Novel Nontoxic 5,9-Disubstituted SN38 Derivatives: Characterization of Their Pharmacological Properties and Interactions with DNA Oligomers
Abstract
1. Introduction
2. Results and Discussion
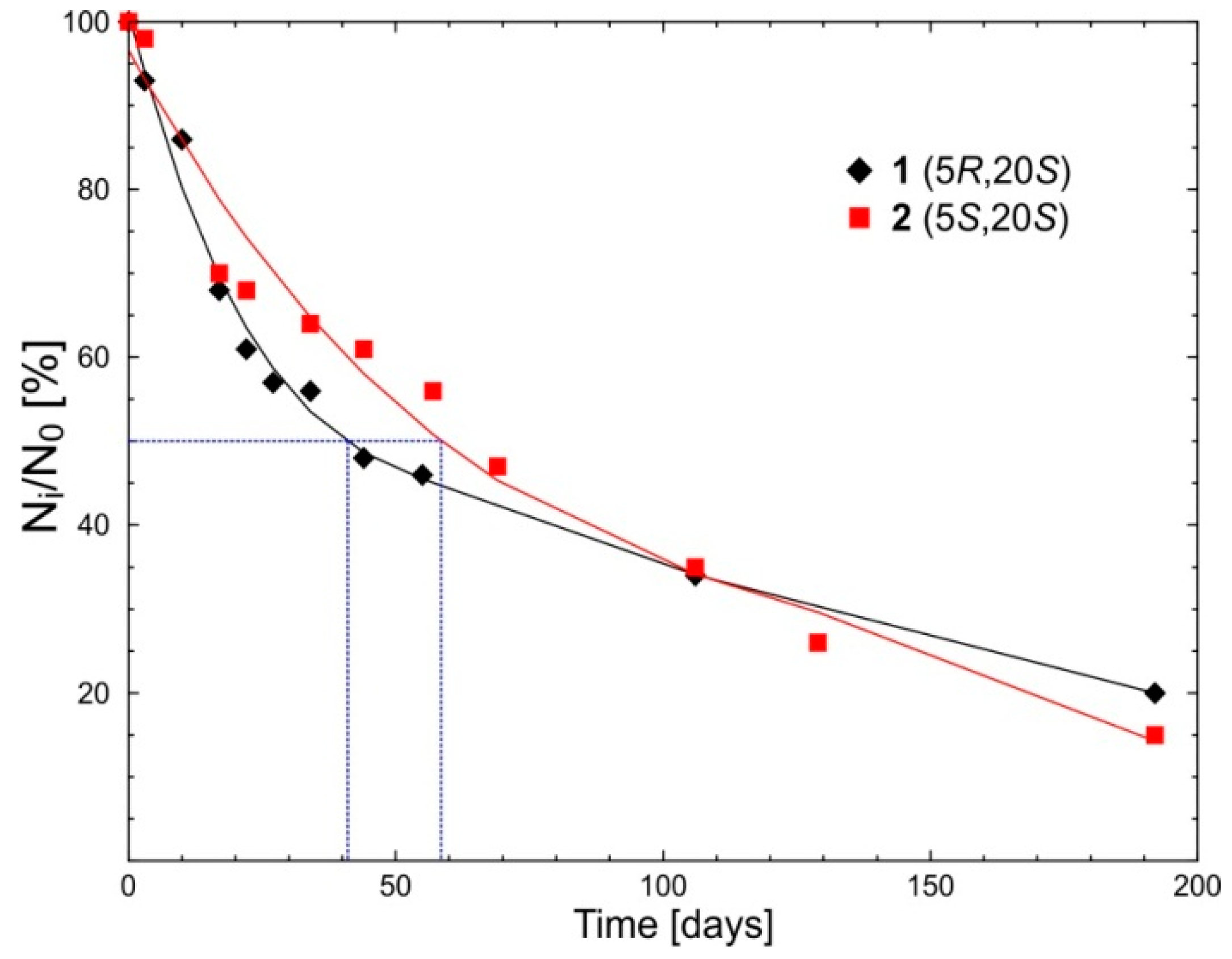
2.1. Chemical and Stereochemical Stability of 1 and 2 in Water
2.2. Aggregation Studies of 1 and 2 in D2O Buffer
2.3. Binding of 1 or 2 with d(GCGATCGC)2, Characterized by DOSY
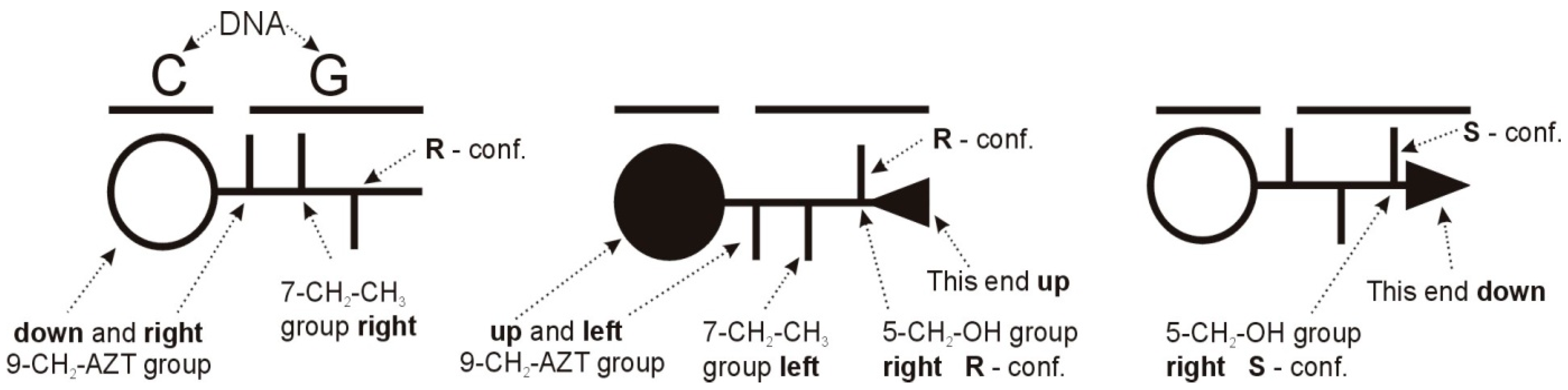
2.4. Mode of Binding of Diastereomers 1 and 2 to d(GCGATCGC)2 Based on 1H NMR Titration
