell Adhesion. Cell Adhes. Migr. 2013, 7, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Png, E.; Hou, A.; Tong, L. Mechanistic Role of Transglutaminase-2 in Focal Adhesions. Sci. Rep. 2018, 8, 12370. [Google Scholar] [CrossRef] [PubMed]
- Almeida, E.A.C.; Ilić, D.; Han, Q.; Hauck, C.R.; Jin, F.; Kawakatsu, H.; Schlaepfer, D.D.; Damsky, C.H. Matrix Survival Signaling. J. Cell Biol. 2000, 149, 741–754. [Google Scholar] [CrossRef]
- Lee, M.-H.; Koria, P.; Qu, J.; Andreadis, S.T. JNK Phosphorylates Β-catenin and Regulates Adherens Junctions. FASEB J. 2009, 23, 3874–3883. [Google Scholar] [CrossRef]
- Lee, M.; Padmashali, R.; Koria, P.; Andreacas, S.T. JNK Regulates Binding of A-catenin to Adherens Junctions and cell-cell Adhesion. FASEB J. 2011, 25, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Zeke, A.; Bastys, T.; Alexa, A.; Garai, Á.; Mészáros, B.; Kirsch, K.; Dosztányi, Z.; Kalinina, O.V.; Reményi, A. Systematic Discovery of Linear Binding Motifs Targeting an Ancient Protein Interaction Surface on MAP Kinases. Mol. Syst. Biol. 2015, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-K.; Yeap, Y.Y.C.; Bogoyevitch, M.A. The JNK1/JNK3 Interactome—Contributions by the JNK3 Unique N-Terminus and JNK Common Docking Site Residues. Biochem. Biophys. Res. Commun. 2014, 453, 576–581. [Google Scholar] [CrossRef]
- Kawasaki, A.; Okada, M.; Tamada, A.; Okuda, S.; Nozumi, M.; Ito, Y.; Kobayashi, D.; Yamasaki, T.; Yokoyama, R.; Shibata, T.; et al. Growth Cone Phosphoproteomics Reveals That GAP-43 Phosphorylated by JNK Is a Marker of Axon Growth and Regeneration. Iscience 2018, 4, 190–203. [Google Scholar] [CrossRef]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Microtubule Minus-End Regulation at a Glance. J. Cell Sci. 2019, 132, jcs227850. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20. [Google Scholar] [CrossRef]
- Escudero, C.A.; Cabeza, C.; Moya-Alvarado, G.; Maloney, M.T.; Flores, C.M.; Wu, C.; Court, F.A.; Mobley, W.C.; Bronfman, F.C. C-Jun N-Terminal Kinase (JNK)-Dependent Internalization and Rab5-Dependent Endocytic Sorting Mediate Long-Distance Retrograde Neuronal Death Induced by Axonal BDNF-P75 Signaling. Sci. Rep. 2019, 9, 6070. [Google Scholar] [CrossRef] [PubMed]
- Verhey, K.J.; Meyer, D.; Deehan, R.; Blenis, J.; Schnapp, B.J.; Rapoport, T.A.; Margolis, B. Cargo of Kinesin Identified as Jip Scaffolding Proteins and Associated Signaling Molecules. J. Cell Biol. 2001, 152, 959–970. [Google Scholar] [CrossRef]
- Geeraert, C.; Ratier, A.; Pfisterer, S.G.; Perdiz, D.; Cantaloube, I.; Rouault, A.; Pattingre, S.; Proikas-Cezanne, T.; Codogno, P.; Poüs, C. Starvation-Induced Hyperacetylation of Tubulin Is Required for the Stimulation of Autophagy by Nutrient Deprivation. J. Biol. Chem. 2010, 285, 24184–24194. [Google Scholar] [CrossRef]
- Perdiz, D.; Lorin, S.; Leroy-Gori, I.; Poüs, C. Stress-Induced Hyperacetylation of Microtubule Enhances Mitochondrial Fission and Modulates the Phosphorylation of Drp1 at 616Ser. Cell Signal. 2017, 39, 32–43. [Google Scholar] [CrossRef]
- Li, D.; Ding, X.; Xie, M.; Huang, Z.; Han, P.; Tian, D.; Xia, L. CAMSAP2-Mediated Noncentrosomal Microtubule Acetylation Drives Hepatocellular Carcinoma Metastasis. Theranostics 2020, 10, 3749–3766. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, L.; Lu, X.; Wang, Y.; Li, R.; Jiang, G. KIF15 Contributes to Cell Proliferation and Migration in Breast Cancer. Hum. Cell 2020, 33, 1218–1228. [Google Scholar] [CrossRef]
- Voelzmann, A.; Okenve-Ramos, P.; Qu, Y.; Chojnowska-Monga, M.; del Caño-Espinel, M.; Prokop, A.; Sanchez-Soriano, N. Tau and Spectraplakins Promote Synapse Formation and Maintenance through Jun Kinase and Neuronal Trafficking. Elife 2016, 5, e14694. [Google Scholar] [CrossRef]
- Dewey, E.B.; Parra, A.S.; Johnston, C.A. Loss of the Spectraplakin Gene Short Stop Induces a DNA Damage Response in Drosophila Epithelia. Sci. Rep. 2020, 10, 20165. [Google Scholar] [CrossRef]
- Ni, Y.; Wang, X.; Yin, X.; Li, Y.; Liu, X.; Wang, H.; Liu, X.; Zhang, J.; Gao, H.; Shi, B.; et al. Plectin Protects Podocytes from Adriamycin-induced Apoptosis and F-actin Cytoskeletal Disruption through the Integrin A6β4/FAK/P38 MAPK Pathway. J. Cell. Mol. Med. 2018, 22, 5450–5467. [Google Scholar] [CrossRef] [PubMed]
- Bendrick, J.L.; Eldredge, L.A.; Williams, E.I.; Haight, N.B.; Dubash, A.D. Desmoplakin Harnesses Rho GTPase and P38 Mitogen-Activated Protein Kinase Signaling to Coordinate Cellular Migration. J. Investig. Dermatol. 2019, 139, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.S.; Fernandez-Rodriguez, L.; Zhao, Y.; Monaco, G.; Trefny, M.P.; Yoshida, N.; Martin, K.; Sharma, A.; Olieric, N.; Shah, P.; et al. GEF-H1 Signaling upon Microtubule Destabilization Is Required for Dendritic Cell Activation and Specific Anti-Tumor Responses. Cell Rep. 2019, 28, 3367–3380.e8. [Google Scholar] [CrossRef] [PubMed]
- Poulton, J.S.; McKay, D.J.; Peifer, M. Centrosome Loss Triggers a Transcriptional Program To Counter Apoptosis-Induced Oxidative Stress. Genetics 2019, 212, 187–211. [Google Scholar] [CrossRef]
- Castro-Torres, R.D.; Busquets, O.; Parcerisas, A.; Verdaguer, E.; Olloquequi, J.; Etchetto, M.; Beas-Zarate, C.; Folch, J.; Camins, A.; Auladell, C. Involvement of JNK1 in Neuronal Polarization During Brain Development. Cells 2020, 9, 1897. [Google Scholar] [CrossRef]
