Arabidopsis G-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Measurement of Gibberellin Content
2.3. Construction of Plasmids and Transgenic Lines
2.4. Extraction of RNA and Analysis of Gene Expression
2.5. Subcellular Localization Analysis
2.6. Yeast Two-Hybrid Assay
2.7. Bimolecular Fluorescence Complementation (BiFC) Assay
2.8. Pull-Down Assays
2.9. Transcriptional Activation Experiment in Yeast
2.10. LUC Assay of MYB62 in Tobacco (N. benthamiana)
2.11. Electrophoretic Mobility Shift Assay (EMSA)
2.12. Low Phosphate-Tolerant Phenotypic Assay of Plants
3. Results
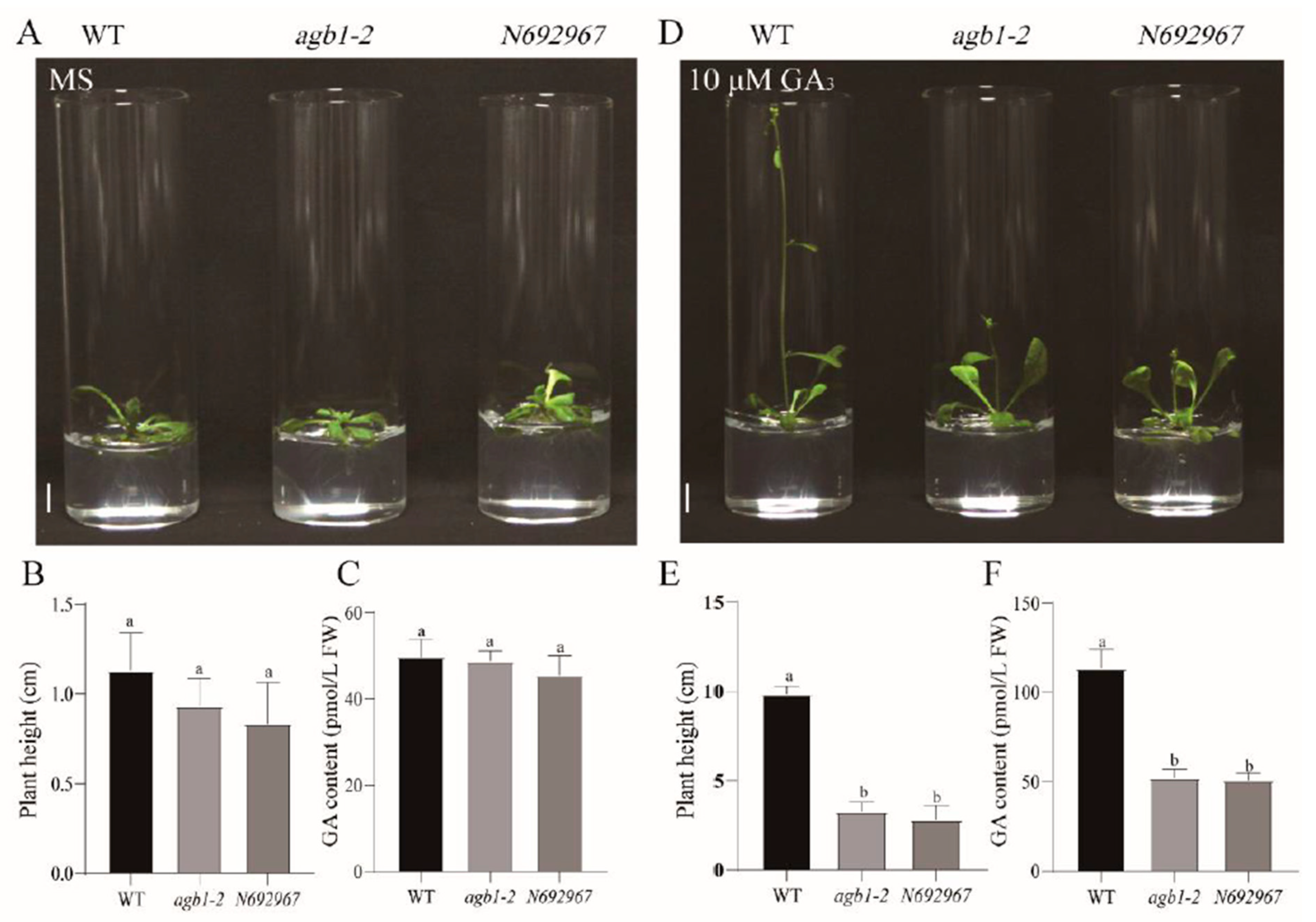
3.1. AGB1 Mutants agb1-2 and N692967 Were Dwarfed Compared with the WT after Exogenous Application of GA3
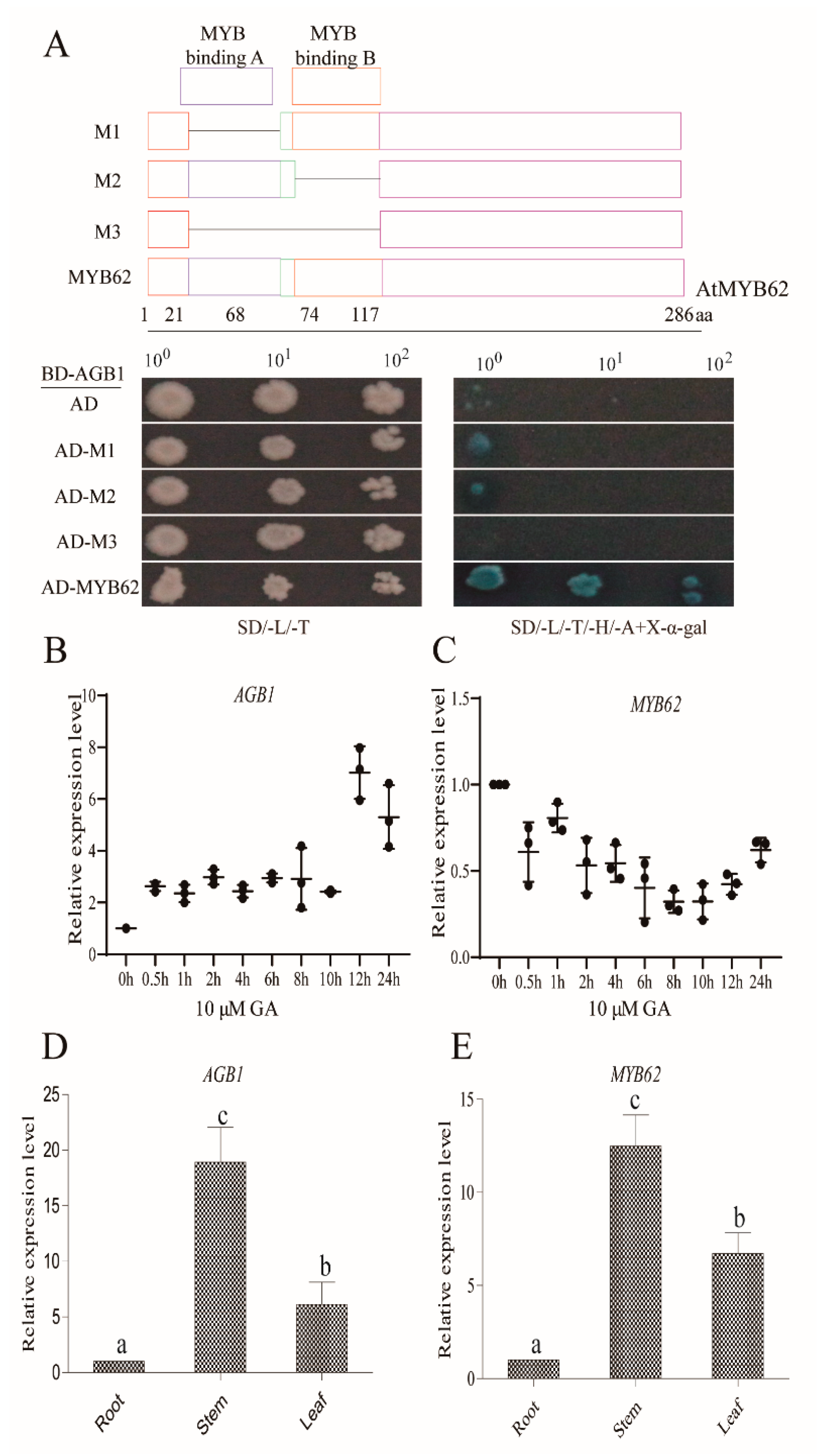
3.2. AGB1 Interacts with the DNA-Binding Region of MYB62, a GA Pathway Suppressor
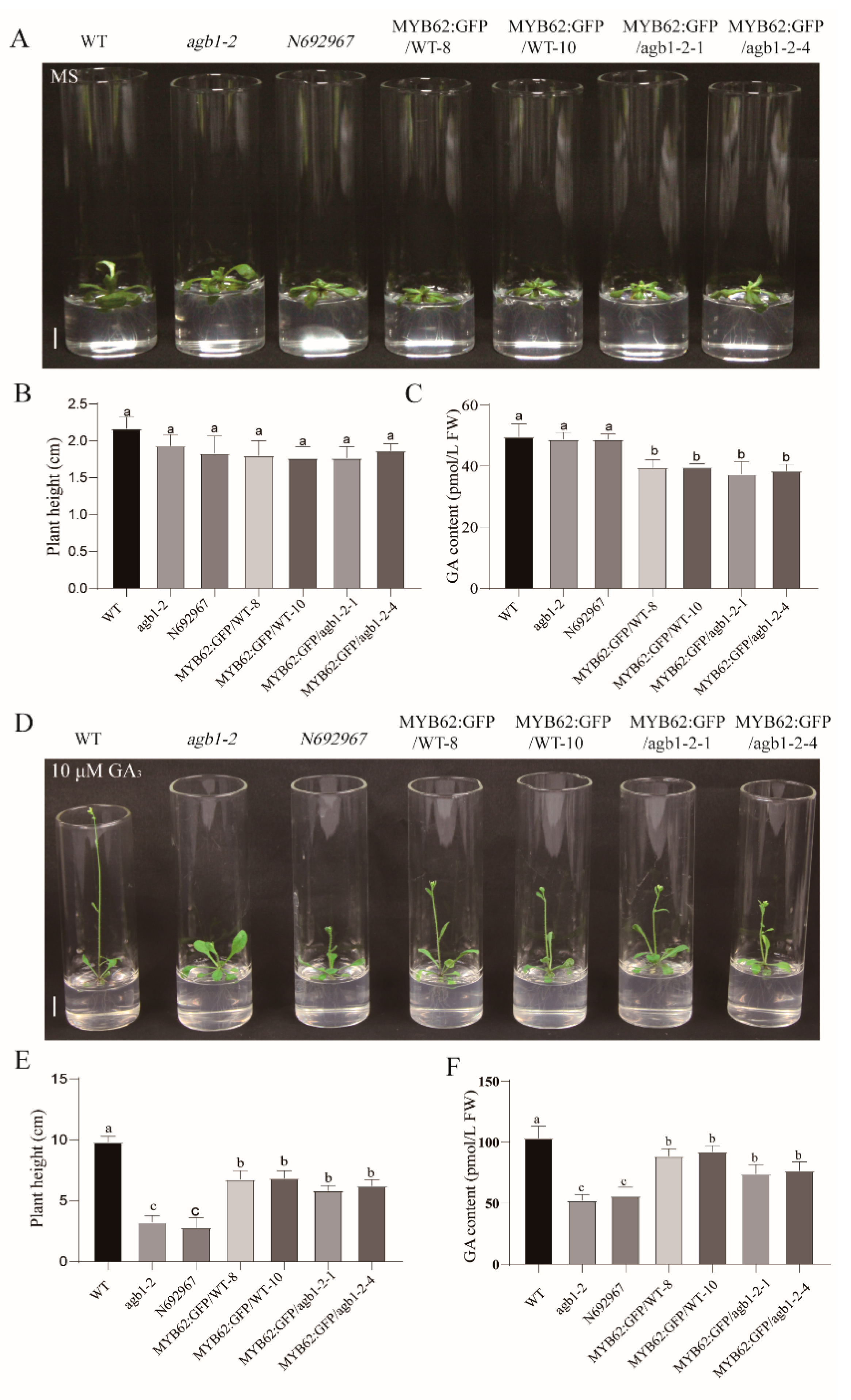
3.3. Genetic Analysis Indicated that MYB62 Was Downstream of AGB1 in the GA Pathway
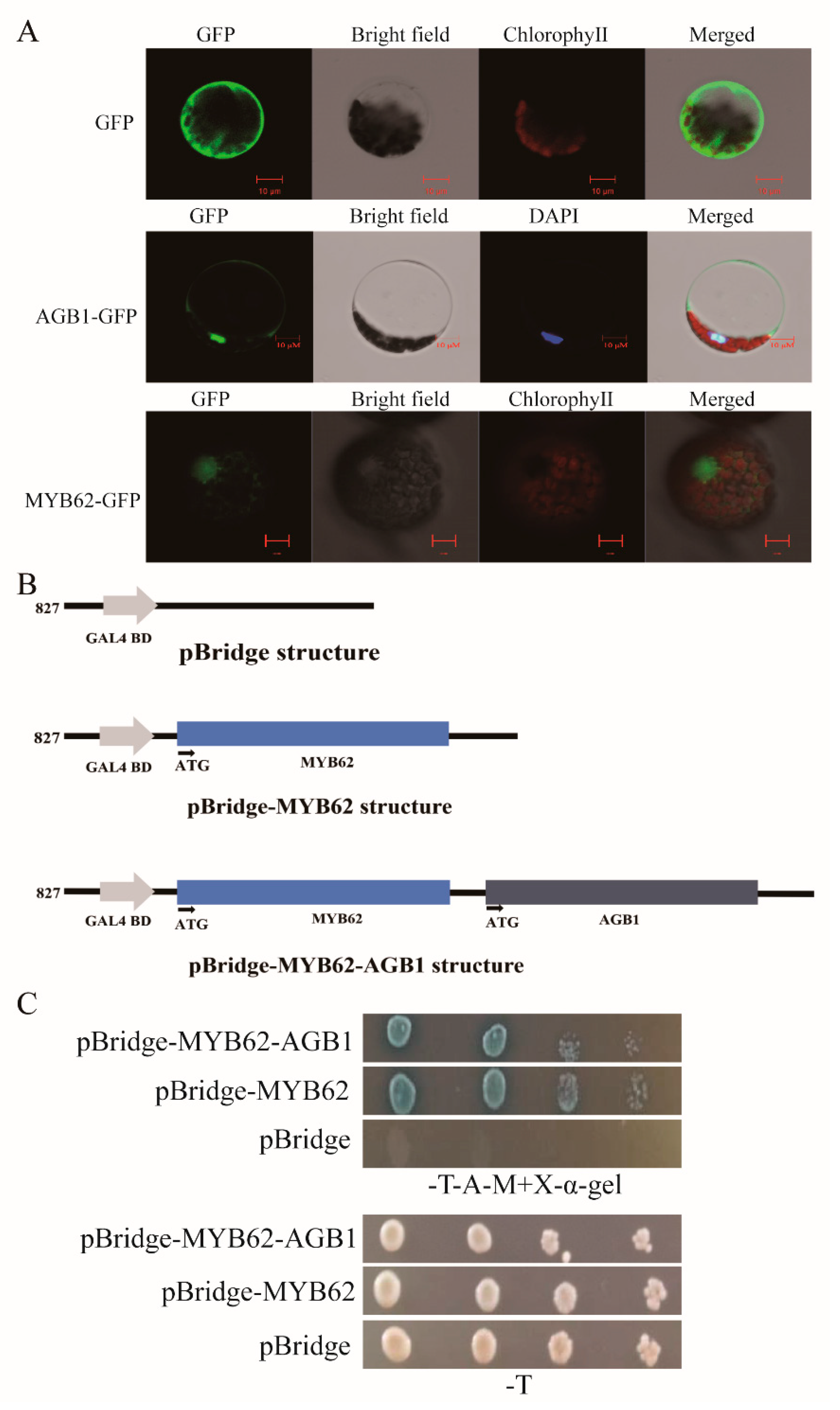
3.4. The Interaction between AGB1 and MYB62 Did Not Affect the Transcriptional Activation of MYB62
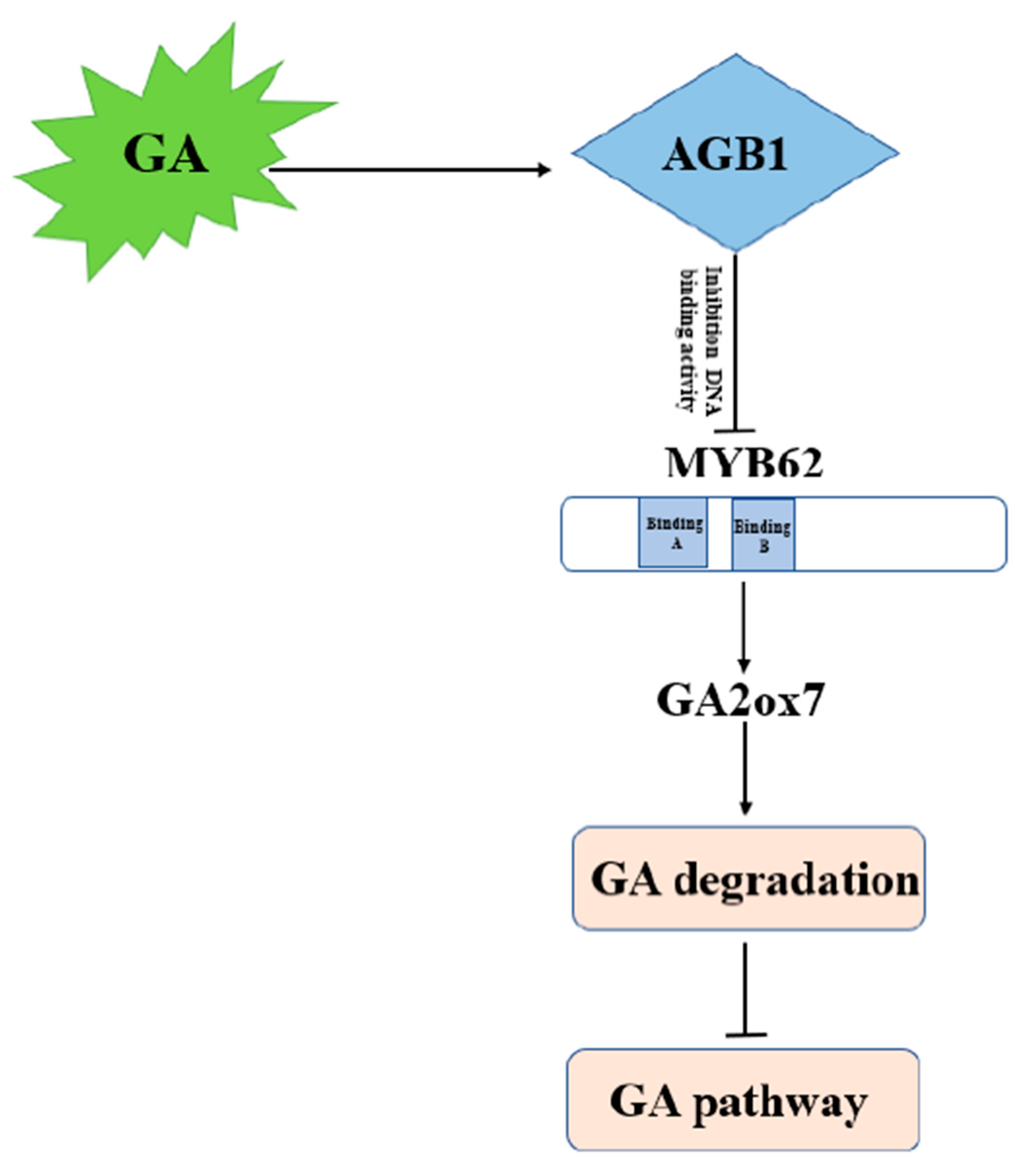
3.5. MYB62 Can Bind the Promoter of GA Degradation Gene GA2ox7 to Enhance Its Expression, and AGB1 Negatively Regulates the DNA-Binding Activity of MYB62 on the Promoter of GA2ox7
4. Discussion