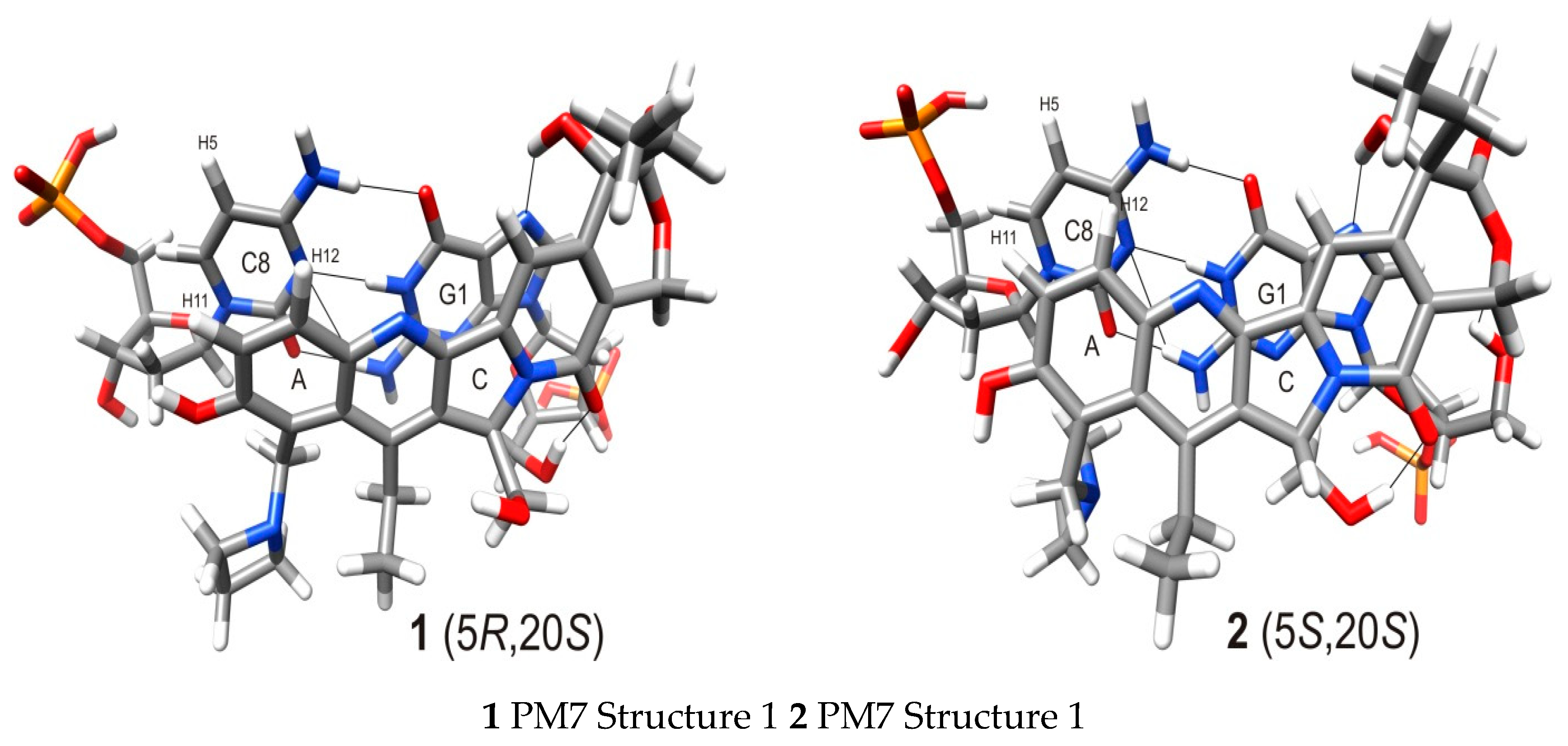
2.5. Molecular Modeling
3. Materials and Methods
3.1. Chemical Substrates
3.2. HPLC Analysis
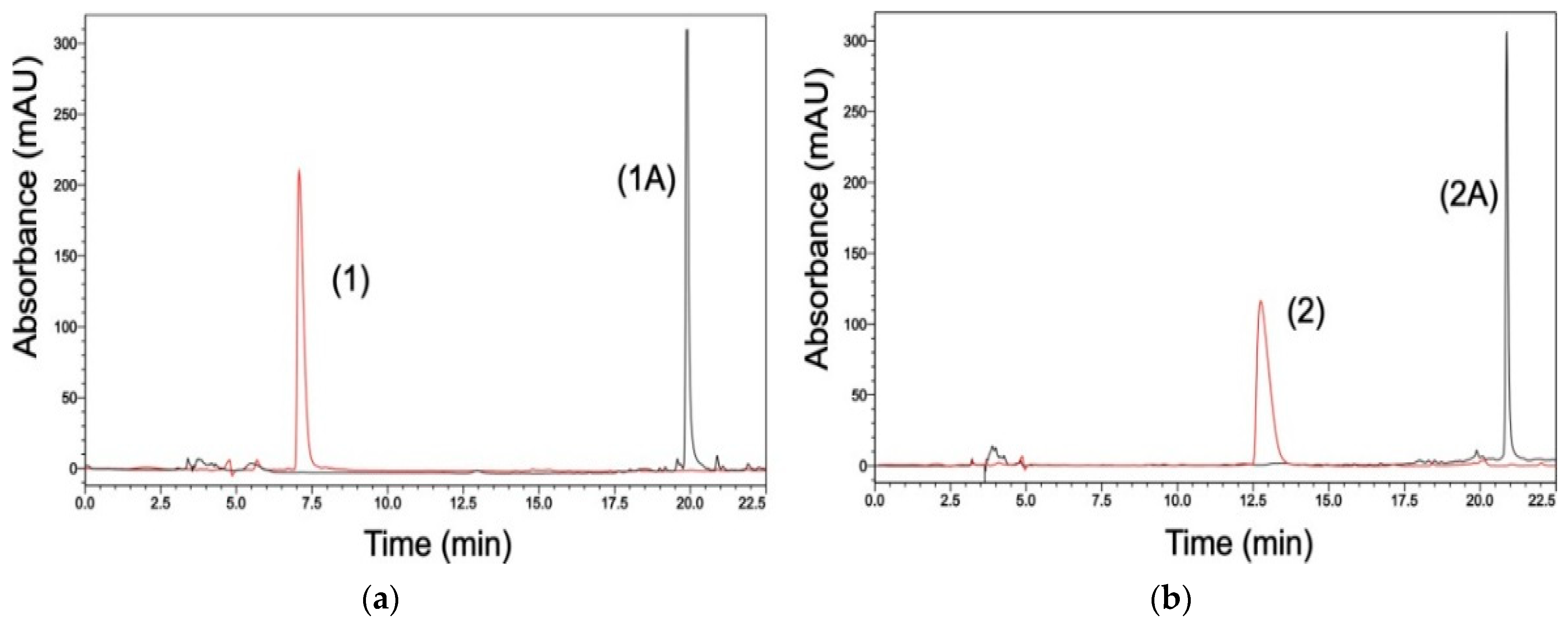
3.2.1. HPLC Separation and Purification
3.2.2. Compounds Purity
3.2.3. HPLC Analysis of Incubation Samples
3.3. Sample Preparation
3.4. NMR Experiments
- 1D 1H NMR spectra: spectral width 8000 Hz, 16–256 scans, 32 K complex points, acquisition time 2 s, relaxation delay 2 s.