- Ramkumar, A.; Jong, B.Y.; Ori-McKenney, K.M. ReMAPping the Microtubule Landscape: How Phosphorylation Dictates the Activities of Microtubule-associated Proteins. Dev. Dyn. 2018, 247, 138–155. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Okada, M.; Honda, A.; Ito, Y.; Tamada, A.; Endo, N.; Igarashi, M. Phosphorylation Sites of Microtubule-Associated Protein 1B (MAP 1B) Are Involved in Axon Growth and Regeneration. Mol. Brain 2019, 12, 93. [Google Scholar] [CrossRef]
- Komulainen, E.; Zdrojewska, J.; Freemantle, E.; Mohammad, H.; Kulesskaya, N.; Deshpande, P.; Marchisella, F.; Mysore, R.; Hollos, P.; Michelsen, K.A.; et al. JNK1 Controls Dendritic Field Size in L2/3 and L5 of the Motor Cortex, Constrains Soma Size, and Influences Fine Motor Coordination. Front. Cell. Neurosci. 2014, 8, 272. [Google Scholar] [CrossRef]
- Gdalyahu, A.; Ghosh, I.; Levy, T.; Sapir, T.; Sapoznik, S.; Fishler, Y.; Azoulai, D.; Reiner, O. DCX, a New Mediator of the JNK Pathway. EMBO J. 2004, 23, 823–832. [Google Scholar] [CrossRef]
- Jin, J.; Suzuki, H.; Hirai, S.; Mikoshiba, K.; Ohshima, T. JNK Phosphorylates Ser332 of Doublecortin and Regulates Its Function in Neurite Extension and Neuronal Migration. Dev. Neurobiol. 2010, 70, 929–942. [Google Scholar] [CrossRef]
- Moslehi, M.; Ng, D.C.H.; Bogoyevitch, M.A. Doublecortin X (DCX) Serine 28 Phosphorylation Is a Regulatory Switch, Modulating Association of DCX with Microtubules and Actin Filaments. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Brown, K.J.; Yap, C.C.; Winckler, B.; Jaiswal, J.K.; Liu, J.S. Doublecortin (Dcx) Family Proteins Regulate Filamentous Actin Structure in Developing Neurons. J. Neurosci. 2013, 33, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Coquelle, F.M.; Levy, T.; Bergmann, S.; Wolf, S.G.; Bar-El, D.; Sapir, T.; Brody, Y.; Orr, I.; Barkai, N.; Eichele, G.; et al. The DCX Superfamily 1: Common and Divergent Roles for Members of the Mouse DCX Superfamily. Cell Cycle 2006, 5, 976–983. [Google Scholar] [CrossRef]
- Shmueli, A.; Gdalyahu, A.; Sapoznik, S.; Sapir, T.; Tsukada, M.; Reiner, O. Site-Specific Dephosphorylation of Doublecortin (DCX) by Protein Phosphatase 1 (PP1). Mol. Cell Neurosci. 2006, 32, 15–26. [Google Scholar] [CrossRef]
- Yoshida, H.; Hastie, C.J.; McLauchlan, H.; Cohen, P.; Goedert, M. Phosphorylation of Microtubule-associated Protein Tau by Isoforms of C-Jun N-terminal Kinase (JNK). J. Neurochem. 2004, 90, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Ploia, C.; Antoniou, X.; Sclip, A.; Grande, V.; Cardinetti, D.; Colombo, A.; Canu, N.; Benussi, L.; Ghidoni, R.; Forloni, G.; et al. JNK Plays a Key Role in Tau Hyperphosphorylation in Alzheimer’s Disease Models. J. Alzheimer’s Dis. 2011, 26, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Gourmaud, S.; Shou, H.; Irwin, D.J.; Sansalone, K.; Jacobs, L.M.; Lucas, T.H.; Marsh, E.D.; Davis, K.A.; Jensen, F.E.; Talos, D.M. Alzheimer-like Amyloid and Tau Alterations Associated with Cognitive Deficit in Temporal Lobe Epilepsy. Brain 2020, 143, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Lagalwar, S.; Guillozet-Bongaarts, A.L.; Berry, R.W.; Binder, L.I. Formation of Phospho-SAPK/JNK Granules in the Hippocampus Is an Early Event in Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2006, 65, 455–464. [Google Scholar] [CrossRef]
- Guillozet-Bongaarts, A.L.; Cahill, M.E.; Cryns, V.L.; Reynolds, M.R.; Berry, R.W.; Binder, L.I. Pseudophosphorylation of Tau at Serine 422 Inhibits Caspase Cleavage: In Vitro Evidence and Implications for Tangle Formation in Vivo. J. Neurochem. 2006, 97, 1005–1014. [Google Scholar] [CrossRef]
- Santa-Catalina, M.O.; Bermejo, M.C.; Argent, R.; Alonso, J.C.; Centeno, F.; Lorenzo, M.J. JNK Signaling Pathway Regulates Sorbitol-Induced Tau Proteolysis and Apoptosis in SH-SY5Y Cells by Targeting Caspase-3. Arch. Biochem. Biophys. 2017, 636, 42–49. [Google Scholar] [CrossRef]
- Lei, Z.; Brizzee, C.; Johnson, G.V.W. BAG3 Facilitates the Clearance of Endogenous Tau in Primary Neurons. Neurobiol. Aging 2015, 36, 241–248. [Google Scholar] [CrossRef]
- Buccarello, L.; Musi, C.A.; Turati, A.; Borsello, T. The Stress C-Jun N-Terminal Kinase Signaling Pathway Activation Correlates with Synaptic Pathology and Presents A Sex Bias in P301L Mouse Model of Tauopathy. Neuroscience 2018, 393, 196–205. [Google Scholar] [CrossRef]
- Grassi, D.; Diaz-Perez, N.; Volpicelli-Daley, L.A.; Lasmézas, C.I. Pα-Syn* Mitotoxicity Is Linked to MAPK Activation and Involves Tau Phosphorylation and Aggregation at the Mitochondria. Neurobiol. Dis. 2019, 124, 248–262. [Google Scholar] [CrossRef]
- Yip, Y.Y.; Yeap, Y.Y.C.; Bogoyevitch, M.A.; Ng, D.C.H. Differences in C-Jun N-Terminal Kinase Recognition and Phosphorylation of Closely Related Stathmin-Family Members. Biochem. Biophys. Res. Commun. 2014, 446, 248–254. [Google Scholar] [CrossRef]
- Tararuk, T.; Östman, N.; Li, W.; Björkblom, B.; Padzik, A.; Zdrojewska, J.; Hongisto, V.; Herdegen, T.; Konopka, W.; Courtney, M.J.; et al. JNK1 Phosphorylation of SCG10 Determines Microtubule Dynamics and Axodendritic Length. J. Cell Biol. 2006, 173, 265–277. [Google Scholar] [CrossRef]
- Birnie, K.A.; Yip, Y.Y.; Ng, D.C.H.; Kirschner, M.B.; Reid, G.; Prêle, C.M.; Musk, A.W.; Lee, Y.C.G.; Thompson, P.J.; Mutsaers, S.E.; et al. Loss of MiR-223 and JNK Signaling Contribute to Elevated Stathmin in Malignant Pleural Mesothelioma. Mol. Cancer Res. 2015, 13, 1106–1118. [Google Scholar] [CrossRef]