4.1. AGB1 Regulates the GA Pathway by Negatively Regulating MYB62 Activity
4.2. The AGB1–MYB62 Pair Is Involved in Regulating the Phosphate Starvation Response in Plants
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Assmann, S.M. G proteins go green: A plant G protein signaling FAQ sheet. Science 2005, 310, 71–73. [Google Scholar] [CrossRef]
- Chen, J.G. Heterotrimeric G-proteins in plant development. Front. Biosci. 2007, 13, 3321. [Google Scholar] [CrossRef][Green Version]
- Assmann, S.M. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 2002, 14 (Suppl. S1), S355–S373. [Google Scholar] [CrossRef]
- Anantharaman, V.; Abhiman, S.; de Souza, R.F.; Aravind, L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 2011, 475, 63–78. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.G.; Wang, S.C.; Jones, A.M. Role of a Heterotrimeric G Protein in Regulation of Arabidopsis Seed Germination. Plant Physiol. 2004, 129, 897–907. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.G.; Temple, B.; Boyes, D.C.; Alonso, J.M.; Davis, K.R.; Jones, A.M. The b-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 2003, 15, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Assmann, S.M.; Yu, Y.Q. The heterotrimeric g-protein beta subunit, agb1, plays multiple roles in the Arabidopsis salinity response. Plant Cell Environ. 2015, 38, 2143–2156. [Google Scholar] [CrossRef]