- NOESY: spectral widths 5000 Hz in both dimensions, 1024 complex points in t2, 512 complex points in t1, 64 scans per increment, relaxation delay 1 s, and mixing time 200 ms.
- 1H-13C HSQC: spectral widths 6000 Hz in F2 and 21,600 Hz in F1, 1024 complex points in t2, 400 complex points in t1, 64 scans per increment, relaxation delay 1s, 1J(C,H) = 146 Hz.
- 1H-13C HMBC: spectral widths 6000 Hz in F2 and 23,880 Hz in F1, 1024 complex points in t2, 400 complex points in t1, 256 scans per increment, relaxation delay 1 s, nJ(C,H) = 5 or 8 Hz.
3.5. Calculating Self-Association Constants of 1 and 2
3.6. Calculating Binding Constants Based on 1H NMR Titration
3.7. Calculating Binding Constants from the Diffusion Coefficients
3.8. Molecular Dynamics Calculations
3.8.1. Calculating Binding Free Energies (Enthalpies) Using MM-PBSA and MM-GBSA Methods
3.8.2. PM7 Semi-Empirical Calculations
3.8.3. Cluster Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stewart, L.; Redinbo, M.R.; Qiu, X.; Hol, W.G.; Champoux, J.J. A model for the mechanism of human topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef]
- Redinbo, M.R.; Stewart, L.; Kuhn, P.; Champoux, J.J.; Hol, W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 1998, 279, 1504–1513. [Google Scholar] [CrossRef]
- Pommier, Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem. Rev. 2009, 109, 2894–2902. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Li, W.Q.; Morris-Natschke, S.L.; Qian, K.; Yang, L.; Zhu, G.X.; Wu, X.B.; Chen, A.L.; Zhang, S.Y.; Nan, X.; et al. Perspectives on biologically active camptothecin derivatives. Med. Res. Rev. 2015, 35, 753–789. [Google Scholar] [CrossRef] [PubMed]
- Zunino, F.; Pratesi, G. Camptothecins in clinical development. Expert Opin. Investig. Drugs 2004, 13, 269–284. [Google Scholar] [CrossRef]
- Thomas, C.J.; Rahier, N.J.; Hecht, S.M. Camptothecin: Current perspectives. Bioorg. Med. Chem. 2004, 12, 1585–1604. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Camptothecins: A SAR/QSAR study. Chem. Rev. 2009, 109, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996, 65, 635–692. [Google Scholar] [CrossRef]
- Naumczuk, B.; Hyz, K.; Kawęcki, R.; Bocian, W.; Bednarek, E.; Sitkowski, J.; Wielgus, E.; Kozerski, L. DOSY NMR and MALDI-TOF evidence of covalent binding the DNA duplex by trimethylammonium salts of topotecan upon near UV irradiation. Magn. Reson. Chem. 2015, 53, 565–571. [Google Scholar] [CrossRef]
- Naumczuk, B.; Kawęcki, R.; Bocian, W.; Bednarek, E.; Sitkowski, J.; Kozerski, L. Preliminary study of mechanism of action of SN38 derivatives. Physicochemical data, evidence of interaction and alkylation of DNA octamer d(GCGATCGC)2. Magn. Reson. Chem. 2017, 55, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Naumczuk, B.; Wiktorska, K.; Lubelska, K.; Kawęcki, R.; Bocian, W.; Bednarek, E.; Sitkowski, J.; Chilmonczyk, Z.; Kozerski, L. New generation of camptothecin derivatives spontaneously alkylating DNA. New J. Chem. 2016, 40, 7978–7985. [Google Scholar] [CrossRef]
- Bocian, W.; Kawęcki, R.; Bednarek, E.; Sitkowski, J.; Pietrzyk, A.; Williamson, M.P.; Hansen, P.E.; Kozerski, L. Multiple binding modes of the camptothecin family to DNA oligomers. Chem. Eur. J. 2004, 10, 5776–5787. [Google Scholar] [CrossRef]
- Bocian, W.; Kawęcki, R.; Bednarek, E.; Sitkowski, J.; Williamson, M.P.; Hansen, P.E.; Kozerski, L. Binding of Topotecan to a Nicked DNA Oligomer in Solution. Chem. Eur. J. 2008, 14, 2788–2794. [Google Scholar] [CrossRef]
- Naumczuk, B.; Górecki, M.; Wiktorska, K.; Urbanowicz, M.; Sitkowski, J.; Lubelska, K.; Milczarek, M.; Bednarek, E.; Bocian, W.; Kozerski, L. New camptothecin derivatives for generalized oncological chemotherapy: Synthesis, stereochemistry and biology. Bioorg. Med. Chem. Lett. 2021, 46, 128146. [Google Scholar] [CrossRef]
- Rivory, L.P.; Robert, J. [Pharmacology of camptothecin and its derivatives]. Bull. Cancer 1995, 82, 265–285. [Google Scholar]
- Kozerski, L.; Mazurek, A.P.; Kawęcki, R.; Bocian, W.; Krajewski, P.; Bednarek, E.; Sitkowski, J.; Williamson, M.P.; Moir, A.J.; Hansen, P.E. A nicked duplex decamer DNA with a PEG(6) tether. Nucleic Acids Res. 2001, 29, 1132–1143. [Google Scholar] [CrossRef][Green Version]
- Pelta, M.D.; Morris, G.A.; Stchedroff, M.J.; Hammond, S.J. A one-shot sequence for high-resolution diffusion-ordered spectroscopy. Magn. Reson. Chem. 2002, 40, 147–152. [Google Scholar] [CrossRef]
- Baxter, N.J.; Williamson, M.P.; Lilley, T.H.; Haslam, E. Stacking interactions between caffeine and methyl gallate. J. Chem. Soc. Faraday Trans. 1996, 92, 231–234. [Google Scholar] [CrossRef]
- Charlton, A.J.; Baxter, N.J.; Khan, M.L.; Moir, A.J.G.; Haslam, E.; Davies, A.P.; Williamson, M.P. Polyphenol/peptide binding and precipitation. J. Agric. Food Chem. 2002, 50, 1593–1601. [Google Scholar] [CrossRef]
- Thordarson, P. Determining association constants from titration experiments in supramolecular chemistry. Chem. Soc. Rev. 2011, 40, 1305–1323. [Google Scholar] [CrossRef] [PubMed]
- Waldeck, A.R.; Kuchel, P.W.; Lennon, A.J.; Chapman, B.E. NMR diffusion measurements to characterize membrane transport and solute binding. Prog. Nucl. Magn. Reson. Spectrosc. 1997, 30, 39–68. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Merz, K.M.; Pearlman, D.A.; Crowley, M.; et al. AMBER 9; University of California: San Francisco, CA, USA, 2006. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision A.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Luo, R.; David, L.; Gilson, M.K. Accelerated Poisson-Boltzmann calculations for static and dynamic systems. J. Comput. Chem. 2002, 23, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J. MOPAC2016, Version: 17181L. Available online: http://OpenMOPAC.net (accessed on 5 April 2021).