- Ng, D.C.H.; Zhao, T.T.; Yeap, Y.Y.C.; Ngoei, K.R.; Bogoyevitch, M.A. C-Jun N-Terminal Kinase Phosphorylation of Stathmin Confers Protection against Cellular Stress. J. Biol. Chem. 2010, 285, 29001–29013. [Google Scholar] [CrossRef]
- Zhao, E.; Amir, M.; Lin, Y.; Czaja, M.J. Stathmin Mediates Hepatocyte Resistance to Death from Oxidative Stress by down Regulating JNK. PLoS ONE 2014, 9, e109750. [Google Scholar] [CrossRef]
- Morfini, G.A.; You, Y.-M.; Pollema, S.L.; Kaminska, A.; Liu, K.; Yoshioka, K.; Björkblom, B.; Coffey, E.T.; Bagnato, C.; Han, D.; et al. Pathogenic Huntingtin Inhibits Fast Axonal Transport by Activating JNK3 and Phosphorylating Kinesin. Nat. Neurosci. 2009, 12, 864–871. [Google Scholar] [CrossRef]
- Bharat, V.; Siebrecht, M.; Burk, K.; Ahmed, S.; Reissner, C.; Kohansal-Nodehi, M.; Steubler, V.; Zweckstetter, M.; Ting, J.T.; Dean, C. Capture of Dense Core Vesicles at Synapses by JNK-Dependent Phosphorylation of Synaptotagmin-4. Cell Rep. 2017, 21, 2118–2133. [Google Scholar] [CrossRef]
- Inomata, H.; Nakamura, Y.; Hayakawa, A.; Takata, H.; Suzuki, T.; Miyazawa, K.; Kitamura, N. A Scaffold Protein JIP-1b Enhances Amyloid Precursor Protein Phosphorylation by JNK and Its Association with Kinesin Light Chain 1. J. Biol. Chem. 2003, 278, 22946–22955. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Chiba, K.; Sobu, Y.; Shiraki, Y.; Okumura, Y.; Hata, S.; Kitamura, A.; Nakaya, T.; Uchida, S.; Kinjo, M.; et al. The Cytoplasmic Region of the Amyloid Β-protein Precursor (APP) Is Necessary and Sufficient for the Enhanced Fast Velocity of APP Transport by Kinesin-1. FEBS Lett. 2018, 592, 2716–2724. [Google Scholar] [CrossRef]
- Chiba, K.; Araseki, M.; Nozawa, K.; Furukori, K.; Araki, Y.; Matsushima, T.; Nakaya, T.; Hata, S.; Saito, Y.; Uchida, S.; et al. Quantitative Analysis of APP Axonal Transport in Neurons- Role of JIP1 in Enhanced APP Anterograde Transport. Mol. Biol. Cell 2014, 25, 3569–3580. [Google Scholar] [CrossRef]
- Taru, H.; Suzuki, T. Facilitation of Stress-Induced Phosphorylation of β-Amyloid Precursor Protein Family Members by X11-like/Mint2 Protein. J. Biol. Chem. 2004, 279, 21628–21636. [Google Scholar] [CrossRef]
- Lei, K.; Davis, R.J. JNK Phosphorylation of Bim-Related Members of the Bcl2 Family Induces Bax-Dependent Apoptosis. Proc. Natl. Acad. Sci. USA 2003, 100, 2432–2437. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Holzbaur, E.L.F. JIP1 Regulates the Directionality of APP Axonal Transport by Coordinating Kinesin and Dynein Motors. J. Cell Biol. 2013, 202, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.Q.; Aumont-Nicaise, M.; Andreani, J.; Velours, C.; Chenon, M.; Vilela, F.; Geneste, C.; Varela, P.F.; Llinas, P.; Ménétrey, J. Characterization of the Binding Mode of JNK-Interacting Protein 1 (JIP1) to Kinesin-Light Chain 1 (KLC1). J. Biol. Chem. 2018, 293, 13946–13960. [Google Scholar] [CrossRef] [PubMed]
- Nihalani, D.; Wong, H.N.; Holzman, L.B. Recruitment of JNK to JIP1 and JNK-Dependent JIP1 Phosphorylation Regulates JNK Module Dynamics and Activation. J. Biol. Chem. 2003, 278, 28694–28702. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, N.; Gupta, S.; Dickens, M.; Davis, R.J. Interaction of a Mitogen-Activated Protein Kinase Signaling Module with the Neuronal Protein JIP3. Mol. Cell. Biol. 2000, 20, 1030–1043. [Google Scholar] [CrossRef]
- Caswell, P.T.; Dickens, M. JIP3 Localises to Exocytic Vesicles and Focal Adhesions in the Growth Cones of Differentiated PC12 Cells. Mol. Cell. Biochem. 2018, 444, 1–13. [Google Scholar] [CrossRef]
- Henrie, H.; Bakhos-Douaihy, D.; Cantaloube, I.; Pilon, A.; Talantikite, M.; Stoppin-Mellet, V.; Baillet, A.; Poüs, C.; Benoit, B. Stress-Induced Phosphorylation of CLIP-170 by JNK Promotes Microtubule Rescue. J. Cell Biol. 2020, 219, e201909093. [Google Scholar] [CrossRef]
- Ayala, I.; Crispino, R.; Colanzi, A. GRASP65 Controls Golgi Position and Structure during G2/M Transition by Regulating the Stability of Microtubules. Traffic 2019, 20, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Lim, N.R.; Yeap, Y.Y.C.; Zhao, T.T.; Yip, Y.Y.; Wong, S.C.; Xu, D.; Ang, C.-S.; Williamson, N.A.; Xu, Z.; Bogoyevitch, M.A.; et al. Opposing Roles for JNK and Aurora A in Regulating the Association of WDR62 with Spindle Microtubules. J. Cell Sci. 2015, 128, 527–540. [Google Scholar] [CrossRef]
- Xu, D.; Yao, M.; Wang, Y.; Yuan, L.; Hoeck, J.D.; Yu, J.; Liu, L.; Yeap, Y.Y.C.; Zhang, W.; Zhang, F.; et al. MEKK3 Coordinates with FBW7 to Regulate WDR62 Stability and Neurogenesis. PLoS Biol. 2018, 16, e2006613. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.J.-K.; Jasper, H. Control of Intestinal Cell Fate by Dynamic Mitotic Spindle Repositioning Influences Epithelial Homeostasis and Longevity. Cell Rep. 2019, 28, 2807–2823.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qin, Y.; Qin, Y.; Xu, B.; Guo, T.; Ke, H.; Chen, M.; Zhang, L.; Han, F.; Li, Y.; et al. Wdr62 Is Involved in Female Meiotic Initiation via Activating JNK Signaling and Associated with POI in Humans. PLoS Genet. 2018, 14, e1007463. [Google Scholar] [CrossRef] [PubMed]
- Padzik, A.; Deshpande, P.; Hollos, P.; Franker, M.; Rannikko, E.H.; Cai, D.; Prus, P.; Mågård, M.; Westerlund, N.; Verhey, K.J.; et al. KIF5C S176 Phosphorylation Regulates Microtubule Binding and Transport Efficiency in Mammalian Neurons. Front. Cell. Neurosci. 2016, 10, 57. [Google Scholar] [CrossRef]
- Toivola, D.M.; Strnad, P.; Habtezion, A.; Omary, M.B. Intermediate Filaments Take the Heat as Stress Proteins. Trends Cell Biol. 2010, 20, 79–91. [Google Scholar] [CrossRef]