- Wang, R.S.; Pandey, S.; Li, S.; Gookin, T.E.; Zhao, Z.X.; Albert, R.; Assmann, S.M. Common and unique elements of the ABA-regulated transcriptome of arabidopsis guard cells. BMC Genom. 2011, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Liu, Y.J.; Zheng, L.Y.; Chen, L.L.; Li, N.; Corke, F.; Bevan, M.W. The plant-specific G protein γ subunit AGG3 influences organ size and shape in arabidopsis thaliana. New Phytol. 2012, 194, 690–703. [Google Scholar] [CrossRef]
- Chen, J.G.; Pandey, S.; Huang, J.; Alonso, J.M.; Ecker, J.R.; Assmann, S.M.; Jones, A.M. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004, 135, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Che, J.G.; Jones, A.M.; Assmann, S.M. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006, 141, 243–256. [Google Scholar] [CrossRef]
- Urano, D.; Jones, A.M. Heterotrimeric G protein–coupled signaling in plants. Annu. Rev. Plant Biol. 2014, 65, 365–384. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Fujisaw, Y.; Kobayashi, M.; Ashikari, M.; Iwasaki, Y.; Kitano, H.; Matsuoka, M. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric g protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 2000, 97, 11638–11643. [Google Scholar] [CrossRef]