- Klamt, A.; Schuurmann, G. Cosmo—A New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient. J. Chem. Soc. Perkin Trans. 2 1993, 5, 799–805. [Google Scholar] [CrossRef]
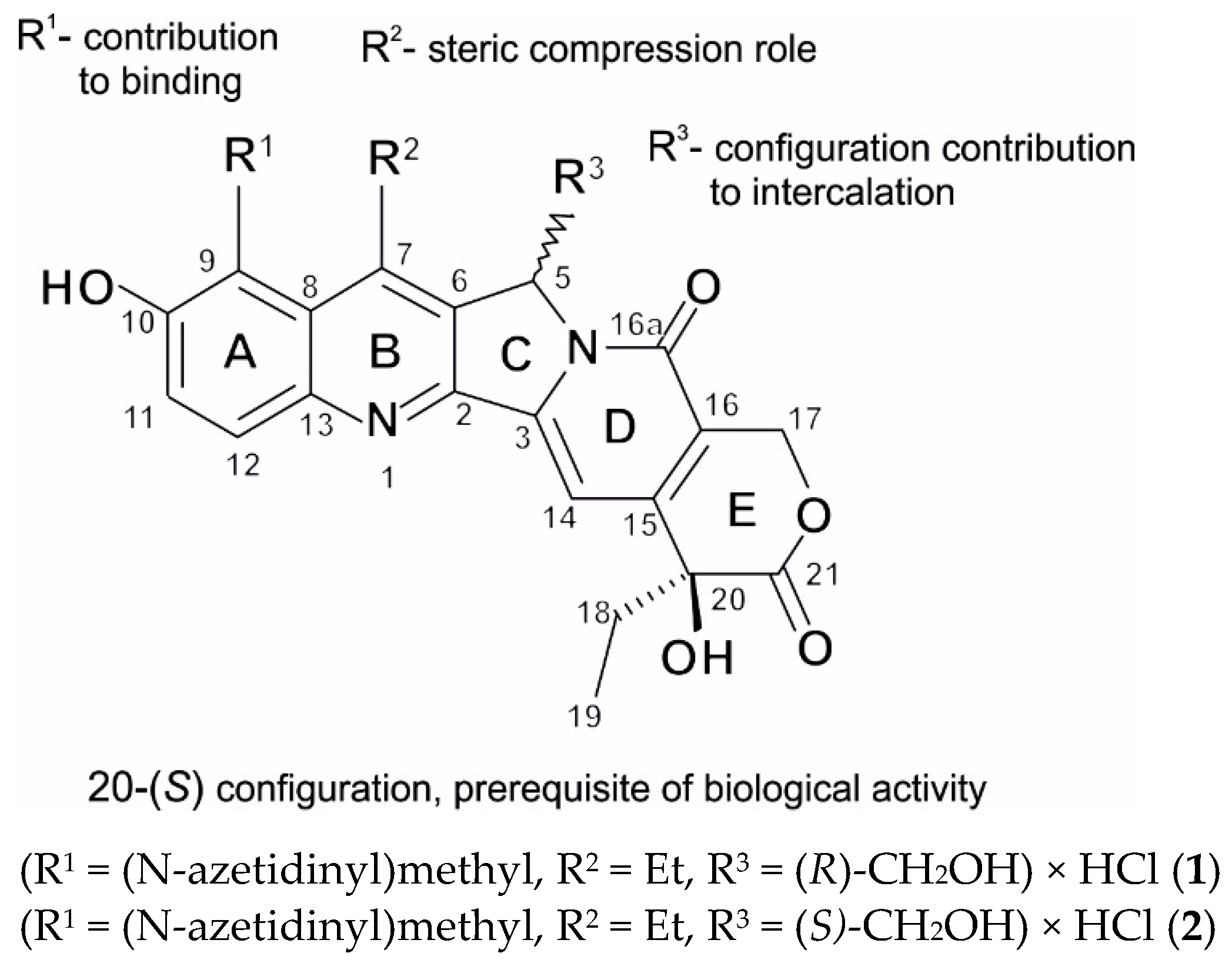
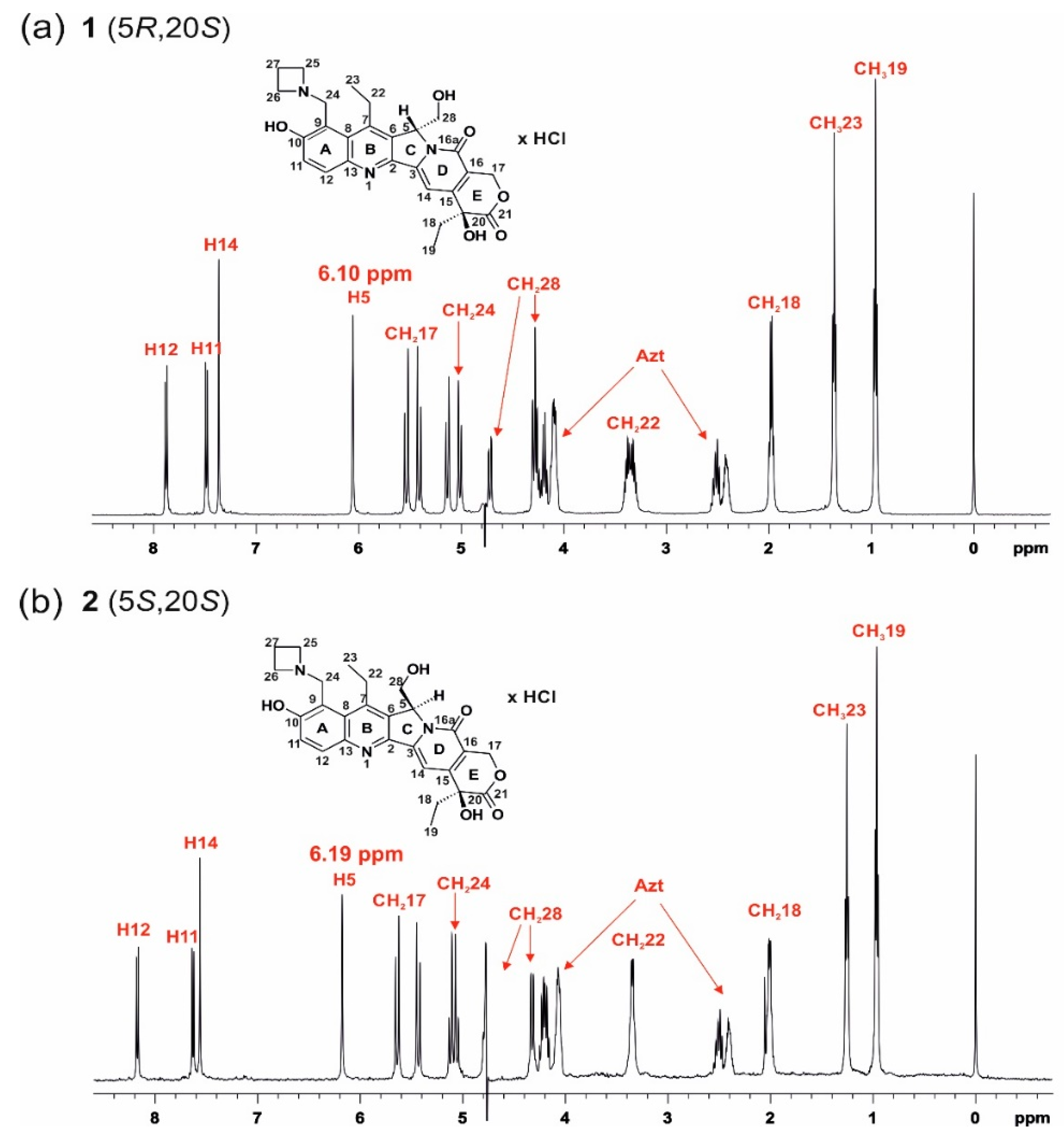

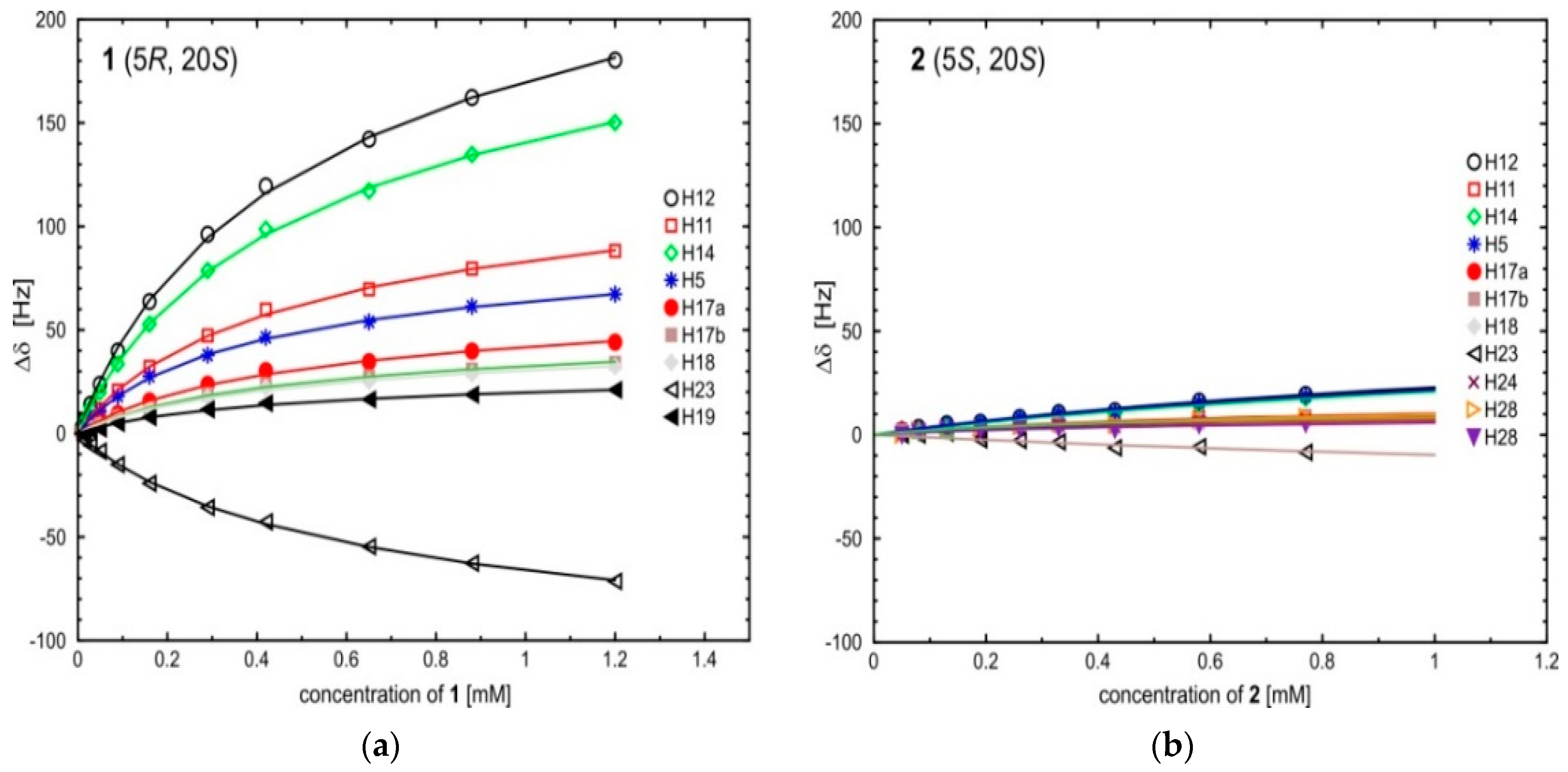
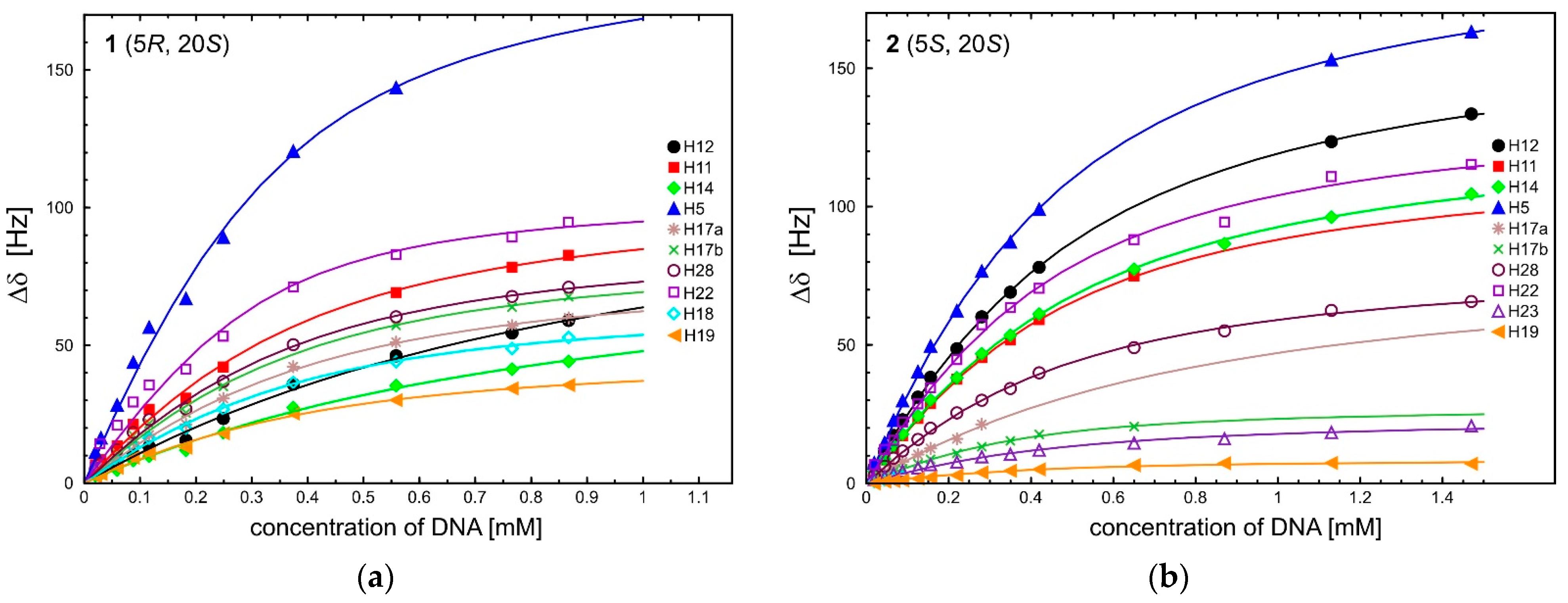
| Δδ for 1 | ||||||||||||
| c1 | Ring A | Ring B | Ring C | Ring D | Ring E | |||||||
| H12 | H11 | H23 | H5 | H14 | H17a | H17b | H18 | H19 | ||||
| 0.005 | (8.15) 2 | (7.60) | (1.26) | (6.16) | (7.59) | (5.65) | (5.50) | (2.05) | (1.02) | |||
| 0.05 | 23.6 | 11.5 | −8.7 | 10.6 | 19.7 | 5.3 | 4.2 | 3.8 | 2.2 | |||
| 0.09 | 39.8 | 20.6 | −15.1 | 17.9 | 33.4 | 9.6 | 7.9 | 7.6 | 4.9 | |||
| 0.29 | 96.1 | 47.2 | −35.8 | 37.7 | 78.7 | 23.6 | 18.8 | 17.4 | 11.5 | |||
| 0.65 | 142.0 | 69.5 | −54.7 | 53.8 | 117.1 | 34.6 | 27.4 | 25.1 | 16.4 | |||
| 1.20 | 180.3 | 88.1 | −71.5 | 67.2 | 150.1 | 44.1 | 34.1 | 32.2 | 20.9 | |||
| Ka Δδmax 3 | 1.61 367.0 | 1.74 174.6 | 1.34 −153.4 | 2.28 121.9 | 1.59 305.7 | 1.64 89.4 | 1.76 67.9 | 1.70 64.3 | 1.76 41.3 | |||
| the average value of Ka | 1.71 ± 0.24 | |||||||||||
| Δδ for 2 | ||||||||||||
| c2 | Ring A | Ring B | Ring C | Ring D | Ring E | |||||||
| H12 | H11 | H24 | H23 | H51 | H28 | H28 | H14 | H17a | H17b | H18 | ||
| 0.003 | (8.15) 2 | (7.60) | (5.10) | (1.26) | (6.20) | (4.81) | (4.35) | (7.57) | (5.66) | (5.46) | (2.02) | |