- Lam, M.; Calvo, F. Regulation of Mechanotransduction: Emerging Roles for Septins. Cytoskeleton 2019, 76, 115–122. [Google Scholar] [CrossRef]
- Spiliotis, E.T.; McMurray, M.A. Masters of Asymmetry—Lessons and Perspectives from 50 Years of Septins. Mol. Biol. Cell 2020, 31, 2289–2297. [Google Scholar] [CrossRef]
- Spiliotis, E.T.; Nakos, K. Cellular Functions of Actin- and Microtubule-Associated Septins. Curr. Biol. 2021, 31, R651–R666. [Google Scholar] [CrossRef]
- Eldirany, S.A.; Lomakin, I.B.; Ho, M.; Bunick, C.G. Recent Insight into Intermediate Filament Structure. Curr. Opin. Cell Biol. 2021, 68, 132–143. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate Filaments: Structure and Assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef]
- Omary, M.B.; Ku, N.-O.; Tao, G.-Z.; Toivola, D.M.; Liao, J. ‘Heads and Tails’ of Intermediate Filament Phosphorylation: Multiple Sites and Functional Insights. Trends Biochem. Sci. 2006, 31, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, M.G.; Bremner, S.N.; Hornberger, T.A.; Meyer, G.A.; Domenighetti, A.A.; Shah, S.B.; Kiss, B.; Kellermayer, M.; Ryan, A.F.; Lieber, R.L. Skeletal Muscle Intermediate Filaments Form a Stress-Transmitting and Stress-Signaling Network. J. Cell Sci. 2015, 128, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Javed, E.; Thangavel, C.; Frara, N.; Singh, J.; Mohanty, I.; Hypolite, J.; Birbe, R.; Braverman, A.S.; Den, R.B.; Rattan, S.; et al. Increased Expression of Desmin and Vimentin Reduces Bladder Smooth Muscle Contractility via JNK2. FASEB J. 2020, 34, 2126–2146. [Google Scholar] [CrossRef]
- He, T.; Stepulak, A.; Holmström, T.H.; Omary, M.B.; Eriksson, J.E. The Intermediate Filament Protein Keratin 8 Is a Novel Cytoplasmic Substrate for C-Jun N-Terminal Kinase. J. Biol. Chem. 2002, 277, 10767–10774. [Google Scholar] [CrossRef] [PubMed]
- Guldiken, N.; Zhou, Q.; Kucukoglu, O.; Rehm, M.; Levada, K.; Gross, A.; Kwan, R.; James, L.P.; Trautwein, C.; Omary, M.B.; et al. Human Keratin 8 Variants Promote Mouse Acetaminophen Hepatotoxicity Coupled with C-jun Amino-terminal Kinase Activation and Protein Adduct Formation. Hepatology 2015, 62, 876–886. [Google Scholar] [CrossRef]
- D’Alessandro, M.; Russell, D.; Morley, S.M.; Davies, A.M.; Lane, E.B. Keratin Mutations of Epidermolysis Bullosa Simplex Alter the Kinetics of Stress Response to Osmotic Shock. J. Cell Sci. 2002, 115, 4341–4351. [Google Scholar] [CrossRef]
- Tang, G.; Xu, Z.; Goldman, J.E. Synergistic Effects of the SAPK/JNK and the Proteasome Pathway on Glial Fibrillary Acidic Protein (GFAP) Accumulation in Alexander Disease. J. Biol. Chem. 2006, 281, 38634–38643. [Google Scholar] [CrossRef]
- Toivola, D.M.; Zhou, Q.; English, L.S.; Omary, M.B. Type II Keratins Are Phosphorylated on a Unique Motif during Stress and Mitosis in Tissues and Cultured Cells. Mol. Biol. Cell 2002, 13, 1857–1870. [Google Scholar] [CrossRef]
- Weerasinghe, S.V.W.; Ku, N.-O.; Altshuler, P.J.; Kwan, R.; Omary, M.B. Mutation of Caspase-Digestion Sites in Keratin 18 Interferes with Filament Reorganization, and Predisposes to Hepatocyte Necrosis and Loss of Membrane Integrity. J. Cell Sci. 2014, 127, 1464–1475. [Google Scholar] [CrossRef]
- Ku, N.-O.; Omary, M.B. A Disease- and Phosphorylation-Related Nonmechanical Function for Keratin 8. J. Cell Biol. 2006, 174, 115–125. [Google Scholar] [CrossRef]
- Park, M.K.; Lee, H.J.; Shin, J.; Noh, M.; Kim, S.Y.; Lee, C.H. Novel Participation of Transglutaminase-2 through c-Jun N-Terminal Kinase Activation in Sphingosylphosphorylcholine-Induced Keratin Reorganization of PANC-1 Cells. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2011, 1811, 1021–1029. [Google Scholar] [CrossRef]
- Giasson, B.I.; Mushynski, W.E. Aberrant Stress-Induced Phosphorylation of Perikaryal Neurofilaments. J. Biol. Chem. 1996, 271, 30404–30409. [Google Scholar] [CrossRef]
- Brownlees, J.; Yates, A.; Bajaj, N.P.; Davis, D.; Anderton, B.H.; Leigh, P.N.; Shaw, C.E.; Miller, C.C. Phosphorylation of Neurofilament Heavy Chain Side-Arms by Stress Activated Protein Kinase-1b/Jun N-Terminal Kinase-3. J. Cell Sci. 2000, 113 Pt 3, 401–407. [Google Scholar] [CrossRef]
- Ackerley, S.; Grierson, A.J.; Banner, S.; Perkinton, M.S.; Brownlees, J.; Byers, H.L.; Ward, M.; Thornhill, P.; Hussain, K.; Waby, J.S.; et al. P38α Stress-Activated Protein Kinase Phosphorylates Neurofilaments and Is Associated with Neurofilament Pathology in Amyotrophic Lateral Sclerosis. Mol. Cell. Neurosci. 2004, 26, 354–364. [Google Scholar] [CrossRef]
- Veeranna; Amin, N.D.; Ahn, N.G.; Jaffe, H.; Winters, C.A.; Grant, P.; Pant, H.C. Mitogen-Activated Protein Kinases (Erk1,2) Phosphorylate Lys-Ser-Pro (KSP) Repeats in Neurofilament Proteins NF-H and NF-M. J. Neurosci. 1998, 18, 4008–4021. [Google Scholar] [CrossRef]
- Boubriak, I.I.; Malhas, A.N.; Drozdz, M.M.; Pytowski, L.; Vaux, D.J. Stress-Induced Release of Oct-1 from the Nuclear Envelope Is Mediated by JNK Phosphorylation of Lamin B1. PLoS ONE 2017, 12, e0177990. [Google Scholar] [CrossRef]
- Kayser, J.; Haslbeck, M.; Dempfle, L.; Krause, M.; Grashoff, C.; Buchner, J.; Herrmann, H.; Bausch, A.R. The Small Heat Shock Protein Hsp27 Affects Assembly Dynamics and Structure of Keratin Intermediate Filament Networks. Biophys. J. 2013, 105, 1778–1785. [Google Scholar] [CrossRef]