- Trusov, Y.; Rookes, J.E.; Tilbrook, K.; Chakravorty, D.; Mason, M.G.; Anderson, D.; Botella, J.R. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in arabidopsis. Plant Cell 2007, 19, 1235–1250. [Google Scholar] [CrossRef]
- Lease, K.A.; Wen, J.Q.; Li, J.; Doke, J.T.; Liscum, E.; Walker, J.C. A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 2001, 13, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.B.; Chen, M.; Ma, Y.N.; Xu, Z.S.; Li, L.C.; Chen, Y.F.; Ma, Y.Z. A G-Protein β Subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS ONE 2015, 10, e0116385. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.B.; Ma, Y.N.; Gao, S.Q.; Wang, X.T.; Feng, L.; Li, L.C.; Ma, Y.Z. The G-protein β subunit AGB1 promotes hypocotyl elongation through inhibiting transcription activation function of BBX21 in Arabidopsis. Mol. Plant 2017, 10, 1206–1223. [Google Scholar] [CrossRef]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Yoshida, T.; Sawada, Y.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein beta-subunit, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011, 67, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Urano, D.; Miura, K.; Wu, Q.Y.; Iwasaki, Y.; Jackson, D.; Jones, A.M. Plant Morphology of Heterotrimeric G protein Mutants. Plant Cell Physiol. 2016, 57, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Jermakow, A.M.; Swain, S.M. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 2002, 14, 3133–3147. [Google Scholar] [CrossRef]