| 0.05 | 2.5 | 1.9 | 1.0 | 0 | 2.5 | 2.1 | 1.2 | 2.3 | 1.6 | 1.6 | 0.5 | |
| 0.08 | 3.4 | 2.1 | 1.9 | 0 | 3.2 | 2.3 | 1.4 | 2.8 | 2.0 | 1.7 | 1.7 | |
| 0.26 | 8.3 | 4.5 | 3.6 | −2.8 | 8.1 | 4.2 | 2.7 | 7.3 | 3.3 | 3.2 | 2.7 | |
| 0.43 | 11.5 | 5.3 | 4.4 | −6.4 | 11.0 | 5.0 | 2.6 | 10.1 | 3.6 | 3.9 | 2.2 | |
| 0.77 | 19.4 | 8.9 | 7.5 | −8.4 | 18.6 | 8.6 | 4.9 | 17.6 | 7.1 | 8.0 | 5.4 | |
| Ka Δδmax 3 | 0.40 98.0 | 0.91 27.7 | 0.68 26.9 | 0.20 −67.3 | 0.42 89.4 | 0.82 27.7 | 1.03 14.8 | 0.35 96.5 | 1.53 15.7 | 0.41 40.9 | 0.98 15.1 | |
| the average value of Ka | 0.70 ± 0.38 | |||||||||||
| 1 | 2 | ||
|---|---|---|---|
| c [mm] | Di × 10−10 [m2s−1] | c [mm] | Di × 10−10 [m2s−1] |
| 1.5 | 1.92 ± 0.05 | 0.77 | 2.18 ± 0.05 |
| 0.29 | 2.13 ± 0.10 | 0.19 | 2.22 ± 0.10 |
| 0.23 | 2.13 ± 0.10 | 0.13 | 2.24 ± 0.10 |
| Concentration [mm] | DOBS | DOBS-3 | MFcomp | Ka ± 0.3 | ||
|---|---|---|---|---|---|---|
| c | c3 | × 10−10 [m2 s−1] | × 10−10 [m2 s−1] | [mm−1] | ||
| 1 | 0.29 | 0.92 | 1.11 ± 0.05 | 0.86 ± 0.05 | 0.80 ± 0.05 | 5.9 |
| 0.45 | 0.92 | 1.14 ± 0.05 | 0.91 ± 0.05 | 0.81 ± 0.05 | 7.7 | |
| 1.89 | 1.26 | 1.34 ± 0.05 | 0.88 ± 0.05 | 0.55 ± 0.05 | 5.5 | |
| 2 | 0.29 | 0.28 | 1.83 ± 0.10 | 0.93 ± 0.10 | 0.29 ± 0.05 | 2.1 |
| 0.29 | 0.42 | 1.67 ± 0.10 | 0.92 ± 0.10 | 0.41 ± 0.05 | 2.4 | |
| 0.29 | 0.87 | 1.45 ± 0.10 | 0.92 ± 0.10 | 0.59 ± 0.05 | 2.0 | |
| 0.9 | 1.47 | 1.35 ± 0.10 | 0.84 ± 0.10 | 0.63 ± 0.05 | 1.8 | |
| Δδ for 1 | ||||||||||