- Ruan, J.; Qi, Z.; Shen, L.; Jiang, Y.; Xu, Y.; Lan, L.; Luo, L.; Yin, Z. Crosstalk between JNK and NF-ΚB Signaling Pathways via HSP27 Phosphorylation in HepG2 Cells. Biochem. Biophys. Res. Commun. 2015, 456, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Harper, M.; Tillit, J.; Kress, M.; Ernoult-Lange, M. Phosphorylation-dependent Binding of Human Transcription Factor MOK2 to Lamin A/C. FEBS J. 2009, 276, 3137–3147. [Google Scholar] [CrossRef]
- Plowman, J.E. The Hair Fibre: Proteins, Structure and Development. Adv. Exp. Med. Biol. 2018, 1054, 21–32. [Google Scholar] [CrossRef]
- Capetanaki, Y.; Papathanasiou, S.; Diokmetzidou, A.; Vatsellas, G.; Tsikitis, M. Desmin Related Disease: A Matter of Cell Survival Failure. Curr. Opin. Cell Biol. 2015, 32, 113–120. [Google Scholar] [CrossRef]
- Mostowy, S.; Cossart, P. Septins: The Fourth Component of the Cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 183–194. [Google Scholar] [CrossRef]
- Froidevaux-Klipfel, L.; Targa, B.; Cantaloube, I.; Ahmed-Zaïd, H.; Poüs, C.; Baillet, A. Septin Cooperation with Tubulin Polyglutamylation Contributes to Cancer Cell Adaptation to Taxanes. Oncotarget 2015, 6, 36063–36080. [Google Scholar] [CrossRef]
- Targa, B.; Klipfel, L.; Cantaloube, I.; Salameh, J.; Benoit, B.; Poüs, C.; Baillet, A. Septin Filament Coalignment with Microtubules Depends on SEPT9_i1 and Tubulin Polyglutamylation, and Is an Early Feature of Acquired Cell Resistance to Paclitaxel. Cell Death Dis. 2019, 10, 54. [Google Scholar] [CrossRef]
- Gonzalez, M.E.; Makarova, O.; Peterson, E.A.; Privette, L.M.; Petty, E.M. Up-Regulation of SEPT9_v1 Stabilizes c-Jun-N-Terminal Kinase and Contributes to Its pro-Proliferative Activity in Mammary Epithelial Cells. Cell Signal. 2009, 21, 477–487. [Google Scholar] [CrossRef]
- Neubauer, K.; Zieger, B. The Mammalian Septin Interactome. Front. Cell Dev. Biol. 2017, 5, 3. [Google Scholar] [CrossRef]
- Gönczi, M.; Dienes, B.; Dobrosi, N.; Fodor, J.; Balogh, N.; Oláh, T.; Csernoch, L. Septins, a Cytoskeletal Protein Family, with Emerging Role in Striated Muscle. J. Muscle Res. Cell Motil. 2020, 1–15. [Google Scholar] [CrossRef]
- Tokhtaeva, E.; Capri, J.; Marcus, E.A.; Whitelegge, J.P.; Khuzakhmetova, V.; Bukharaeva, E.; Deiss-Yehiely, N.; Dada, L.A.; Sachs, G.; Fernandez-Salas, E.; et al. Septin Dynamics Are Essential for Exocytosis. J. Biol. Chem. 2015, 290, 5280–5297. [Google Scholar] [CrossRef]
- Tanaka, T.; Iino, M.; Goto, K. Knockdown of Sec8 Enhances the Binding Affinity of C-Jun N-terminal Kinase (JNK)-interacting Protein 4 for Mitogen-activated Protein Kinase Kinase 4 (MKK4) and Suppresses the Phosphorylation of MKK4, P38, and JNK, Thereby Inhibiting Apoptosis. FEBS J. 2014, 281, 5237–5250. [Google Scholar] [CrossRef]
- Estey, M.P.; Ciano-Oliveira, C.D.; Froese, C.D.; Bejide, M.T.; Trimble, W.S. Distinct Roles of Septins in Cytokinesis: SEPT9 Mediates Midbody Abscission. J. Cell Biol. 2010, 191, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Biggi, S.; Buccarello, L.; Sclip, A.; Lippiello, P.; Tonna, N.; Rumio, C.; Marino, D.D.; Miniaci, M.C.; Borsello, T. Evidence of Presynaptic Localization and Function of the C-Jun N-Terminal Kinase. Neural Plast. 2017, 2017, 1–14. [Google Scholar] [CrossRef]
- Tsang, C.W.; Estey, M.P.; DiCiccio, J.E.; Xie, H.; Patterson, D.; Trimble, W.S. Characterization of Presynaptic Septin Complexes in Mammalian Hippocampal Neurons. Biol. Chem. 2011, 392, 739–749. [Google Scholar] [CrossRef]
- Marttinen, M.; Kurkinen, K.M.; Soininen, H.; Haapasalo, A.; Hiltunen, M. Synaptic Dysfunction and Septin Protein Family Members in Neurodegenerative Diseases. Mol. Neurodegener. 2015, 10, 16. [Google Scholar] [CrossRef]
- Wasik, A.A.; Dumont, V.; Tienari, J.; Nyman, T.A.; Fogarty, C.L.; Forsblom, C.; Lehto, M.; Lehtonen, E.; Groop, P.-H.; Lehtonen, S. Septin 7 Reduces Nonmuscle Myosin IIA Activity in the SNAP23 Complex and Hinders GLUT4 Storage Vesicle Docking and Fusion. Exp. Cell Res. 2017, 350, 336–348. [Google Scholar] [CrossRef]
- Farrugia, A.J.; Rodríguez, J.; Orgaz, J.L.; Lucas, M.; Sanz-Moreno, V.; Calvo, F. CDC42EP5/BORG3 Modulates SEPT9 to Promote Actomyosin Function, Migration, and Invasion. J. Cell Biol. 2020, 219, e201912159. [Google Scholar] [CrossRef]
- Joberty, G.; Perlungher, R.R.; Macara, I.G. The Borgs, a New Family of Cdc42 and TC10 GTPase-Interacting Proteins. Mol. Cell. Biol. 1999, 19, 6585–6597. [Google Scholar] [CrossRef]
- Carim, S.C.; Kechad, A.; Hickson, G.R.X. Animal Cell Cytokinesis: The Rho-Dependent Actomyosin-Anilloseptin Contractile Ring as a Membrane Microdomain Gathering, Compressing, and Sorting Machine. Front. Cell Dev. Biol. 2020, 8, 575226. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, T.; Masters, T.A.; Buss, F. The MYO6 Interactome Reveals Adaptor Complexes Coordinating Early Endosome and Cytoskeletal Dynamics. EMBO Rep. 2018, 19, e44884. [Google Scholar] [CrossRef]
- Schauer, M.; Kleinwort, K.J.H.; Degroote, R.L.; Wiedemann, C.; Kremmer, E.; Hauck, S.M.; Deeg, C.A. Interaction of Septin 7 and DOCK8 in Equine Lymphocytes Reveals Novel Insights into Signaling Pathways Associated with Autoimmunity. Sci. Rep. 2018, 8, 12332. [Google Scholar] [CrossRef]
- De Jonge, J.J.; Batters, C.; O’Loughlin, T.; Arden, S.D.; Buss, F. The MYO6 Interactome: Selective Motor-cargo Complexes for Diverse Cellular Processes. FEBS Lett. 2019, 593, 1494–1507. [Google Scholar] [CrossRef]