- Lange, B.M.; Ghassemian, M. Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 2003, 51, 925–948. [Google Scholar] [CrossRef]
- Morrone, D.; Chambers, J.; Lowry, L.; Kim, G.; Anterola, A.; Bender, K.; Peters, R.J. Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in bradyrhizobium japonicum. FEBS Lett. 2009, 583, 475–480. [Google Scholar] [CrossRef]
- Davidson, S.E.; Elliott, R.C.; Helliwell, C.A.; Poole, A.T.; Reid, J.B. The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol. 2003, 131, 335–344. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miyura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Miyao, A. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Hedde, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Ashikari, M.; Sasaki, A.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan Datta, S.; Khush, S.G. Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox), led to the rice ‘green revolution’. Breed. Sci. 2002, 52, 143–150. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Chandler, P.M.; Poole, A.; Dennis, E.S.; Peacock, W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 2065–2070. [Google Scholar] [CrossRef]
- Fambrini, M.; Mariotti, L.; Parlanti, S.; Picciarelli, P.; Salvini, M.; Ceccarelli, N.; Pugliesi, C. The extreme dwarf phenotype of the GA-sensitive mutant of sunflower, dwarf2, is generated by a deletion in the ent-kaurenoic acid oxidase1 (HaKAO1) gene sequence. Plant Mol. Biol. 2011, 75, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Matsuoka, M. Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor gid1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Ueguchi-Tanaka, M.; Nakatsu, T.; Nakajima, M.; Naoe, Y.; Ohmiya, H.; Matsuoka, M. Structural basis for gibberellin recognition by its receptor DID1. Nature 2008, 456, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and Gibberellic Acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 2010, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.S.; Cho, Y.H.; Sheen, J. Arabidopsi mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, P.B.; Wang, W.X.; Wang, S.; Caruana, J.C.; Yang, H.Q.; Hongli Lian, H.L. Arabidopsis g-protein β subunit agb1 interacts with BES1 to regulate brassinosteroid signaling and cell elongation. Front. Plant Sci. 2018, 8, 2225. [Google Scholar] [CrossRef]
- Wu, T.Y.; Krishnamoorthi, S.; Goh, H.; Leong, R.; Sanson, A.C.; Urano, D. Crosstalk between heterotrimeric G protein-coupled signaling pathways and WRKY transcription factors modulating plant responses to suboptimal micronutrient conditions. J. Exp. Bot. 2020, 71, 3227–3239. [Google Scholar] [CrossRef]