| c3 | Ring A | Ring B | Ring C | Ring D | Ring E | |||||
| H12 | H11 | H22 | H5 | H28 | H14 | H17a | H17b | H18 | H19 | |
| 0 | (7.95) 2 | (7.51) | (3.36) | (6.10) | (4.76) | (7.44) | 5.60 | (5.46) | (2.01) | (0.99) |
| 0.02 | 3.9 | 6.6 | 10.1 | 11.2 | 5.0 | 2.6 | 4.3 | 4.3 | 3.6 | 2.4 |
| 0.06 | 6.2 | 13.5 | 19.3 | 28.3 | 12.1 | 4.7 | 8.7 | 10.9 | 8.2 | 5.5 |
| 0.12 | 13.2 | 26.8 | 31.3 | 56.5 | 23.0 | 9.8 | 17.5 | 21.6 | 16.2 | 10.7 |
| 0.25 | 23.4 | 42.1 | 49.3 | 89.2 | 36.9 | 18.3 | 30.7 | 35.1 | 26.8 | 18.1 |
| 0.56 | 46.2 | 69.2 | 80.7 | 143.5 | 60.4 | 35.4 | 51.2 | 57.2 | 44.1 | 30.1 |
| 0.87 | 59.0 | 82.7 | 93.2 | - | 71.1 | 44.2 | 60.0 | 67.6 | 52.9 | 35.6 |
| Ka Δδmax 3 | 1.37 4 118.8 | 5.34 105.7 | 7.57 111.4 | 7.72 197.6 | 5.89 89.4 | 1.54 4 85.0 | 4.76 79.4 | 5.56 85.7 | 5.38 66.9 | 4.89 47.0 |
| the average value of Ka | 5.00 ± 2.02 (5.89 ± 1.07) 4 | |||||||||
| Δδ for 2 | ||||||||||
| c4 | Ring A | Ring B | Ring C | Ring D | Ring E | |||||
| H12 | H11 | H22 | H23 | H5 | H28 | H14 | H17a | H17b | H19 | |
| 0.00 | (8.16) 2 | (7.62) | (3.37) | (1.27) | (6.19) | (4.81) | (7.57) | (5.65) | (5.46) | (0.99) |
| 0.02 | 5.7 | 4.1 | 6.7 | 1.4 | 7.3 | 2.8 | 4.3 | 2.1 | 1.5 | 0 |
| 0.07 | 17.3 | 12.9 | 15.5 | 3.4 | 22.7 | 9.1 | 13.6 | 6.1 | 4.1 | −0.9 |
| 0.13 | 31.1 | 23.4 | 28.7 | 5.5 | 40.3 | 15.8 | 24.3 | 10.4 | 7.0 | −1.7 |
| 0.28 | 60.2 | 45.4 | 57.3 | 9.6 | 76.7 | 30.0 | 46.8 | 21.3 | 13.2 | −3.9 |
| 0.65 | - | 74.9 | 88.1 | 14.5 | - | 49.0 | 77.4 | - | 20.5 | −6.5 |