- Gadea, G.; Blangy, A. Dock-Family Exchange Factors in Cell Migration and Disease. Eur. J. Cell Biol. 2014, 93, 466–477. [Google Scholar] [CrossRef]
- Guimbal, S.; Morel, A.; Guérit, D.; Chardon, M.; Blangy, A.; Vives, V. Dock5 Is a New Regulator of Microtubule Dynamic Instability in Osteoclasts. Biol. Cell 2019, 111, 271–283. [Google Scholar] [CrossRef]
- Morishita, K.; Ozasa, F.; Eguchi, K.; Yoshioka, Y.; Yoshida, H.; Hiai, H.; Yamaguchi, M. Drosophila DOCK Family Protein Sponge Regulates the JNK Pathway during Thorax Development. Cell Struct. Funct. 2014, 39, 113–124. [Google Scholar] [CrossRef]
- Sobczak, M.; Chumak, V.; Pomorski, P.; Wojtera, E.; Majewski, Ł.; Nowak, J.; Yamauchi, J.; Rędowicz, M.J. Interaction of Myosin VI and Its Binding Partner DOCK7 Plays an Important Role in NGF-Stimulated Protrusion Formation in PC12 Cells. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 1589–1600. [Google Scholar] [CrossRef]
- Barrett, A.; Pellet-Many, C.; Zachary, I.C.; Evans, I.M.; Frankel, P. P130Cas: A Key Signalling Node in Health and Disease. Cell Signal. 2013, 25, 766–777. [Google Scholar] [CrossRef]
- Spiliotis, E.T. Spatial Effects − Site-Specific Regulation of Actin and Microtubule Organization by Septin GTPases. J. Cell Sci. 2018, 131, jcs207555. [Google Scholar] [CrossRef] [PubMed]
- Kesisova, I.A.; Robinson, B.P.; Spiliotis, E.T. A Septin GTPase Scaffold of Dynein–Dynactin Motors Triggers Retrograde Lysosome Transport. J. Cell Biol. 2021, 220, e202005219. [Google Scholar] [CrossRef]
- Zander, S.; Baumann, S.; Weidtkamp-Peters, S.; Feldbrügge, M. Endosomal Assembly and Transport of Heteromeric Septin Complexes Promote Septin Cytoskeleton Formation. J. Cell Sci. 2016, 129, 2778–2792. [Google Scholar] [CrossRef] [PubMed]
- Sreeja, J.S.; John, R.; Dharmapal, D.; Nellikka, R.K.; Sengupta, S. A Fresh Look at the Structure, Regulation, and Functions of Fodrin. Mol. Cell. Biol. 2020, 40, e00133-20. [Google Scholar] [CrossRef]
- Lorenzo, D.N. Cargo Hold and Delivery: Ankyrins, Spectrins, and Their Functional Patterning of Neurons. Cytoskeleton 2020, 77, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, B.; Grochowalska, R.; Bogusławska, D.M.; Sikorski, A.F. The Role of Spectrin in Cell Adhesion and Cell–Cell Contact. Exp. Biol. Med. 2019, 244, 1303–1312. [Google Scholar] [CrossRef]
- Goodman, S.R.; Johnson, D.; Youngentob, S.L.; Kakhniashvili, D. The Spectrinome: The Interactome of a Scaffold Protein Creating Nuclear and Cytoplasmic Connectivity and Function. Exp. Biol. Med. 2019, 244, 1273–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Simon, D.J.; Wu, Z.; Belsky, D.M.; Heller, E.; O’Rourke, M.K.; Hertz, N.T.; Molina, H.; Zhong, G.; Tessier-Lavigne, M.; et al. Structural Plasticity of Actin-Spectrin Membrane Skeleton and Functional Role of Actin and Spectrin in Axon Degeneration. Elife 2019, 8, e38730. [Google Scholar] [CrossRef]
- Massaro, C.M.; Pielage, J.; Davis, G.W. Molecular Mechanisms That Enhance Synapse Stability despite Persistent Disruption of the Spectrin/Ankyrin/Microtubule Cytoskeleton. J. Cell Biol. 2009, 187, 101–117. [Google Scholar] [CrossRef]
- Ursitti, J.A.; Petrich, B.G.; Lee, P.C.; Resneck, W.G.; Ye, X.; Yang, J.; Randall, W.R.; Bloch, R.J.; Wang, Y. Role of an Alternatively Spliced Form of AII-Spectrin in Localization of Connexin 43 in Cardiomyocytes and Regulation by Stress-Activated Protein Kinase. J. Mol. Cell. Cardiol. 2007, 42, 572–581. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, D.; Chadha, G.K.; Feygin, A.; Ivanov, A.I. F-Actin Binding Protein, Anillin, Regulates Integrity of Intercellular Junctions in Human Epithelial Cells. Cell. Mol. Life Sci. 2015, 72, 3185–3200. [Google Scholar] [CrossRef][Green Version]
- Vietri, M.; Radulovic, M.; Stenmark, H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef]
- Schöneberg, J.; Lee, I.-H.; Iwasa, J.H.; Hurley, J.H. Reverse-Topology Membrane Scission by the ESCRT Proteins. Nat. Rev. Mol. Cell Bio. 2017, 18, 5–17. [Google Scholar] [CrossRef] [PubMed]
- West, R.J.H.; Ugbode, C.; Gao, F.-B.; Sweeney, S.T. The Pro-Apoptotic JNK Scaffold POSH/SH3RF1 Mediates CHMP2BIntron5-Associated Toxicity in Animal Models of Frontotemporal Dementia. Hum. Mol. Genet. 2018, 27, 1382–1395. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Seong, K.-H.; Aigaki, T. POSH, a Scaffold Protein for JNK Signaling, Binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 2006, 580, 3296–3300. [Google Scholar] [CrossRef] [PubMed]
- Rodahl, L.M.; Haglund, K.; Sem-Jacobsen, C.; Wendler, F.; Vincent, J.-P.; Lindmo, K.; Rusten, T.E.; Stenmark, H. Disruption of Vps4 and JNK Function in Drosophila Causes Tumour Growth. PLoS ONE 2009, 4, e4354. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.; Takase, H.; Nunome, M.; Enomoto, H.; Ito, J.; Gong, J.-S.; Michikawa, M. Amyloid-β Reduces Exosome Release from Astrocytes by Enhancing JNK Phosphorylation. J. Alzheimer’s Dis. 2016, 53, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, M.J.; Liu, J.; Lavoie, B.D.; Wilde, A. Anillin-Dependent Organization of Septin Filaments Promotes Intercellular Bridge Elongation and Chmp4B Targeting to the Abscission Site. Open Biol. 2014, 4, 130190. [Google Scholar] [CrossRef] [PubMed]
- Goliand, I.; Adar-Levor, S.; Segal, I.; Nachmias, D.; Dadosh, T.; Kozlov, M.M.; Elia, N. Resolving ESCRT-III Spirals at the Intercellular Bridge of Dividing Cells Using 3D STORM. Cell Rep. 2018, 24, 1756–1764. [Google Scholar] [CrossRef]