- Chakraborty, N.; Singh, N.; Kaur, K.; Raghuram, N. G-protein signaling components GCR1 and GPA1 mediate responses to multiple abiotic stresses in arabidopsis. Front. Plant Sci. 2015, 6, 1000. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, X.; Tang, W.; Li, W.; He, Z.; Xu, W.; Fan, Z.; Zhou, Y.; Wang, C.; Xu, Z.; Chen, J.; et al. Arabidopsis G-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway. Int. J. Mol. Sci. 2021, 22, 8270. https://doi.org/10.3390/ijms22158270
Qi X, Tang W, Li W, He Z, Xu W, Fan Z, Zhou Y, Wang C, Xu Z, Chen J, et al. Arabidopsis G-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway. International Journal of Molecular Sciences. 2021; 22(15):8270. https://doi.org/10.3390/ijms22158270
Chicago/Turabian StyleQi, Xin, Wensi Tang, Weiwei Li, Zhang He, Weiya Xu, Zhijin Fan, Yongbin Zhou, Chunxiao Wang, Zhaoshi Xu, Jun Chen, and et al. 2021. "Arabidopsis G-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway" International Journal of Molecular Sciences 22, no. 15: 8270. https://doi.org/10.3390/ijms22158270
APA StyleQi, X., Tang, W., Li, W., He, Z., Xu, W., Fan, Z., Zhou, Y., Wang, C., Xu, Z., Chen, J., Gao, S., Ma, Y., & Chen, M. (2021). Arabidopsis G-Protein β Subunit AGB1 Negatively Regulates DNA Binding of MYB62, a Suppressor in the Gibberellin Pathway. International Journal of Molecular Sciences, 22(15), 8270. https://doi.org/10.3390/ijms22158270