| 1.13 | 123.5 | - | 110.9 | 18.4 | 153.0 | 62.5 | 96.2 | - | - | −7.3 |
| 1.47 | 133.5 | - | 115.3 | 20.7 | 163.2 | 65.7 | 104.6 | - | - | −7.0 |
| Ka Δδmax 3 | 3.11 167.5 | 3.52 119.9 | 3.93 138.0 | 3.86 23.8 | 3.64 199.3 | 3.40 89.4 | 3.09 130.4 | 1.66 5 81.2 | 6.48 5 28.0 | 4.88 −8.85 |
| the average value of Ka | 3.76 ± 1.19 | |||||||||
| Δδ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1-H8 | G1-H1′ | G1-H5′ | C2-H6 | C2-H5 | A4-H8 | A4-H2 | A4-H1′ | T5-H6 | C8-H1′ | |
| (7.98) 2 | (5.99) | (3.72) | (7.42) | (5.38) | (8.24) | (7.79) | (6.28) | (7.19) | (6.19) | |
| 1 | 60.2 | 36.4 | −28.8 | 20.3 | 28.3 | 11.4 | 10.5 | 9.9 | 10.3 | 31.9 |
| 2 | 11.3 | 26.4 | −16.3 | 5.0 | 9.6 | 1.4 | 1.4 | 3.0 | 1.9 | 3.5 |
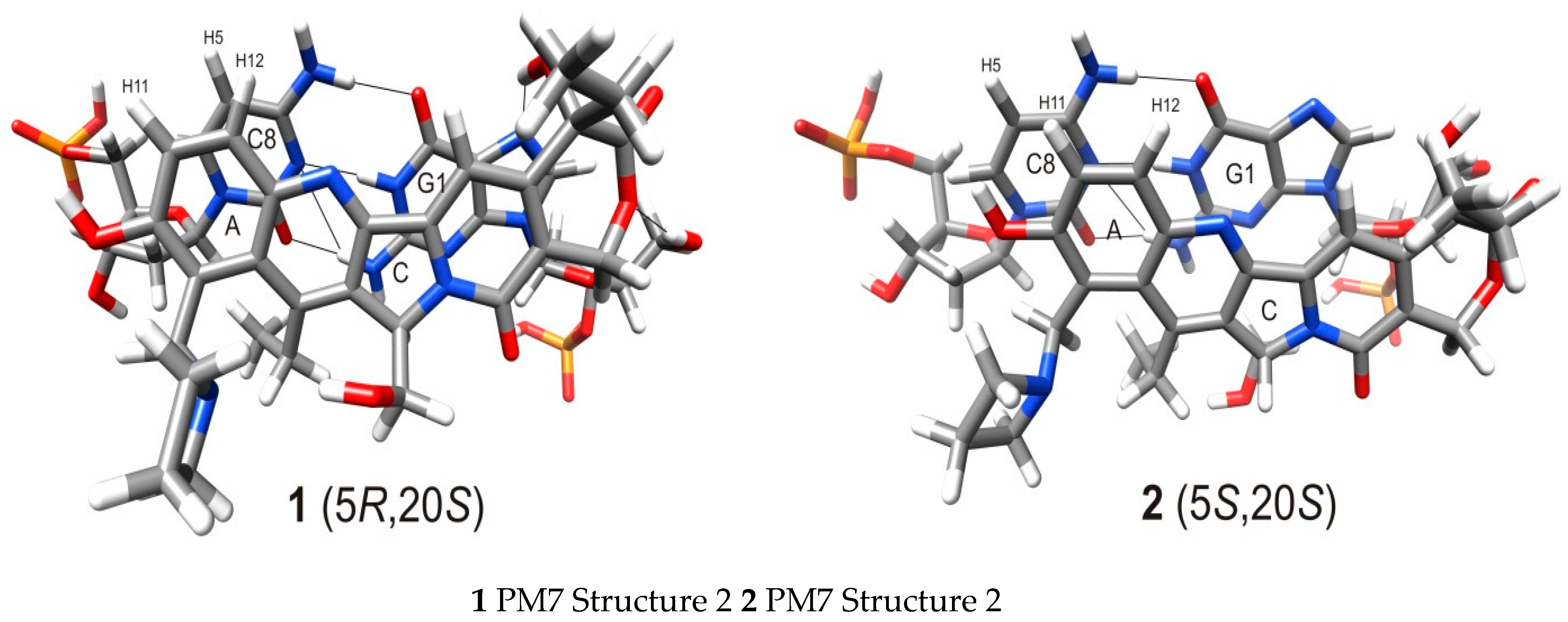
| No. | Structure | PM7 ENERGY [kcal/mol] | % Population | |
|---|---|---|---|---|
| Clust. Average | 10% Clust. Min. | |||
| 1 |  | −1174.25 ± 3.79 | −1181.15 | 20.39 |
| 2 |  | −1179.37 ± 3.39 | −1185.18 | 17.14 |
| 3 |  | −1177.11 ± 3.94 | −1184.58 | 14.25 |
| No. | Structure | PM7 ENERGY [kcal/mol] | % Population | |
|---|---|---|---|---|
| Clust. Average | 10% Clust. Min. | |||
| 1 |  | −1178.76 ± 4.77 | −1186.83 | 20.19 |
| 2 |  | −1177.52 ± 4.07 | −1183.82 | 16.28 |
| Atoms in | Distances [Å] | ||||
|---|---|---|---|---|---|
| Cytidine | 1, 2 | 1 struct. no. 1 * | 2 struct. no. 1 # | 1 struct. no. 2 * | 2 struct. no. 2 # |