- Busquets, O.; Parcerisas, A.; Verdaguer, E.; Ettcheto, M.; Camins, A.; Beas-Zarate, C.; Castro-Torres, R.D.; Auladell, C. C-Jun N-Terminal Kinases in Alzheimer’s Disease: A Possible Target for the Modulation of the Earliest Alterations. J. Alzheimer’s Dis. 2021, 82, S127–S139. [Google Scholar] [CrossRef]
- Yasuda, J.; Whitmarsh, A.J.; Cavanagh, J.; Sharma, M.; Davis, R.J. The JIP Group of Mitogen-Activated Protein Kinase Scaffold Proteins. Mol. Cell. Biol. 1999, 19, 7245–7254. [Google Scholar] [CrossRef]
- Ikonomov, O.C.; Fligger, J.; Sbrissa, D.; Dondapati, R.; Mlak, K.; Deeb, R.; Shisheva, A. Kinesin Adapter JLP Links PIKfyve to Microtubule-Based Endosome-to-Trans-Golgi Network Traffic of Furin. J. Biol. Chem. 2009, 284, 3750–3761. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Ishikawa, M.; Mochizuki, M.; Ohta, M.; Ohkura, M.; Nakai, J.; Takamatsu, N.; Yoshioka, K. JSAP1/JIP3 and JLP Regulate Kinesin-1-Dependent Axonal Transport to Prevent Neuronal Degeneration. Cell Death Differ. 2015, 22, 1260–1274. [Google Scholar] [CrossRef]
- Nagata, K.; Puls, A.; Futter, C.; Aspenstrom, P.; Schaefer, E.; Nakata, T.; Hirokawa, N.; Hall, A. The MAP Kinase Kinase Kinase MLK2 Co-localizes with Activated JNK along Microtubules and Associates with Kinesin Superfamily Motor KIF3. EMBO J. 1998, 17, 149–158. [Google Scholar] [CrossRef]
- Cohen-Katsenelson, K.; Wasserman, T.; Khateb, S.; Whitmarsh, A.J.; Aronheim, A. Docking Interactions of the JNK Scaffold Protein WDR62. Biochem. J. 2011, 439, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Hadad, M.; Aviram, S.; Darlyuk-Saadon, I.; Cohen-Katsenelson, K.; Whitmarsh, A.J.; Aronheim, A. The Association of the JNK Scaffold Protein, WDR62, with the Mixed Lineage Kinase 3, MLK3. MAP Kinase 2015, 4. [Google Scholar] [CrossRef]
- Zhang, F.; Yu, J.; Yang, T.; Xu, D.; Chi, Z.; Xia, Y.; Xu, Z. A Novel C-Jun N-Terminal Kinase (JNK) Signaling Complex Involved in Neuronal Migration during Brain Development. J. Biol. Chem. 2017, 291, 11466–11475. [Google Scholar] [CrossRef]
- Xu, L.; Wang, W.; Meng, T.; Ma, L.-P.; Tong, L.-J.; Shen, J.-K.; Wang, Y.-Q.; Miao, Z.-H. New Microtubulin Inhibitor MT189 Suppresses Angiogenesis via the JNK-VEGF/VEGFR2 Signaling Axis. Cancer Lett. 2018, 416, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.; Zhou, X.; Yu, J.; Yuan, L.; Zhang, H.; Ng, D.C.H.; Xu, Z.; Xu, D. Pathophysiological Significance of WDR62 and JNK Signaling in Human Diseases. Front. Cell Dev. Biol. 2021, 9, 640753. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, T.; Katsenelson, K.; Daniliuc, S.; Hasin, T.; Choder, M.; Aronheim, A. A Novel C-Jun N-Terminal Kinase (JNK)-Binding Protein WDR62 Is Recruited to Stress Granules and Mediates a Nonclassical JNK Activation. Mol. Biol. Cell 2010, 21, 117–130. [Google Scholar] [CrossRef]
- DeWire, S.M.; Ahn, S.; Lefkowitz, R.J.; Shenoy, S.K. β-Arrestins and Cell Signaling. Physiology 2007, 69, 483–510. [Google Scholar] [CrossRef]
- Hanson, S.M.; Cleghorn, W.M.; Francis, D.J.; Vishnivetskiy, S.A.; Raman, D.; Song, X.; Nair, K.S.; Slepak, V.Z.; Klug, C.S.; Gurevich, V.V. Arrestin Mobilizes Signaling Proteins to the Cytoskeleton and Redirects Their Activity. J. Mol. Biol. 2007, 368, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V.; Gurevich, E.V. Overview of Different Mechanisms of Arrestin-Mediated Signaling. Curr. Protoc. Pharmacol. 2014, 67, 2.10.1–2.10.9. [Google Scholar] [CrossRef]
- Kook, S.; Vishnivetskiy, S.A.; Gurevich, V.V.; Gurevich, E.V. Cleavage of Arrestin-3 by Caspases Attenuates Cell Death by Precluding Arrestin-Dependent JNK Activation. Cell Signal. 2019, 54, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Crépieux, P.; Poupon, A.; Langonné-Gallay, N.; Reiter, E.; Delgado, J.; Schaefer, M.H.; Bourquard, T.; Serrano, L.; Kiel, C. A Comprehensive View of the β-Arrestinome. Front. Endocrinol. 2017, 8, 32. [Google Scholar] [CrossRef]
- Sharma, A.; Batra, J.; Stuchlik, O.; Reed, M.S.; Pohl, J.; Chow, V.T.K.; Sambhara, S.; Lal, S.K. Influenza A Virus Nucleoprotein Activates the JNK Stress-Signaling Pathway for Viral Replication by Sequestering Host Filamin A Protein. Front. Microbiol. 2020, 11, 581867. [Google Scholar] [CrossRef] [PubMed]
- Janoštiak, R.; Pataki, A.C.; Brábek, J.; Rösel, D. Mechanosensors in Integrin Signaling: The Emerging Role of P130Cas. Eur. J. Cell Biol. 2014, 93, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Tan, S.H.; Machiyama, H.; Kawauchi, K.; Araki, K.; Hirata, H.; Sawada, Y. Association between Tensin 1 and P130Cas at Focal Adhesions Links Actin Inward Flux to Cell Migration. Biol. Open 2016, 5, 499–506. [Google Scholar] [CrossRef][Green Version]
- Defilippi, P.; Stefano, P.D.; Cabodi, S. P130Cas: A Versatile Scaffold in Signaling Networks. Trends Cell Biol. 2006, 16, 257–263. [Google Scholar] [CrossRef]
- Li, J.; Wang, R.; Tang, D.D. Vimentin Dephosphorylation at Ser-56 Is Regulated by Type 1 Protein Phosphatase in Smooth Muscle. Respir. Res. 2016, 17, 91. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, D.; Thomason, D.B.; Jarrett, H.W. Laminin-Induced Activation of Rac1 and JNKp46 Is Initiated by Src Family Kinases and Mimics the Effects of Skeletal Muscle Contraction. Biochemistry 2007, 46, 14907–14916. [Google Scholar] [CrossRef][Green Version]