| C8-H5 | H11 | 4.14 | 3.83 | 3.28 | 3.24 |
| C8-H5 | H12 | 4.69 | 3.87 | 3.05 | 5.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bednarek, E.; Bocian, W.; Urbanowicz, M.; Sitkowski, J.; Naumczuk, B.; Kozerski, L. Novel Nontoxic 5,9-Disubstituted SN38 Derivatives: Characterization of Their Pharmacological Properties and Interactions with DNA Oligomers. Int. J. Mol. Sci. 2021, 22, 8190. https://doi.org/10.3390/ijms22158190
Bednarek E, Bocian W, Urbanowicz M, Sitkowski J, Naumczuk B, Kozerski L. Novel Nontoxic 5,9-Disubstituted SN38 Derivatives: Characterization of Their Pharmacological Properties and Interactions with DNA Oligomers. International Journal of Molecular Sciences. 2021; 22(15):8190. https://doi.org/10.3390/ijms22158190
Chicago/Turabian StyleBednarek, Elżbieta, Wojciech Bocian, Magdalena Urbanowicz, Jerzy Sitkowski, Beata Naumczuk, and Lech Kozerski. 2021. "Novel Nontoxic 5,9-Disubstituted SN38 Derivatives: Characterization of Their Pharmacological Properties and Interactions with DNA Oligomers" International Journal of Molecular Sciences 22, no. 15: 8190. https://doi.org/10.3390/ijms22158190
APA StyleBednarek, E., Bocian, W., Urbanowicz, M., Sitkowski, J., Naumczuk, B., & Kozerski, L. (2021). Novel Nontoxic 5,9-Disubstituted SN38 Derivatives: Characterization of Their Pharmacological Properties and Interactions with DNA Oligomers. International Journal of Molecular Sciences, 22(15), 8190. https://doi.org/10.3390/ijms22158190