- Kumar, B.; Dutta, D.; Iqbal, J.; Ansari, M.A.; Roy, A.; Chikoti, L.; Pisano, G.; Veettil, M.V.; Chandran, B. ESCRT-I Protein Tsg101 Plays a Role in the Post-Macropinocytic Trafficking and Infection of Endothelial Cells by Kaposi’s Sarcoma-Associated Herpesvirus. PLoS Pathog. 2016, 12, e1005960. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, W.; Wang, J.; Feng, J.; Wang, Q.; Jin, J.; Lv, M.; Li, X.; Li, Y.; Ma, Y.; et al. Receptor for Activated C Kinase 1 Promotes Hepatocellular Carcinoma Growth by Enhancing Mitogen-activated Protein Kinase Kinase 7 Activity. Hepatology 2013, 57, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Ai, E.; Poole, D.S.; Skop, A.R. RACK-1 Directs Dynactin-Dependent RAB-11 Endosomal Recycling during Mitosis in Caenorhabditis Elegans. Mol. Biol. Cell 2009, 20, 1629–1638. [Google Scholar] [CrossRef][Green Version]
- Neasta, J.; Fiorenza, A.; He, D.-Y.; Phamluong, K.; Kiely, P.A.; Ron, D. Activation of the CAMP Pathway Induces RACK1-Dependent Binding of β-Actin to BDNF Promoter. PLoS ONE 2016, 11, e0160948. [Google Scholar] [CrossRef]
- Myklebust, L.M.; Horvli, O.; Raae, A.J. RACK1 (Receptor for Activated C-kinase 1) Interactions with Spectrin Repeat Elements. J. Mol. Recognit. 2015, 28, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Zhou, Y.; Long, R.; Chen, C.; Zhao, J.; Cui, P.; Guo, M.; Liang, G.; Xu, L. DUSP8 Phosphatase: Structure, Functions, Expression Regulation and the Role in Human Diseases. Cell Biosci. 2019, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Habelhah, H.; Shah, K.; Huang, L.; Burlingame, A.L.; Shokat, K.M.; Ronai, Z. Identification of New JNK Substrate Using ATP Pocket Mutant JNK and a Corresponding ATP Analogue. J. Biol. Chem. 2001, 276, 18090–18095. [Google Scholar] [CrossRef]
- Allen, J.J.; Li, M.; Brinkworth, C.S.; Paulson, J.L.; Wang, D.; Hübner, A.; Chou, W.-H.; Davis, R.J.; Burlingame, A.L.; Messing, R.O.; et al. A Semisynthetic Epitope for Kinase Substrates. Nat. Methods 2007, 4, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Depry, C.; Mehta, S.; Li, R.; Zhang, J. Visualization of Compartmentalized Kinase Activity Dynamics Using Adaptable BimKARs. Chem. Biol. 2015, 22, 1470–1479. [Google Scholar] [CrossRef]
- Hollos, P.; John, J.M.; Lehtonen, J.V.; Coffey, E.T. Optogenetic Control of Spine-Head JNK Reveals a Role in Dendritic Spine Regression. Eneuro 2020, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gambarotto, D.; Zwettler, F.U.; Guennec, M.L.; Schmidt-Cernohorska, M.; Fortun, D.; Borgers, S.; Heine, J.; Schloetel, J.-G.; Reuss, M.; Unser, M.; et al. Imaging Cellular Ultrastructures Using Expansion Microscopy (U-ExM). Nat. Methods 2019, 16, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Park, C.E.; Cho, Y.; Cho, I.; Jung, H.; Kim, B.; Shin, J.H.; Choi, S.; Kwon, S.-K.; Hahn, Y.K.; Chang, J.-B. Super-Resolution Three-Dimensional Imaging of Actin Filaments in Cultured Cells and the Brain via Expansion Microscopy. ACS Nano 2020, 14, 14999–15010. [Google Scholar] [CrossRef] [PubMed]
- Wassie, A.T.; Zhao, Y.; Boyden, E.S. Expansion Microscopy: Principles and Uses in Biological Research. Nat. Methods 2019, 16, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yao, L.; Zhang, L.; Fei, Y.; Mi, L.; Ma, J. Applications of Super Resolution Expansion Microscopy in Yeast. Front. Phys. 2021, 9, 136. [Google Scholar] [CrossRef]
- Bain, J.; Plater, L.; Elliott, M.; Shpiro, N.; Hastie, C.J.; Mclauchlan, H.; Klevernic, I.; Arthur, J.S.C.; Alessi, D.R.; Cohen, P. The Selectivity of Protein Kinase Inhibitors: A Further Update. Biochem. J. 2007, 408, 297–315. [Google Scholar] [CrossRef]
- Wu, Y.; Honegger, A.; Batyuk, A.; Mittl, P.R.E.; Plückthun, A. Structural Basis for the Selective Inhibition of C-Jun N-Terminal Kinase 1 Determined by Rigid DARPin–DARPin Fusions. J. Mol. Biol. 2018, 430, 2128–2138. [Google Scholar] [CrossRef]
- Jemaà, M.; Abassi, Y.; Kifagi, C.; Fezai, M.; Daams, R.; Lang, F.; Massoumi, R. Reversine Inhibits Colon Carcinoma Cell Migration by Targeting JNK1. Sci. Rep. 2018, 8, 11821. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, Z.; Shah, F.A.; Shafique, S.; Alattar, A.; Ali, T.; Alvi, A.M.; Rashid, S.; Li, S. Repurposing FDA Approved Drugs as JNK3 Inhibitor for Prevention of Neuroinflammation Induced by MCAO in Rats. J. Inflamm. Res. 2020, 13, 1185–1205. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.; Jo, M.H.; Riaz, M.; Alam, S.I.; Saeed, K.; Ali, W.; Rehman, I.U.; Ikram, M.; Kim, M.O. Glycine, the Smallest Amino Acid, Confers Neuroprotection against d-Galactose-Induced Neurodegeneration and Memory Impairment by Regulating c-Jun N-Terminal Kinase in the Mouse Brain. J. Neuroinflamm. 2020, 17, 303. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benoit, B.; Baillet, A.; Poüs, C. Cytoskeleton and Associated Proteins: Pleiotropic JNK Substrates and Regulators. Int. J. Mol. Sci. 2021, 22, 8375. https://doi.org/10.3390/ijms22168375
Benoit B, Baillet A, Poüs C. Cytoskeleton and Associated Proteins: Pleiotropic JNK Substrates and Regulators. International Journal of Molecular Sciences. 2021; 22(16):8375. https://doi.org/10.3390/ijms22168375
Chicago/Turabian StyleBenoit, Béatrice, Anita Baillet, and Christian Poüs. 2021. "Cytoskeleton and Associated Proteins: Pleiotropic JNK Substrates and Regulators" International Journal of Molecular Sciences 22, no. 16: 8375. https://doi.org/10.3390/ijms22168375
APA StyleBenoit, B., Baillet, A., & Poüs, C. (2021). Cytoskeleton and Associated Proteins: Pleiotropic JNK Substrates and Regulators. International Journal of Molecular Sciences, 22(16), 8375. https://doi.org/10.3390/ijms22168375





